Crystallization and preliminary X-ray diffraction of the ZO-binding domain of human occludin.
Keywords: occludin, tight junction
Abstract
Occludin is a tight-junction protein controlling the integrity of endothelial and epithelial cell layers. It forms complexes with the cytoplasmic proteins ZO-1, ZO-2 and ZO-3. The ZO-binding domain in the C-terminal cytoplasmic region of human occludin has previously been isolated and identified. This domain, as expressed in a bacterial system or isolated from native cellular occludin, maintains its ability to bind ZO-1 and ZO-2. The crystallization conditions of the human ZO-binding domain are reported here. The crystals diffract to 2.3 Å resolution and were shown to belong to the orthorhombic space group P212121, with unit-cell parameters a = 33.3, b = 35.4, c = 107.3 Å.
1. Introduction
A major function of occludin, in addition to its intercellular sealing role in the tight junction, is the recruitment of cytoplasmic proteins in the formation and maintenance of tight junctions at the apical cell–cell junction region of the endothelium or epithelium (McCarthy et al., 1996 ▶). These junctions provide both a diffusion barrier in the paracellular space and restrict mobile membrane proteins to physiological compartments appropriate to their specific functions. The blood–brain barrier (BBB) is formed by a continuous monolayer of microvascular endothelial cells sealed by tight junctions on the luminal side and supported by basement membranes and astrocyte foot processes on the other side. Blood substances (proteins, infectious agents, inflammatory factors etc.) are restricted to the lumens of the blood vessels unless specific transporting systems are present in the endothelial cells. Dysfunction of the tight junctions leads to leakage of blood-borne substances through the BBB, often resulting in injury to the brain tissue (Dallasta et al., 1999 ▶; Farkas et al., 1998 ▶; Petito & Cash, 1992 ▶; Sharief et al., 1992 ▶). In addition to brain microvessels, tight junctions are also important functional components of the endothelium of organs such as the lungs and testes, as well as in the absorptive and secretory epithelium in many organs, including the kidneys, exocrine glands, liver and gastrointestinal tract (Saitou et al., 1997 ▶).
Occludin, the first transmembrane tight-junction protein identified, was isolated from chick brain more than a decade ago (Furuse et al., 1993 ▶). Chicken occludin is composed of 504 amino acids with a molecular weight of 55.9 kDa. The C-terminal 150-amino-acid fragment was shown to be essential for binding to the GUK domain of ZO-1, ZO-2 or ZO-3 (Furuse et al., 1994 ▶; Haskins et al., 1998 ▶; Itoh et al., 1999 ▶) and these scaffolding proteins then help link the tight junction to the cytoskeleton (Fanning et al., 1998 ▶; Wittchen et al., 1999 ▶). It has not yet been determined how occludin binds to the ZO proteins. However, computational prediction has shown that the secondary structure of occludin is conserved between human, chick, dog, mouse and rat kangaroo, suggesting a similar binding motif in this ZO-binding domain across species (Ando-Akatsuka et al., 1996 ▶). Our previous studies have isolated and identified the functional binding domain to the ZO proteins as the last 119 residues of human occludin (Peng et al., 2003 ▶).
Here, we report the crystallization and preliminary X-ray diffraction results of this domain.
2. Materials and methods
2.1. Expression and purification
We cloned the ZO-binding domain of occludin into a pRSET-based expression plasmid as previously described (Peng et al., 2003 ▶). Expression of the fusion protein was carried out in BL21 bacteria after induction by 0.1 mM IPTG for 5 h at 310 K. Bacteria were lyzed in sodium phosphate buffer (20 mM sodium phosphate, 500 mM sodium chloride pH 7.4) using the Piranha Press (TESLA, Paxton, IL, USA) at 69 MPa. The fusion protein was then purified using a nickel column and eluted with 400 mM imidazole in sodium phosphate buffer. After 15% SDS–PAGE and Coomassie Blue staining, fractions containing the fusion protein were combined together and dialyzed against EK buffer (50 mM Tris–HCl, 1 mM CaCl2 and 100 mM NaCl pH 7.8) to remove imidazole and to switch buffer for the subsequent enzyme-digestion step. The resultant protein was concentrated and incubated with enterokinase (0.4 U per milligram of protein; Invitrogen, Carlsbad, CA, USA) at 310 K for at least 12 h. The His tag was then separated from the ZO-binding domain by gel filtration (Sephacryl 100-HR; Sigma, St Louis, MO, USA) in EK buffer. The fractions containing the ZO-binding domain were pooled together and concentrated to a high concentration (>10 mg ml−1) for crystallization. This final ZO-binding domain (G388R389S390K391R392 to D518R519Q520K521T522) includes an additional five amino acids (DRWGS) from the fusion tag at the N-terminal end.
