Thioredoxin reductase 1 (Trr1) from S. cerevisiae is a component of the thioredoxin system, which is involved in several biological processes, including the reduction of disulfide bonds and response to oxidative stress. The expression, purification, crystallization and preliminary X-ray crystallographic studies of yeast Trr1 are reported.
Keywords: thioredoxin reductase I (TRR1), Saccharomyces cerevisiae
Abstract
Thioredoxin reductase 1 (Trr1) from Saccharomyces cerevisiae is a member of the family of pyridine nucleotide-disulfide oxidoreductases capable of reducing the redox-active disulfide bond of the cytosolic thioredoxin 1 (Trx1) and thioredoxin 2 (Trx2). NADPH, Trr1 and Trx1 (or Trx2) comprise the thioredoxin system, which is involved in several biological processes, including the reduction of disulfide bonds and response to oxidative stress. Recombinant Trr1 was expressed in Escherichia coli as a His6-tagged fusion protein and purified by nickel-affinity chromatography. The protein was crystallized using the hanging-drop vapour-diffusion method in the presence of PEG 3000 as precipitant after treatment with hydrogen peroxide. X-ray diffraction data were collected to a maximum resolution of 2.4 Å using a synchrotron-radiation source. The crystal belongs to the centred monoclinic space group C2, with unit-cell parameters a = 127.97, b = 135.41, c = 75.81 Å, β = 89.95°. The crystal structure was solved by molecular-replacement methods and structure refinement is in progress.
1. Introduction
In the cytoplasm of the majority of cell types, proteins are in a reduced state, in contrast to extracellular proteins where disulfide bonds are commonly found and help to maintain their structure in a very harsh environment. Thioredoxins and glutaredoxins are heat-stable proteins capable of reducing disulfide bonds in target proteins. These oxidoreductases contain two vicinal cysteines at their active sites that after reduction of the target disulfide become oxidized to a disulfide bond. The thioredoxin disulfide is reduced by nicotinamide adenine dinucleotide phosphate (NADPH) in a reaction catalyzed by thioredoxin reductase (Ritz & Beckwith, 2001 ▶).
Thioredoxin reductases are important flavoenzymes that belong to a family of proteins that includes lipoamide dehydrogenase and glutathione reductase. These proteins play a critical role in maintaining the redox status of cytoplasm. All these flavoenzymes contain two redox centres: a flavin adenine dinucleotide (FAD) and a dithiol/disulfide group (reviewed by Williams et al., 2000 ▶).
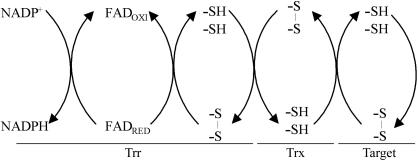
Because the reduction of disulfide bonds is a biochemical event involved in several processes, the thioredoxin system (comprised of NADPH, thioredoxin reductase and thioredoxin) is implicated in several phenomena, such as synthesis of deoxyribonucleotides, activation of transcription factors, regulation of the cell cycle, reduction of methionine sulfoxide, assimilation of sulfur and photosynthesis (Mustacich & Powis, 2000 ▶; Williams et al., 2000 ▶) (Fig. 1 ▶).
Figure 1.
Electron transfer between Trr, Trx and protein targets. Reducing equivalents from NADPH are transferred to the Trr FAD, then to enzyme disulfide. These equivalents are transferred to oxidized Trx, which finally reduces the target proteins.
Another important aspect of the thioredoxin system is its capacity to regenerate the reduced form of thiol-dependent peroxidases belonging to the peroxiredoxin family. Because of their enzymatic activity, these enzymes were named thioredoxin peroxidases (Chae et al., 1994 ▶). Thioredoxin peroxidases are able to reduce H2O2 or organic peroxides to water and alcohols, respectively (Chae, Chung et al., 1994 ▶; Chae, Robison et al., 1994 ▶; Park et al., 2000 ▶; Nordberg & Arner, 2001 ▶; Yoshida et al., 2003 ▶). This antioxidant system is conserved from prokaryotes to mammals and plants. In eukaryotes there are several counterparts in the cytosol and in organelles such as chloroplasts, mitochondria and even in the nucleus (Waksman et al., 1994 ▶; Dai et al., 1996 ▶; Park et al., 2000 ▶; Hofmann et al., 2002 ▶). The fact that the Saccharomyces cerevisiae trr1 null mutant is not viable (Giaever et al., 2002 ▶) probably reflects the fact that thioredoxin reductase 1 is implicated in several processes.
