Abstract
Forkhead box O (Foxo) transcription factors induce muscle atrophy by upregulating the muscle-specific E3 ubiquitin ligases MuRF-1 and atrogin-1/MAFbx, but other than Akt, the upstream regulators of Foxos during muscle atrophy are largely unknown. To examine the involvement of the dystrophin glycoprotein complex (DGC) in regulation of Foxo activities and muscle atrophy, we analyzed the expression of DGC members during tail suspension, a model of unloading-induced muscle atrophy. Among several DGC members, only neuronal NOS (nNOS) quickly dislocated from the sarcolemma to the cytoplasm during tail suspension. Electron paramagnetic resonance spectrometry revealed production of NO in atrophying muscle. nNOS-null mice showed much milder muscle atrophy after tail suspension than did wild-type mice. Importantly, nuclear accumulation of dephosphorylated Foxo3a was not evident in nNOS-null muscle, and neither MuRF-1 nor atrogin-1/MAFbx were upregulated during tail suspension. Furthermore, an nNOS-specific inhibitor, 7-nitroindazole, significantly prevented suspension-induced muscle atrophy. The NF-κB pathway was activated in both wild-type and nNOS-null muscle during tail suspension. We also show that nNOS was involved in the mechanism of denervation-induced atrophy. We conclude that nNOS/NO mediates muscle atrophy via regulation of Foxo transcription factors and is a new therapeutic target for disuse-induced muscle atrophy.
Introduction
Reduced muscle activity such as bed rest, limb immobilization, denervation, or unloading (e.g., tail suspension or space flight) leads to significant muscle atrophy (1, 2). In these conditions, the atrophying muscles show increased rates of protein degradation mainly through activation of the ubiquitin proteasome system (3, 4), and the muscle-specific E3 ubiquitin ligases muscle-specific RING finger protein 1 (MuRF-1) and atrogin-1/muscle atrophy F-box protein (atrogin-1/MAFbx) are commonly upregulated (3, 4). Recent studies further showed that muscle inactivity results in suppression of the IGF-1/PI3K/Akt pathway (5–8) and activation of transcription factors such as the forkhead box O (Foxo) family and NF-κB (9–11).
Foxo family of forkhead transcription factors regulates a variety of biological process such as metabolism, cell proliferation and death, tumor growth, response to stress, and longevity (12–15). Mammals have 4 members, Foxo1a, Foxo3a, Foxo4, and Foxo6 (12). Among them, Foxo1a is activated in almost all forms of muscle atrophy (16). Importantly, several experiments indicate that forced expression of Foxo1a or Foxo3a upregulates the expression of a variety of atrophy-related genes including MuRF-1 and atrogin-1/MAFbx genes and induces muscle atrophy both in vitro and in vivo (4, 10, 16, 17). These observations indicate that Foxo1a and Foxo3a are central players in atrophy signaling.
The activities of Foxo factors are thought to be regulated mainly at posttranslational levels by a serine/threonine kinase Akt. In the absence of muscle activities, Akt is inactivated, leading to dephosphorylation of Foxo transcription factors. As a result, dephosphorylated Foxos enter into the nucleus and activate atrophy-inducing genes (10, 18). Conversely, when Akt is activated, Foxos are phosphorylated and bound by 14-3-3 proteins and move from nucleus to cytoplasm (13, 19).
NF-κB is another key signaling molecule in muscle atrophy. In particular, crucial roles for NF-κB in both cachexia- and inactivity-induced muscle atrophy have been well investigated (9, 20, 21).
Several molecules are proposed as a mechanical sensor or a trigger of disuse atrophy. Recently it has been reported that muscles from tumor-bearing mice exhibited reduced levels of dystrophin, the protein that is mutated in Duchenne muscular dystrophy, together with reduced levels of dystrophin-associated glycoprotein (22, 23). Furthermore, forced expression of dystrophin in transgenic mice counteracted cachexia-induced muscle atrophy (22). The role of the dystrophin glycoprotein complex (DGC) in inactivity-induced muscle atrophy is not known yet, but these results raise the possibility that the DGC works as a regulator of muscle atrophy or serves as a scaffold for anti-atrophic signal transduction.
