A conserved hypothetical protein XC6422 from X. campestris pv. campestris has been overexpressed in E. coli, purified and crystallized. Crystals obtained from the purified recombinant protein showed a variety of forms that diffracted to at least 1.6 Å resolution.
Keywords: conserved hypothetical proteins, Xanthomonas campestris, structural genomics
Abstract
Xanthomonas campestris pv. campestris is a Gram-negative yellow-pigmented pathogenic bacterium that causes black rot, one of the major worldwide diseases of cruciferous crops. Its genome contains approximately 4500 genes, roughly one third of which have no known structure and/or function. However, some genes of unknown function are highly conserved among several different bacterial genuses. XC6422 is one such conserved hypothetical protein and has been overexpressed in Escherichia coli, purified and crystallized in a variety of forms using the hanging-drop vapour-diffusion method. Crystals grew to approximately 2 × 1.5 × 0.4 mm in size after one week and diffracted to at least 1.6 Å resolution. They belong to the monoclinic space group C2, with one molecule per asymmetric unit and unit-cell parameters a = 75.8, b = 79.3, c = 38.2 Å, β = 109.4°. Determination of this structure may provide insights into the protein’s function.
1. Introduction
Structural genomics is a new and rapidly developing field in biology (Edwards et al., 2004 ▶; Zhang & Kim, 2004 ▶) and has been under active investigation worldwide. The goal of this frontier research is to discover novel protein folds and to solve the structures of a representative sample of protein molecules in order to obtain a more thorough understanding of biology from a structural perspective. We have joined the efforts and initiated a structural genomics program of the local plant pathogen Xanthomonas campestris pv. campestris strain 17 (Xcc). We have focused on studying its unique regulatory pathway towards pathogenicity. Xcc Clp (cAMP-receptor like protein) has been found to be a global transcription factor that is homologous to the cAMP receptor of Escherichia coli. It forms a complex with cAMP and is involved in the regulation of over 100 important cellular functions in Enterobacteriaceae and Bacillus (Ebright, 1993 ▶; Kolb et al., 1993 ▶). In Xcc, however, no cAMP-mediated signal transduction pathway was discovered and Clp was found to be solely responsible for regulating a wide variety of genes necessary for the synthesis of exopolyaccharides, extracellular enzyme and components of the apparatus for type II protein secretion, all of which are collectively required for its pathogenicity (de Crecy-Lagard et al., 1990 ▶; Chen & Tseng, 2005 ▶). Xcc is thus a peculiar phytopathogen that deserves more thorough studies in structural terms.
XC6422 has been classified as a conserved hypothetical protein by a bioinformatics approach (http://xcc.life.nthu.edu.tw/). It contains 220 amino acids and shares 94% sequence identity with a similar protein from the Xanthomonas genus (X. campestris pv. campestris strain ATCC 33913; gi|21111227; da Silva et al., 2002 ▶), 71% identity with a protein from Xylella fastidiosa (gi|9106911; Simpson et al., 2000 ▶) and 38% identity with a protein from Ralstonia solanacearum (gi|17427337; Salanoubat et al., 2002 ▶). To date, no homologous structure for the XC6422 protein has been reported in the PDB, although it is classified as a putative hydrolase belonging to the α/β-superfamily in the COG database (Tatusov et al., 2001 ▶). In this report, we describe the cloning, purification, crystallization and initial X-ray analysis of XC6422.
2. Materials and methods
2.1. Cloning, expression and purification
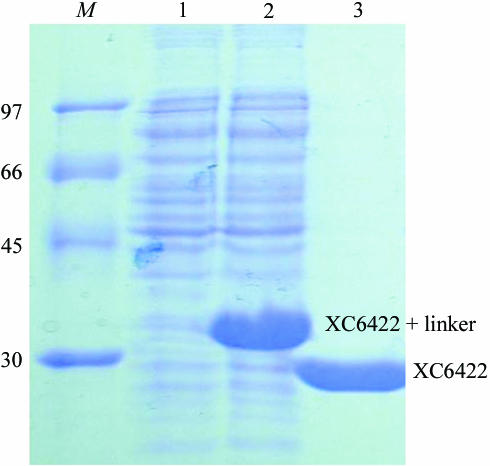