Two types of thioredoxin reductases appeared during evolution (Mustacich & Powis, 2000 ▶; Williams et al., 2000 ▶). Both are homodimeric proteins that catalyze the transfer of electrons through their FAD and a redox-active disulfide. In prokaryotes, lower eukaryotes and plants, each thioredoxin reductase subunit has a molecular weight of about 35 kDa. Thioredoxin reductases from higher eukaryotes possess a molecular weight of about 55 kDa and the active site contains a selenocysteine (Luthman & Holmgren, 1982 ▶; Buettner et al., 1999 ▶; Zhong & Holmgren, 2000 ▶).
Crystallographic studies have revealed that the location of the NADPH-binding and FAD-binding domains differ significantly in these two enzyme types. In higher eukaryotes, the distance between these domains and their orientation allows electron transport from NADPH to the disulfide of Trx without the need for a large alteration in protein conformation (Sandalova et al., 2001 ▶). On the other hand, Waksman et al. (1994 ▶) showed that in Escherichia coli Trr, the NADPH-binding and FAD-binding domains are located on distinct sides of the molecule, with the nicotinamide ring at a distance of more than 17 Å from the flavin ring. Moreover, the reactive cysteines of thioredoxin reductase are inaccessible for interaction with thioredoxin.
Lennon et al. (2000 ▶) explained the catalytic mechanism of this enzyme by describing the structure of a complex formed by E. coli Trr, Trx and AADP+ (3-aminopyridine dinucleotide phosphate), which is an analogue of NADPH. These authors showed that a large conformational change takes place to permit electron transport. The complex possesses a disulfide connecting Cys32 of Trx to Cys138 of Trr. In order to stabilize the complex, Cys35 of Trx and Cys135 of Trr were replaced by serines. Under these conditions, NADPH and FAD domains undergo a large rotation of 67° with respect to each other, which is necessary for electron transfer. This rotation places the nicotinamide ring of AADP+ and the disulfide bond close to the flavin ring and exposes the cysteine residues to reaction with thioredoxin. This twist of the two domains permits FAD reduction by NADPH and oxidation of the enzyme dithiol by thioredoxin (Lennon et al., 2000 ▶).
To date, Arabidopsis thaliana Trr (Dai et al., 1996 ▶) and E. coli Trr are the only low-molecular-weight thioredoxin reductases to have had their three-dimensional structures reported. Here, we report the preliminary X-ray diffraction analysis of Trr1 from S. cerevisiae in an oxidized state that represents the first Trr structure described in yeasts. The yeast Trr1 shares a sequence identity of 50 and 63% with its counterparts in E. coli and A. thaliana, respectively.
The structure was solved by molecular-replacement methods using the atomic coordinates of A. thaliana Trr as the search model. Analysis of the Trr1 structure should provide insights into the evolution and enzymatic mechanism of this protein and may also provide reasons for the specificity of the protein towards cytosolic thioredoxins.
2. Methods
2.1. Cloning
The 960-base-pair trr1 gene (YDR353W) was amplified by PCR from genomic DNA of S. cerevisiae (Invitrogen, catalogue No. 40802) and cloned in pPROEX-1 vector (Invitrogen) using BamHI-NdeI restriction sites. The resulting pPROEX/trr1 was sequenced in an Applied Biosystems ABI Prism 377 96 to confirm that the construction was correct.
2.2. Expression and purification
E. coli DH5α strain harbouring the pPROEX/trr1 plasmid was grown (50 ml) overnight in Luria–Bertani (LB) medium containing 50 µg ml−1 ampicillin at 310 K and transferred to 1 l of fresh LB/amp medium and cultured further at 310 K until the OD600 reached 0.6–0.8. Expression was induced with 1 mM IPTG and the cells were harvested after 4 h incubation. The cell pellet was resuspended in the starting buffer (20 mM sodium phosphate pH 7.4). The cells were lysed by sonication and the cell extract was kept on ice during streptomycin sulfate (1%) treatment for 15 min. The suspension was centrifuged at 31 500g for 30 min at 277 K to remove nucleic acid precipitate. Finally, the resulting supernatant was applied onto a nickel-affinity column (Hi-Trap from GE Healthcare). Bound protein was eluted with a linear gradient of 0–0.5 M imidazole. Protein purity was confirmed by SDS–PAGE. The purified protein was concentrated to 10 mg ml−1 in 5 mM Tris–HCl pH 7.5 for crystallization trials.