To clarify the roles for the DGC in muscle atrophy, we examined the expression and function of the members of DGC in skeletal muscle during tail suspension, a model of unloading. Here we demonstrate that neuronal NOS (nNOS), a peripheral member of the DGC, is activated in unloading conditions, regulates Foxo3a, and promotes muscle atrophy. We also show that a nNOS-specific inhibitor, 7-nitroindazole (7NI), significantly attenuates suspension-induced muscle atrophy. Furthermore, we show the involvement of nNOS in denervation-induced muscle atrophy process. Thus nNOS and NO are to our knowledge new therapeutic targets for disuse-induced muscle atrophy.
Results
nNOS disappears from the sarcolemma during tail suspension.
To elucidate molecular mechanisms of unloading-induced muscle atrophy, we performed tail suspension (14 days) and reloading (7 days) experiments using wild-type C57BL/6 mice. The weights of the soleus and gastrocnemius (Figure 1A) muscles were decreased to 50%–70% of those of the control mice after tail suspension. The mice showed weakened grasping power and less endurance in running on the rotarod test after tail suspension (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI30654DS1). The diameter of myofiber was also drastically decreased (Figure 1B). The expression patterns of the components of DGC, dystrophin, β-dystroglycan, α-sarcoglycan, dystrobrevin, laminin-α2, α1-syntrophin, and caveolin-3 were not changed during tail suspension (data not shown). The serum creatine kinase level was not elevated, and Evans blue dye uptake by myofibers was not evident in atrophied muscles (Supplemental Figure 2), indicating that the sarcolemmal integrity was maintained during tail suspension. nNOS mRNA levels were not significantly reduced (Figure 1F), and total nNOS protein was slightly decreased during tail suspension (Figure 1D). Importantly, immunohistochemistry revealed that nNOS was lost from the sarcolemma during tail suspension (Figure 1C). The sarcolemmal expression was gradually restored during the reloading process (Figure 1C). eNOS expression was not changed (Figure 1D), and iNOS was not expressed (data not shown) during the course.
Figure 1. nNOS disappears from the sarcolemma during tail suspension.
(A) Weights of soleus (Sol) and gastrocnemius (Gc) muscles from tail-suspended (sus) wild-type mice are normalized to body weight and are expressed as percentage of ground control (gro) (n = 15/group). *P < 0.05, Student’s t test. (B) Box and whiskers plot of myofiber diameter size of soleus. Boxes represent the middle 50% of the data, lines represent the median, and whiskers represent the range. More than 200 fibers were measured on laminin-α2 chain–stained cross sections. (C) Immunofluorescent staining for nNOS and laminin-α2 chain. Transverse muscle sections from ground control mice, tail-suspended mice, and mice after reloading for 7 days (reload 7d) were stained with anti-nNOS (green) and anti–laminin-α2 chain antibodies (red). Scale bar: 50 μm. (D) Western blot using anti-nNOS antibody on subcellular fractions of muscle extracts. P indicates insoluble pellet after sequential extraction of skeletal muscle homogenates with 100 mM NaCl (S1), 500 mM NaCl (S2), and 0.5% Triton X-100 (S3). GAPDH signals in S1 and Na/K-ATPase signals in P confirmed that our fractionation was correctly done. Fractionation and western blotting were repeated 5 times, and representative data are presented. Note the slight increase of nNOS levels in S1 fraction and loss of nNOS signal in insoluble P fraction during tail suspension. (E) Quantification of nNOS signals in S1 and P fractions of muscle extracts shown in D (n = 5 /group). The signals in S1 and P fractions were normalized to GAPDH or Na/K-ATPase, respectively. Mann-Whitney, *P < 0.05. (F) Levels of nNOS mRNA in muscles from ground control mice and tail-suspended mice for 2 weeks were evaluated by real-time PCR (n = 5/group). No significant difference was found by Mann-Whitney test. (G) Immunoprecipitation with caveolin-3 antibody and immunoblot with nNOS antibody, and vice versa, for ground control and tail suspension groups. (H) Immunoprecipitation with α1-syntrophin Ab and immunoblot with nNOS antibody, and vice versa. (I) Immunoprecipitation of dysferlin antibody and immunoblot with nNOS Ab, and vice versa. In G–I, flow-through fraction was also examined by western blotting with anti–α-tubulin antibody.