The XC6422 gene fragment was PCR-amplified directly from a local Xcc genome (X. campestris pv. campestris strain 17) with a forward 5′-TACTTCCAATCCAATGCTATGTCCAATCCCTTATTCCCCACC primer and a backward 5′-TTATCCACTTCCAATGTCAGGGCGTGGCCGGTAGCCAG primer. A ligation-independent cloning (LIC) approach (Aslanidis & de Jong, 1990 ▶) was carried out to obtain the desired construct. A pTBSG1 vector (F. P. Gao, unpublished results) was cut to completion with SspI (Novagen). For LIC, 100 ng of the linearized vector and 100 ng of the PCR product were treated with 2 units of T4 DNA polymerase (Novagen) in separate reactions in the presence of 2.5 mM dGTP or dCTP and 5 mM DTT, respectively. The reactions were carried out at 298 K for 30 min. The enzyme was subsequently heat-inactivated at 348 K for 20 min. The vector and the PCR product were then mixed and heated at 301 K for 3 min before cooling to room temperature. The mixture was directly transformed into the E. coli BL21 (DE3) host without ligation. The final construct codes for an N-terminal His6 tag, a 17-amino-acid linker and an XC6422 protein (220 amino acids) under the control of a T7 promoter. Transformed E. coli BL21 (DE3) host cells were grown in LB medium at 310 K until an OD600 of 0.8 was attained. Overexpression was induced by the addition of 1 mM IPTG at 310 K for 3.5 h. The cells were harvested, resuspended in equilibration buffer (20 mM Na2HPO4, 70 mM NaCl pH 8.0) and lysed using a microfluidizer (Microfluidics). Most tagged target proteins were in the soluble fraction (Fig. 1 ▶). After centrifugation, the target protein was purified by immobilized metal-affinity chromatography (IMAC) on a nickel column (Sigma), which was eluted with 20 mM Tris pH 8.0, 70 mM NaCl and a gradient of 50–300 mM imidazole. The fractions containing XC6422 were monitored by SDS–PAGE and recombined and dialyzed repeatedly against 50 mM Na2HPO4 pH 8.0, 10% glycerol and 500 mM NaCl. After buffer exchange, the His6 tag was cleaved from XC6422 by TEV (tobacco etch virus) protease at 283 K for 12 h to obtain the cleaved product. Upon rechromatography on the nickel column, the target XC6422 protein was in the flowthrough fractions, with the His6 tag and the tagged TEV protease retained on the nickel column. The purified protein was then dialyzed against 20 mM Tris pH 8.0 and 70 mM NaCl. For crystallization, XC6422 was further purified on an anion-exchange column (AKTA, Pharmacia Inc.). The fractions eluted with 20 mM Tris pH 8.0, 750 mM NaCl were combined and dialyzed against 20 mM Tris pH 8.0 and 70 mM NaCl. The final target protein (220 amino acids) has greater than 99% purity (Fig. 1 ▶) and contains only an extra tripeptide (SNA) at the N-terminal end with an expected MW of 24 251 Da, which was confirmed by mass-spectrometric analysis. The overexpression and purification of XC6422 was monitored by SDS–PAGE as shown in Fig. 1 ▶.
Figure 1.
SDS–PAGE monitoring of the overexpression and purification of XC6422. Lane M, molecular-weight markers in kDa; lane 1, soluble fraction before IPTG induction; lane 2, soluble fraction after IPTG induction; lane 3, purified XC6422 after TEV cleavage. The positions of fused and free XC6422 are also marked.
2.2. Crystallization
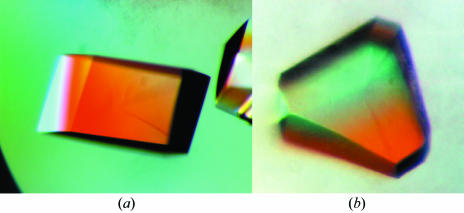
For crystallization, the protein was concentrated to 35.5 mg ml−1 in 20 mM Tris pH 8.0 and 70 mM NaCl using an Amicon Ultra-10 (Millipore). Crystallization screening was performed using sitting-drop vapour diffusion in 96-well plates (Hampton Research) at 295 K by mixing 0.5 µl protein solution with 0.5 µl reagent solution. Initial screens included the Hampton sparse-matrix Crystal Screens 1 and 2, a systematic PEG–pH screen and the PEG/Ion Screen and were performed using a Gilson C240 crystallization workstation. Parallelepiped-shaped and prism-shaped crystals appeared in 1 d from a reservoir solution comprising 0.1 M HEPES buffer pH 7.5, 2.0 M (NH4)2SO4 and 2% PEG 400. This initial condition was then optimized by varying the concentration of ammonium sulfate. Crystals suitable for diffraction experiments were grown by mixing 1.5 µl protein solution with 1.5 µl reagent solution and reached maximum dimensions of 2.0 × 1.5 × 0.4 mm after one week (Fig. 2 ▶).
Figure 2.
Two different crystal forms of XC6422 grown by the sitting-drop vapour-diffusion method. Crystallization conditions were optimized to 0.1 M HEPES buffer pH 7.5, 1.6 M (NH4)2SO4 and 2% PEG 400 (a) and 0.1 M Tris buffer pH 8.5, 2.2 M (NH4)2SO4 and 2% PEG 400 (b). The approximate dimensions of these crystals were 2 × 1.5 × 0.4 mm.