2.3. Crystallization and data collection
After H2O2 treatment (1 mM) at 310 K for 1 h, the samples were used to perform crystallization experiments using the hanging-drop vapour-diffusion method. Initial screenings were performed at 293 K using Crystal Screen and Crystal Screen 2 from Hampton Research. The drops, containing equal volumes (2.0 µl) of protein solution (10 mg ml−1 in 5 mM Tris–HCl pH 7.5) and reservoir solution, were equilibrated against 0.3 ml reservoir solution. From the initial screenings, several conditions produced thin plate-shaped crystals. Promising crystals were identified in five conditions: condition Nos. 10, 20 and 37 from Crystal Screen and condition Nos. 12 and 47 from Crystal Screen 2. All initial hits contained low-pH buffers and PEG as precipitant. Crystals suitable for diffraction experiments were obtained by slight variation of these conditions, such as the temperature of the assays (293 or 277 K). An additive search was also performed using Additive Screens 1, 2 and 3 from Hampton Research.
The best crystals, cryoprotected with the reservoir solution supplemented with 25% glycerol, were cooled to 110 K in a nitrogen-gas stream and X-ray diffraction data were collected using synchrotron radiation at the protein crystallography beamline D03B of the Laboratório Nacional de Luz Síncrotron (LNLS), Campinas, Brazil. LNLS D03B is a monochromatic beamline with a maximum photon flux between 1.3 and 1.6 Å. The wavelength of the incident X-ray was set to 1.431 Å and a MAR CCD detector was used to record the oscillation data with Δϕ = 1.0°, covering a total oscillation range of 240°. The data set was processed using the program MOSFLM (Leslie, 1992 ▶) and the resulting intensities were scaled and merged using the program SCALA (Evans, 1993 ▶) from the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
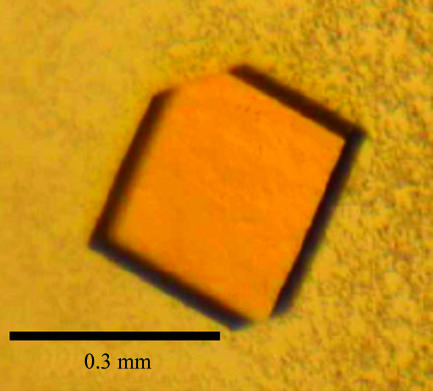
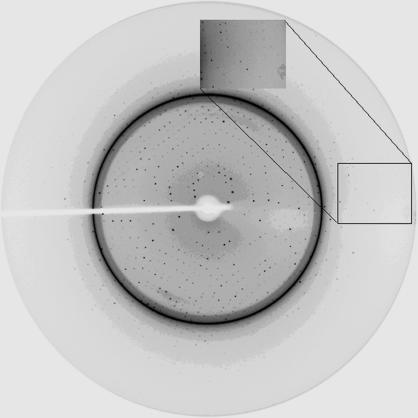
The optimal yeast Trr1 crystallization condition was obtained with a drop volume of 6.0 µl. 2.7 µl reservoir solution (sodium citrate pH 4.0, 12% PEG 3000) was mixed with an equal volume of protein solution and 0.6 µl 0.1 M trimethylamine hydrochloride was used as an additive. The best Trr1 crystals reached dimensions of 0.3 × 0.3 × 0.05 mm after four weeks (Fig. 2 ▶). Despite the presence of ice rings in the diffraction pattern (Fig. 3 ▶), data processing yielded a good quality data set to 2.4 Å resolution. A total of 297 954 measured reflections were merged into 48 181 unique reflections with an R sym of 9.9%. The crystal belongs to the monoclinic space group C2, with unit-cell parameters a = 127.97, b = 135.41, c = 75.81 Å, β = 89.95°. Table 1 ▶ summarizes the data-collection statistics.
Figure 2.
Trr1 crystal from S. cerevisiae. Crystals were obtained by vapour-diffusion equilibration against a reservoir consisting of sodium citrate pH 4.0, 12% PEG 3000 and 0.1 M trimethylamine hydrochloride.
Figure 3.
X-ray diffraction pattern obtained from a Trr1 crystal on beamline D03B at LNLS. The exposure time was 45 s and the oscillation range per frame was 1°. Despite the presence of ice rings, data processing yielded a good-quality data set to 2.4 Å resolution.
Table 1. Data-collection parameters and crystallographic data statistics.
Values in parentheses refer to the highest resolution shell.
| Temperature (K) | 110 |
| Wavelength (Å) | 1.431 |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 127.97, b = 135.41, c = 75.81, β = 89.95 |
| Resolution limits (Å) | 46.6–2.4 (2.53–2.4) |
| Total No. of reflections | 297954 |
| No. of unique reflections | 48181 |
| Completeness (%) | 99.6 (99.7) |
| Multiplicity | 6.1 (6.0) |
| Rsym (%) | 9.9 (32.5) |
| 〈I/σ(I)〉 | 17.6 (4.3) |
The protein structure was solved by molecular-replacement methods with the program AMoRe (Navaza, 2001 ▶) using the atomic coordinates of A. thaliana Trr (Dai et al., 1996 ▶) as the search model (63% sequence identity; PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1vdc). The molecular-replacement solution shows four monomers in the asymmetric unit, in agreement with the Matthews coefficient calculation (Matthews, 1968 ▶), the best result giving V M = 2.3 Å3 Da−1 and a solvent content of 45.5%. The molecular-replacement protocol resulted in an R factor and correlation factor of 51.0 and 33.3%, respectively, for the first solution and 53.1 and 27.7%, respectively, for the second solution. Initial rigid-body refinement was carried out using AMoRe, yielding an R factor of 0.459. Model completion and refinement are currently in progress.