2.2. Crystallization
2.2.1. Initial crystallization
Initial crystallization screens were performed using the hanging-drop vapor-diffusion technique (McPherson, 1982 ▶) with 0.5 ml reservoir solution taken from Hampton screening kits (Index, SaltRx, PEG/Ion Screen and Grid Screen Sodium Malonate; Aliso Viejo, CA, USA) and 4 µl of mixed solution (2 µl reservoir solution plus 2 µl concentrated protein in EK buffer) in the drops. Crystals appeared after two weeks in a solution consisting of 1.9 M sodium malonate in water (pH 6.0) at 293 K.
2.2.2. Refinement of crystallization
A set of 24 sodium malonate solutions (concentration from 1.5 to 2.1 M; pH from 5.4 to 6.4) was then prepared for the subsequent refinement of crystallization conditions.
2.2.3. Final crystallization condition
The optimized crystallization reservoir solution for this protein was determined to be 1.9 M sodium malonate in water at pH 6.2. Crystals from larger drops (5 µl reservoir solution plus 5 µl concentrated protein in EK buffer and 1 ml reservoir solution) were used for diffraction experiments.
2.3. X-ray diffraction and data processing
Crystals were harvested using 0.1–0.3 mm nylon loops (Hampton) and transferred to 10 µl 3.4 M sodium malonate pH 6.2. After 15 min soaking, crystals were immediately flash-cooled by plunging into liquid nitrogen. Initial diffraction experiments were performed at 100 K with an oscillation range of 1° per frame over 10 min using a MacScience MO6XHF X-ray generator and a MacScience DIP2030H-VLM dual 30 cm diameter imaging-plate detector at the X-ray Crystallography Core Facility at the University of Texas Medical Branch. Further experiments were carried out using the synchrotron beamline 14-BM-C at the Advanced Photon Source (APS, Chicago, IL, USA) and the Center for Advanced Microstructures and Devices (CAMD, Baton Rouge, LA, USA). Collected data were processed using DENZO/SCALEPACK (Otwinowski & Minor, 1997 ▶) or MOSFLM (Leslie, 1992 ▶) and CCP4 (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
Crystals grown in 1.9 M sodium malonate pH 6.2 (Fig. 1 ▶) diffracted to 2.7 Å resolution using either Fomblin or mineral oil as a cryoprotectant, but were highly mosaic (mosaicity >2°). However, the mosaicity of these crystals was substantially improved (mosaicity ≃ 0.5°) when using 3.4 M sodium malonate as the cryoprotectant (McPherson, 2001 ▶).
Figure 1.
Crystals of the cytoplasmic ZO-binding domain of human occludin. The ZO-binding domain of human occludin was expressed in a bacterial system and purified using a nickel column. Pure concentrated (13.7 mg ml−1) ZO-binding domain was crystallized using the hanging-drop vapor-diffusion technique. The average size of these crystals is 0.1 × 0.2 × 0.1 mm.
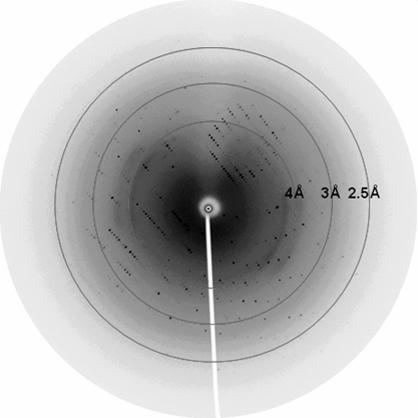
Typical diffraction images of a native crystal are shown in Fig. 2 ▶. The crystals belong to space group P212121 (systematic absences are shown in the supplementary material 1), with unit-cell parameters a = 33.3, b = 35.4, c = 107.4 Å. The Matthews coefficient indicates that there is only one molecule in the asymmetric unit and the solvent content is 40%. Statistics of a native data set collected at the APS are shown in Table 1 ▶. Data were collected to a resolution range of 1.8 Å and processed to 2.3 Å, which corresponds to I/σ(I) ≤ 2. Because of detector saturation for some reflections in the low-resolution shell, the overall completeness and redundancy for the data set are less than for the highest resolution shell (see supplementary material).
Figure 2.