2.3. Data collection
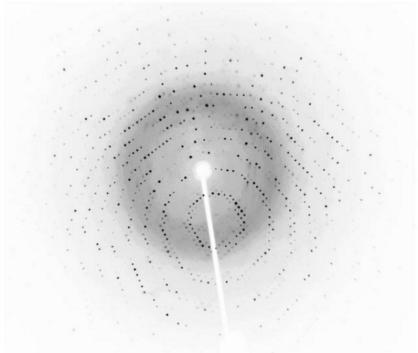
Crystals were soaked in a cryoprotectant solution comprising reservoir solution plus 20%(v/v) glycerol and were then flash-cooled at 100 K in a stream of cold nitrogen. X-ray diffraction data were collected using Cu Kα radiation from a Rigaku RU-300 rotating-anode generator equipped with Osmic mirror optics and an R-AXIS IV++ image plate. A native data set was obtained to a resolution of at least 1.6 Å; the data were indexed and integrated using the HKL software suite (Otwinowski & Minor, 1997 ▶), giving a data set that was 99.1% complete (91.3% in the last shell) with an overall R merge of 4.1% on intensities. The crystals belong to the monoclinic space group C2, with one molecule in each asymmetric unit, and contain 45.1% solvent. The data-collection statistics are summarized in Table 1 ▶. An X-ray diffraction image collected in-house is shown in Fig. 3 ▶.
Table 1. Data-collection statistics for native XC6422 crystals.
Values in parentheses are for the highest resolution shell.
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 75.8, b = 79.3, c = 38.2, β = 109.4 |
| Data-collection temperature (K) | 100 |
| Wavelength (Å) | 1.5418 |
| Resolution range (Å) | 24.8–1.60 (1.66–1.60) |
| Mosaicity (°) | 0.3 |
| Unique reflections | 27824 (2547) |
| Redundancy | 2.9 (2.4) |
| Completeness (%) | 99.1 (91.3) |
| Rmerge (%) | 4.1 (14.4) |
| Mean I/σ(I) | 14.7 (4.6) |
| Solvent content (%) | 45.12 |
Figure 3.
Diffraction pattern of XC6422 collected in-house from a crystal flash-frozen with 20% glycerol cryoprotectant. The exposure time was 10 min, with an oscillation range of 1.0° and a crystal-to-detector distance of 100 mm.
3. Results and discussion
The gene sequence of XC6422 was confirmed after cloning and consists of 663 bp coding for a protein of 220 amino-acid residues, with a calculated isoelectric point of 4.83. The purified XC6422 showed a single band of approximately 24 kDa on SDS–PAGE, with a greater than 99% purity (Fig. 1 ▶). Such a high purity possibly accounts for its straightforward crystallization; good crystals of up to 2.0 × 1.5 × 0.4 mm in size with high diffraction resolution of at least 1.60 Å were readily obtained in one week (Fig. 2 ▶).
We have chosen proteins with unknown structure and/or unknown functions as our targets to increase the possibility of discovering novel protein folds. High-resolution diffraction data were obtained for the native XC6422 crystals (one of which is shown in Fig. 3 ▶), indicating their suitability for further detailed X-ray structural analysis. We now plan to solve the structure of XC6422 using either the multiple isomorphous replacement (MIR) method (Ke, 1997 ▶) or the multiwavelength anomalous diffraction (MAD) method using selenomethionine-substituted protein (Hendrickson & Ogata, 1997 ▶), given that XC6422 contains one cysteine and four methionines, respectively. Heavy-atom positions and phases will be solved using automated Patterson analysis as described in Terwilliger & Berendzen (1999 ▶).
Acknowledgments
This work was supported by an Academic Excellence Pursuit grant from the Ministry of Education and by the National Science Council, Taiwan to S-HC and P-CL. We also thank the Core Facility for Protein Production of the Academia Sinica, Taiwan for providing us with the original vectors used in this study, and the Core Facility for Protein X-ray Crystallography of the Academia Sinica, Taiwan for analyzing the preliminary X-ray data.
References
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res.18, 6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.-C. & Tseng, Y.-H. (2005). Submitted.
- Crecy-Lagard, V. de, Glaser, P., Lejeune, P., Sismeiro, O., Barber, C. E., Daniels, M. J. & Danchin, A. (1990). J. Bacteriol.172, 5877–5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright, R. H. (1993). Mol. Microbiol.8, 797–802. [DOI] [PubMed] [Google Scholar]
- Edwards, A. F., Yee, A., Savchenko, A., Edwards, A. M. & Arrowsmith, C. H. (2004). Curr. Opin. Chem. Biol.8, 42–48. [DOI] [PubMed] [Google Scholar]
- Hendrickson, W. A. & Ogata, C. M. (1997). Methods Enzymol.276, 494–523. [DOI] [PubMed]
- Ke, H. (1997). Methods Enzymol.276, 448–461. [DOI] [PubMed]
- Kolb, A., Busby, S., Buc, H., Garges, S. & Adhya, S. (1993). Annu. Rev. Biochem.62, 749–795. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Salanoubat, M. et al. (2002). Nature (London), 415, 497–502. [Google Scholar]
- Silva, A. C. R. da et al. (2002). Nature (London), 417, 459–463. [Google Scholar]
- Simpson, A. J. G. et al. (2000). Nature (London), 406, 151–156. [Google Scholar]
- Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., Kiryutin, B., Galperin, M. Y., Fedorova, N. D. & Koonin, E. V. (2001). Nucleic Acids Res.29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. & Kim, S.-H. (2004). Curr. Opin. Chem. Biol.7, 28–32.