Next we performed subcellular fractionation of muscle homogenates as described previously (24). In ground control mice, nNOS remained in the insoluble pellet (Figure 1D). In tail-suspended mice, nNOS was largely extracted with 100 mM NaCl and barely detected in the pellet fraction (Figure 1D). Quantification of nNOS bands clearly showed that nNOS dislocated from the sarcolemma to the cytoplasm during tail suspension (Figure 1E). These results indicate that nNOS exists mainly in the cytoplasm during tail suspension. Immunoprecipitation experiments also revealed that nNOS completely dissociated from α1-syntrophin and caveolin-3 during tail suspension (Figure 1, G and H). In contrast, a considerable amount of dysferlin was immunoprecipitated with nNOS antibody during tail suspension (Figure 1I).
nNOS-null mice represent tolerance to disuse-induced muscle atrophy.
To examine the roles of dislocated nNOS in muscle atrophy, we then performed tail suspension experiments using wild-type, nNOS-null, eNOS-null, and α1-syntrophin–null mice. The body weight (Figure 2A), average muscle fiber diameter (Figure 2B), and total number of muscle fibers (data not shown) of nNOS-null mice in the ground condition were similar to those of wild-type mice. However, after tail suspension for 14 days, reduction of muscle weight (Figure 2A), muscle size (Figure 2B), and muscle power (Supplemental Figure 1) were significantly less severe in the nNOS-null mice. Except for nNOS, dystrophin and other components of DGC were expressed in nNOS-null muscle at the same level as in wild-type muscle in both ground and suspended conditions (Figure 2C). eNOS-null muscle revealed atrophy during tail suspension similar to that seen in wild-type muscle (Figure 2, A and B), indicating that eNOS is not essential for atrophy signaling. We previously reported that disruption of the α1-syntrophin gene resulted in dislocation of nNOS from the sarcolemma to the cytoplasm without dystrophic phenotypes (25, 26). Suspension of α1-syntrophin–null mice induced severe muscle atrophy as it did in wild-type muscle (Figure 2, A and B).
Figure 2. nNOS-null mice show partial tolerance to disuse-induced muscle atrophy.
(A) Soleus muscle weight from ground control and tail-suspended wild-type (n = 20), nNOS-null (n = 20), eNOS-null (n = 10), and α1-syntrophin–null (n = 10) mice after 2-week tail suspension is shown as percent of wild-type ground controls. **P < 0.01, Student’s t test. (B) Box and whiskers plot of diameter of myofiber in soleus. Diameters were measured on H&E-stained cross sections of soleus muscles. n = 200 fibers in each experiment. *P < 0.05, Student’s t test. (C) Immunoblots of mouse gastrocnemius muscle extracts for DGC components from wild-type ground control (lane 1), wild-type tail-suspended (lane 2), nNOS-null ground control (lane 3), and nNOS-null tail-suspended (lane 4) mice. All lanes contain 30 μg of total protein. The experiments were performed 5 times, and representative pictures are presented.
Production of NO by nNOS in tail-suspended skeletal muscle.
To directly measure the levels of NO in skeletal muscle during tail suspension, we employed electron paramagnetic resonance (EPR) spectrometry with N-methyl-D-glucamine-dithiocarbamate (MGD) and Fe2+ (27, 28). We injected MGD and Fe2+ into the wild-type and nNOS-null mice at the final stage of 2-week suspension, sacrificed the mice 30 minutes after the injection, and immediately measured the NO levels in muscle tissues. In EPR, the signal height, which is proportional to the amount of NO, was obtained by measuring the peak-to-peak height of the lower field side signal (arrow in Figure 3A) in the characteristic 3-line NO spectrum (29). NO levels in skeletal muscles increased during tail suspension, and the increase was inhibited by daily administration of 7NI to the mice. We used 50 mg/kg of 7NI to selectively inhibit nNOS (30, 31). The increase of NO levels in skeletal muscle during tail suspension was also inhibited by daily administration of 10 mg/kg of N-nitro- l-arginine methylester ( l-NAME; pan-NOS inhibitor) (30) to the same extent (data not shown). NO levels in the skeletal muscle of nNOS-null mice after tail suspension were not different from those of ground control mice (Figure 3B), indicating that nNOS is mainly responsible for elevated levels of NO in muscle during tail suspension. Assay of the catalytic activity of NOS showed a higher level in the soluble (cytoplasmic) fraction than in the particulate fraction of suspended wild-type mice (Figure 3C).
Figure 3. Measurement of NO in muscle and NOS activity during tail suspension.