Diffraction pattern of a native crystal.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Synchrotron radiation | 14-BM-C, APS |
| Detector | MAR345 CCD |
| Wavelength (Å) | 0.9 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 33.3, b = 35.4, c = 107.4 |
| Resolution range (Å) | 20–2.3 |
| Unique reflections | 5971 (392) |
| Completeness (%) | 98.0 (100.0) |
| Redundancy | 6.8 (7.5) |
| Rmerge (%) | 7.1 (41.1) |
| 〈I/σ(I)〉 | 42.9 (8.4) |
| Unit-cell volume (Å3) | 126605.3 |
| Solvent content (%) | 40.8 |
Owing to the lack of a homologous structure for molecular replacement, phasing of the crystal structure is currently in progress using multiple isomorphous replacement and selenomethionine multiwavelength anomalous diffraction techniques. The final structure of this domain will facilitate further investigation of the interactions between this domain and the GUK domain of the ZO proteins and will presumably apply to all species.
Supplementary Material
Supplementary material file. DOI: 10.1107/S1744309105007475/en5099sup1.pdf
Acknowledgments
Diffraction data used in this study were collected using the facilities of the Advanced Photon Source, which is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38. We thank Dr Henry Bellamy for diffraction screening using The Gulf Coast Protein Crystallography Consortium (GCPCC) PX1 beamline at The Center for Advanced Microstructures and Devices. This beamline is supported by NSF grant DBI-9871464 with co-funding from The National Institute for General Medical Sciences. The work is supported by NIH grant GM-45579 to JCL. RBS is supported (in part) by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
Footnotes
Supplementary material is available from Crystallography Journals Online (Reference: EN5099).
References
- Ando-Akatsuka, Y., Saitou, M., Hirase, T., Kishi, M., Sakakibara, A., Itoh, M., Yonemura, S., Furuse, M. & Tsukita, S. (1996). J. Cell. Biol.133, 43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Dallasta, L. M., Pisarov, L. A., Esplen, J. E., Werley, J. V., Moses, A. V., Nelson, J. A. & Achim, C. L. (1999). Am. J. Pathol.155, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning, A. S., Jameson, B. J., Jesaitis, L. A. & Anderson, J. M. (1998). J. Biol. Chem.273, 29745–29753. [DOI] [PubMed] [Google Scholar]
- Farkas, G., Marton, J., Nagy, Z., Mandi, Y., Takacs, T., Deli, M. A. & Abraham, C. S. (1998). Neurosci. Lett.242, 147–150. [DOI] [PubMed] [Google Scholar]
- Furuse, M., Hirase, T., Itoh, M., Nagafuchi, A., Yonemura, S., Tsukita, S. & Tsukita, S. (1993). J. Cell Biol.123, 1777–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse, M., Itoh, M., Hirase, T., Nagafuchi, A., Yonemura, S., Tsukita, S. & Tsukita, S. (1994). J. Cell Biol.127, 1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins, J., Gu, L., Wittchen, E. S., Hibbard, J. & Stevenson, B. R. (1998). J. Cell Biol.141, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, M., Morita, K. & Tsukita, S. (1999). J. Biol. Chem.274, 5981–5986. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1992). Jnt CCP4–ESF/EACBM Newsl. Protein Crystallogr.26
- McCarthy, K. M., Skare, I. B., Stankewich, M. C., Furuse, M., Tsukita, S., Rogers, R. A., Lynch, R. D. & Schneeberger, E. E. (1996). J. Cell Sci.109, 2287–2298. [DOI] [PubMed] [Google Scholar]
- McPherson, A. (1982). Preparation and Analysis of Protein Crystals. New York: John Wiley & Sons.
- McPherson, A. (2001). Protein Sci.10, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Peng, B., Lee, J. C. & Campbell, G. A. (2003). J. Biol. Chem.278, 49644–49651. [DOI] [PubMed] [Google Scholar]
- Petito, C. K. & Cash, K. S. (1992). Ann. Neurol.32, 658–666. [DOI] [PubMed] [Google Scholar]
- Saitou, M., Ando-Akatsuka, Y., Itoh, M., Furuse, M., Inazawa, J., Fujimoto, K. & Tsukita, S. (1997). Eur. J. Cell Biol.73, 222–231. [PubMed] [Google Scholar]
- Sharief, M. K., Ciardi, M. & Thompson, E. J. (1992). J. Infect. Dis.166, 350–358. [DOI] [PubMed] [Google Scholar]
- Wittchen, E. S., Haskins, J. & Stevenson, B. R. (1999). J. Biol. Chem.274, 35179–35185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material file. DOI: 10.1107/S1744309105007475/en5099sup1.pdf