Here, we report the X-ray diffraction data collection from yeast thioredoxin reductase crystals. Trr enzymes play important roles in cellular redox homeostasis and are present from archaea to mammals. Because the protein structure and mechanism of catalysis of Trr from higher eukaryotes differ substantially from those of its counterparts in plants and lower eukaryotes, an evolutionary divergence is suggested (Sandalova et al., 2001 ▶; Waksman et al., 1994 ▶). At present, only two Trr1 structures from lower organisms have been described: E. coli Trr and A. thaliana Trr (Waksman et al., 1994 ▶; Dai et al., 1996 ▶). The comparison of the yeast Trr1 structure with the bacterial and plant homologues should provide valuable information concerning the evolution of Trr and its enzymatic mechanism.
Acknowledgments
This work was supported by grant 01/07539-5, the Structural Molecular Biology Network (SMOLBnet), from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and by the Brazilian Synchrotron Light Laboratory (LNLS) under proposal D03B-CPR-2197.
References
- Buettner, C., Harney, J. W. & Barry, M. J. (1999). J. Biol. Chem.274, 21598–21602. [DOI] [PubMed] [Google Scholar]
- Chae, H. Z., Chung, S. J. & Rhee, S. G. (1994). J. Biol. Chem.269, 27670–27678. [PubMed] [Google Scholar]
- Chae, H. Z., Robison, K., Poole, L. B., Church, G., Storz, G. & Rhee, S. G. (1994). Proc. Natl Acad. Sci. USA, 91, 7017–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Dai, S., Saarinen, M., Ramaswamy, S., Meyer, Y., Jacquot, J. P. & Eklund. H. (1996). J. Mol. Biol.264, 1044–1057. [DOI] [PubMed] [Google Scholar]
- Evans, P. R. (1993). Proceedings of the CCP4 Study Weekend. Data Collection and Processing, edited by L. Sawyer, N. Isaacs & S. Bailey, pp. 114–122. Warrington: Daresbury Laboratory.
- Giaever, G. et al. (2002). Nature (London), 418, 387–391. [Google Scholar]
- Hofmann, B., Hecht, H.-J. & Flohe, L. (2002). Biol. Chem.383, 347–364. [DOI] [PubMed] [Google Scholar]
- Lennon, B. W., Williams, C. H. Jr & Ludwig, M. L. (2000). Science, 289, 1190–1194. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Luthman, M. & Holmgren A. (1982). Biochemistry, 26, 6628–6633. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Mustacich, D. & Powis, G. (2000). Biochem. J.346, 1–8. [PMC free article] [PubMed] [Google Scholar]
- Navaza, J. (2001). Acta Cryst. D57, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Nordberg, J. & Arner, E. S. (2001). Free Radic. Biol. Med.31, 1287–1312. [DOI] [PubMed] [Google Scholar]
- Park, S. G., Cha, M. K., Jeong, W. & Kim, I. H. (2000). J. Biol. Chem.275, 5723–5732. [DOI] [PubMed] [Google Scholar]
- Ritz, D. & Beckwith, J. (2001). Annu. Rev. Microbiol.55, 21–48. [DOI] [PubMed] [Google Scholar]
- Sandalova, T., Zhong, L., Lindqvist, Y., Holmgren, A. & Schneider, G. (2001). Proc. Natl Acad. Sci. USA, 98, 9533–9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman, G., Krishna, T. S. R., Williams, C. H. Jr & Kuriyan, J. (1994). J. Mol. Biol.236, 800–816. [PubMed] [Google Scholar]
- Williams, C. H., Arscott, L. D., Muller, S., Lennon, B. W., Ludwig, M. L., Wang, P. F., Veine, D. M., Becker, K. & Schirmer, R. H. (2000). Eur. J. Biochem.267, 6110–6117. [DOI] [PubMed] [Google Scholar]
- Yoshida, T., Oka, S., Masutani, H., Nakamura, H. & Yodoi, J. (2003). Antioxid. Redox Signal.5, 563–570. [DOI] [PubMed] [Google Scholar]
- Zhong, L. & Holmgren, A. (2000). J. Mol. Biol.275, 18121–18128. [DOI] [PubMed]