(A) EPR spectra of the NO adduct of Fe-MGD complex observed in skeletal muscle at room temperature. The NO-trapping agent was injected 30 minutes before measurements were taken. The EPR spectra of ground control and tail-suspended wild-type and nNOS-null mice were shown. Each spectrum represents the average of 5 accumulations. The signal height was obtained by measuring the peak-to-peak height of the lower field side signal (vertical arrows) in the 3-line spectrum. (B) NO levels of the skeletal muscle with and without tail suspension for 7 or 14 days were analyzed in EPR spectrometry (n = 6; *P < 0.05, Mann-Whitney). Note that NO level in muscle is elevated in wild-type tail-suspended mice at 14 days but not in nNOS-null tail-suspended mice. 7NI was used as a selective nNOS inhibitor. (C) NOS catalytic activity in soluble and particulate fractions (n = 5; Mann-Whitney). [3H]-citrulline, converted from [3H]-arginine in vitro by NOS, was quantified by liquid scintillation spectroscopy. Note higher NOS activities in the soluble fraction than in the particulate fraction for suspended wild-type mice.
Production of NO by nNOS is upstream of Foxo3a pathway.
Foxo transcription factors are reported to upregulate many atrophy-related genes and promote muscle atrophy (9, 10). We found that Foxo3a was dephosphorylated and accumulated in the myonuclei of wild-type mice but not nNOS-null mice in western blotting (Figure 4, A and B) and in immunohistochemistry (Supplemental Figure 3). Foxo1 and Foxo4 were not changed during tail suspension in both wild-type and nNOS-null mice (Figure 4B). Recently, several groups have pointed out that the ubiquitin proteasome pathway is largely involved in selective protein degradation during the muscle atrophy process (3, 4). Consistent with this, mRNA levels of 2 muscle-specific E3 ubiquitin ligases, MuRF-1 and atrogin-1/MAFbx, increased during tail suspension in wild-type mice (Figure 4C) (9, 10). Remarkably, the upregulation of these E3 ligases was modest in nNOS-null mice. These observations suggest that nNOS regulates Foxo3a via NO production and thereby upregulates MuRF-1 and atrogin-1/MAFbx. To further examine whether nNOS regulates Foxo3a, we overexpressed nNOS in myotubes by a retrovirus vector (Supplemental Figure 4). Overexpressed nNOS increased both total and nuclear Foxo3a protein levels and decreased phosphorylated Foxo3a in myotubes (Supplemental Figure 4).
Figure 4. Participation of nNOS in regulation of Foxo3a and upregulation of MuRF-1 and atrogin-1/MAFbx.
(A) The amounts of total Foxo3a, phosphorylated Foxo3a (p-Foxo3a), and intranuclear Foxo3a in wild-type and nNOS-null muscle during tail suspension were quantified (n = 5). Note that Foxo3a was dephosphorylated and accumulated in the myonuclei of wild-type mice but not of nNOS-null mice. *P < 0.05, Mann-Whitney. (B) Representative immunoblot analysis for Foxo3a, phosphorylated Foxo3a, Foxo1, and Foxo4 in total muscle extract, and Foxo3a in nuclear extracts in wild-type ground control (lane 1), wild-type tail-suspended (lane 2), nNOS-null ground control (lane 3), and nNOS-null tail-suspended (lane 4) muscles. α-Tubulin was used as a loading control. (C) mRNA levels of ubiquitin ligases (MuRF-1 and atrogin-1/MAFbx) (n = 5) were quantified by real-time RT-PCR. *P < 0.05, Mann-Whitney. (D) EMSA of NF-κB. Biotin-labeled double-stranded oligonucleotides containing NF-κB binding sites were incubated with nuclear extracts prepared from ground control and suspended muscles. An arrow indicates the DNA-protein complex. LPS was injected intraperitoneally into mice, and the muscle was used as a positive control for NF-κB binding activity. 5× cold, 5-fold excess of nonlabeled competitors.
NF-κB pathway is activated during muscle atrophy in nNOS-null mice.
NF-κB has been shown to be a major regulator of tail suspension–induced muscle atrophy (20, 21). EMSA showed that binding activity of NF-κB to its authentic binding sequence is increased by tail suspension in both wild-type and nNOS-null mice (Figure 4D). Importantly, there was no difference between tail-suspended wild-type and nNOS-null mice in the NF-κB binding activity (Figure 4D). In addition, western blotting revealed that p50 is increased by tail suspension (data not shown). These results suggest that NF-κB pathway was activated during tail suspension in the absence of nNOS. Whether the NF-κB activities mediate the residual muscle atrophy that occurred in nNOS-null mice during tail suspension remains to be clarified in a future study.
Inhibitor of NF-κB kinase β is nitrosylated during tail-suspension.
Foxo3a is known to be phosphorylated by Akt in skeletal muscle (10, 18). In contrast to our expectation, there was no difference between wild-type and nNOS-null mice in the phosphorylation levels of Akt (Figure 5, A and B). There was no difference between the levels of S6k1 and mammalian target of rapamycin (mTOR), which are under regulation by PI3K/Akt signaling and positively regulate protein synthesis between wild-type and nNOS-null mice during tail suspension (5) (Figure 5A). A recent study reported that NO S-nitrosylates inhibitor of NF-κB kinase β (IKKβ) and thereby inhibits its activity (32). Other reports described inhibition of Foxo3a by IKKβ (33). Intriguingly, we found that tyrosine residues of IKKβ were nitrosylated during tail suspension in wild-type mice but not in nNOS-null mice (Figure 5C). However, whether S-nitrosylation of IKKβ detected during tail suspension contributes to activation of Foxo3a remains to be determined.
Figure 5. Phosphorylation of Akt and nitrosylation of IKKβ during 2-week tail suspension.
(A) Immunoblots for protein synthesis pathway components (p-Akt, Akt, p-S6k1, S6k1, and p-mTOR) in wild-type ground control (lane 1), wild-type tail-suspended (lane 2), nNOS-null ground control (lane 3), and nNOS-null tail-suspended (lane 4) muscles. Densities of the bands were normalized to α-tubulin. (B) The ratio of phosphorylated Akt to total Akt is shown (n = 5; *P < 0.05, Mann-Whitney). (C) Detection of nitrosylated IKKβ. Muscle proteins were immunoprecipitated with anti–3-nitrotyrosine antibody (upper panel) or with anti-IKKβ antibody (lower panel) and immunoblotted with anti-IKKβ or anti–3-nitrotyrosine antibodies, respectively. α-Tubulin signals in flow-through fractions are also shown.
7NI alleviates tail suspension–induced muscle atrophy.
To further examine the effect of inhibition of nNOS activity on muscle atrophy, 7NI, a nNOS-selective inhibitor, was injected daily into the peritoneal space of the wild-type mice during the 2-week tail suspension (Figure 6). This treatment significantly prevented muscle atrophy during tail suspension but did not increase muscle mass of ground control mice (Figure 6A). 7NI considerably increased phosphorylated Foxo3a and inhibited the increase in dephosphorylated and nuclear Foxo3a during tail suspension (Figure 6C). We also found that upregulation in mRNA levels of MuRF-1 and atrogin-1/MAFbx was abolished by 7NI during tail suspension (Figure 6B). These data imply that nNOS-specific inhibitor is a potential therapeutic strategy for disuse-induced muscle atrophy.
Figure 6. 7NI alleviates muscle atrophy during tail suspension.
(A) Soleus muscle weight of wild-type ground control and wild-type tail-suspended mice with (+7NI) or without 7NI (+oil). Oil or 7NI (50 mg/kg/d) was injected daily into the abdominal cavity of the mice during 2-week tail suspension. Values (muscle weight/body weight) are expressed as percent of wild-type oil-injected muscles after 14-day tail suspension (n = 5–10 per group; *P < 0.05, Mann-Whitney). (B) mRNA levels of MuRF-1 and atrogin-1/MAFbx (n = 4) in muscle from wild-type ground control and wild-type tail-suspended mice with or without 7NI were quantified by real-time RT-PCR (*P < 0.05, Mann-Whitney). (C) The amount of total Foxo3a, dephosphorylated Foxo3a, and intranuclear Foxo3a (n = 4) were analyzed by western blotting, and band densities were normalized to α-tubulin (*P < 0.05, Mann-Whitney).
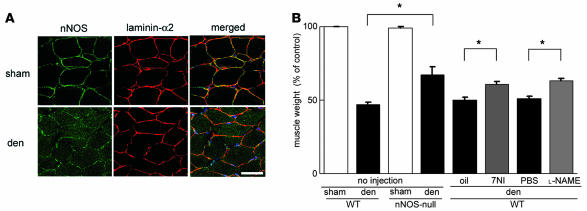
nNOS-null mice show milder muscle atrophy than wild-type mice after cutting of the sciatic nerve.
We next examined the role of nNOS in denervation-induced muscle atrophy. Cutting the sciatic nerve on the denervated side resulted in greatly reduced muscle weight 14 days after operation (Figure 7A). Importantly, nNOS had already disappeared from the sarcolemma 3 days after denervation (Figure 7A). We observed much milder muscle atrophy in denervated muscle of nNOS-null mice than wild-type mice (Figure 7B), suggesting that nNOS is also involved in denervation-induced muscle atrophy. We then tested whether an nNOS inhibitor (7NI) or a pan-NOS inhibitor ( l-NAME) counteracts denervation-induced muscle atrophy (Figure 7B). These 2 inhibitors limited the muscle atrophy (Figure 7B), indicating that NO is indeed a mediator and therefore a therapeutic target for denervation-induced muscle atrophy.
Figure 7. Inhibition of nNOS activities counteracts denervation-induced muscle atrophy.
(A) Sarcolemmal expression of nNOS in control and denervated muscles. Transverse muscle sections from denervated (den) and sham-operated mice were stained with anti-nNOS (green) and anti–laminin-α2 chain (red) antibodies 3 days after sciatic nerve excision. Scale bar: 50 μm. (B) Reduction in soleus muscle weight of wild-type and nNOS-null mice 2 weeks after sham operation or denervation. Weight of wild-type denervated muscle was also measured after 2-week administration of oil, 7NI, PBS, or l-NAME. Values (muscle weight/body weight) were expressed as percentage of the values of sham-operated wild-type muscles (n = 5–10 per group). *P < 0.05, Mann-Whitney.
Discussion
Dislocation of nNOS is a major step in tail suspension–induced muscle atrophy.
Involvement of the DGC in cachexia-induced muscle atrophy was recently reported (22). In our report, we demonstrate for what we believe to be the first time that nNOS is dislocated from the sarcolemma to the cytoplasm during tail suspension, whereas other members of the DGC are normally expressed at the sarcolemma. This observation implies that different mechanisms are involved in unloading-induced muscle atrophy and muscle atrophy seen in cachexia. Sarcolemmal nNOS is reported to be a versatile molecule that modulates satellite cell activation (34), formation of neuromuscular junction (35), glucose uptake (36), muscle contraction, and vasodilation (37). To clarify the mechanisms of nNOS translocation, we examined the effects of clenbuterol, streptomycin, and nifedipine on nNOS dislocation during tail suspension. After administration of these drugs, however, we still observed dislocation of nNOS during tail suspension (data not shown). These results suggest that sympathetic nerves, stretch-activated channels, and L-type calcium channels are not involved in the dissociation of nNOS from the DGC.
nNOS is anchored at the sarcolemma by interaction with α1-syntrophin (25), a member of the DGC; interestingly, however, α1-syntrophin remains at the sarcolemma during tail suspension (data not shown). To examine whether modification of α1-syntrophin is involved in the dissociation of nNOS from α1-syntrophin, we performed 2-dimensional PAGE and western blotting with anti–α1-syntrophin antibody as previously described (38). The first dimensional isoelectric focusing reveals posttranslational modifications of α1-syntrophin. The results showed slight changes in mobility pattern of α1-syntrophin during tail suspension (data not shown), suggesting that some posttranslational modifications of α1-syntrophin may cause dissociation of nNOS from α1-syntrophin.
Dislocated nNOS leads to production of NO and regulates Foxo/E3 ubiquitin ligases pathway.
EPR spectrometry confirmed that cytoplasmic nNOS led to production of NO during tail suspension (Figure 3, A and B). Tail suspension–induced muscle atrophy was blunted in nNOS-null mice (Figure 2, A and B) and 7NI-treated mice (Figure 6A) but not in eNOS-null mice (Figure 2, A and B). iNOS protein was not detected by western blotting in skeletal muscle during tail suspension (data not shown). These data indicate that dislocated nNOS, but neither eNOS nor iNOS, is involved in tail suspension–induced muscle atrophy.
We clearly show that 2 atrophy-related E3 ubiquitin ligases, MuRF-1 and atrogin-1/MAFbx, are not upregulated in nNOS-null muscle during tail suspension (Figure 4C and Figure 6B). Therefore, the induction of these genes is a downstream event of dislocation of nNOS in tail suspension–induced muscle atrophy.
Foxo transcription factors are reported to induce skeletal muscle atrophy by upregulating MuRF-1 or atrogin-1/MAFbx (4, 10, 16, 17). For example, transgenic mice overexpressing Foxo1 in skeletal muscle display a decrease in size of muscle fibers (17). Importantly, Foxo3a remained phosphorylated in nNOS-null mice during tail suspension, and total Foxo3a protein was not increased in tail-suspended nNOS null mice. Moreover, Foxo3a accumulated in the myonuclei of wild-type but not in nNOS-null mice during tail suspension (Figure 4B and Figure 6C).
When we overexpressed nNOS in muscle cells using a retrovirus vector, we found that Foxo3a was activated in nNOS-overexpressing myotubes (Supplemental Figure 4).
How does nNOS/NO regulate Foxo3a pathways?
Our observations suggest that nNOS/NO is an upstream regulator of Foxo3a in the tail suspension–induced muscle atrophy process. There are at least 2 possible explanations for how Foxo3a regulates nNOS/NO. First, NO produced by nNOS might inhibit protein kinases, which phosphorylate Foxo3a, thereby protecting Foxo3a from degradation and promoting its translocation from the cytoplasm to the nucleus. There are several kinases that can phosphorylate Foxo, including Akt (10), IKKβ (33), BCR (39), and CDK2 (40). In the present study, we show that Akt activities in nNOS-null muscle were not different from those in wild-type muscle during tail suspension. A fraction of IKKβ was nitrosylated (inactivated) during tail suspension (Figure 5C), but NF-κB was activated in both wild-type and nNOS-null muscle during suspension. This observation suggests that IKKβ activity was not meaningfully disrupted (Figure 4D). Involvement of other kinases remains to be investigated in a future study.
Second, nNOS/NO signal might decrease nuclear export of Foxo3a, resulting in accumulation of Foxo3a in myonuclei and protecting it from phosphorylation by Akt. Foxo is exported from the nucleus in a 14-3-3 protein–dependent process (19), and these molecular interactions remain to be examined.
Dislocation of nNOS and production of NO have no obvious effects on the activity of NF-κB.
It has been reported that reduced muscle activity induces muscle atrophy via activation of both Foxo and NF-κB transcription factors (9), at the same time suppressing the Akt pathway (5, 6), resulting in activation of the transcription of MuRF-1 and atrogin-1/MAFbx genes. In this report we demonstrated that dephosphorylation and nuclear accumulation of Foxo3a were largely attenuated in nNOS-null muscle during tail suspension. In contrast, our EMSA assay suggested that the NF-κB pathway was activated in nNOS-null mice to a similar extent as in wild-type mice in tail suspension experiments. This observation raises the possibility that NF-κB mediated the residual atrophy that occurred in nNOS-null mice, but further investigation is needed to correctly answer this question.
nNOS and other muscle atrophies.
Many conditions induce muscle atrophy, including space flight, immobilization, denervation, cancer cachexia, motor neuron diseases, starvation, and aging (41). Recently it has been reported that muscles of tumor-bearing mice exhibited membrane abnormalities accompanied by reduced levels of dystrophin and increased glycosylation on DGC proteins (22, 23). It was also shown that the DGC could counteract atrophic signaling in cancer cachexia when overexpressed at the sarcolemma (22). In the tail suspension model, we observed dislocation of nNOS but no changes in the sarcolemmal expression of other members of the DGC (Figure 2C). Therefore it is possible that dystrophin deficiency in cancer cachexia induces nNOS dislocation, which results in activation of nNOS and its downstream effectors.
We also found nNOS dislocation in denervation-induced muscle atrophy (Figure 7A). Remarkably, denervation-induced muscle atrophy was modestly blunted in nNOS-null mice or selective nNOS inhibitor–treated mice (Figure 7B). Although iNOS was induced during denervation (data not shown), both 7NI and l-NAME showed a similar effect on muscle atrophy, suggesting that iNOS does not contribute to denervation-induced muscle atrophy.
In conclusion, we demonstrate that nNOS dislocated from the sarcolemma to the cytoplasm in 2 models of disuse-induced muscle atrophy, tail suspension and denervation. We also show that dislocated nNOS led to the production of NO and regulated Foxo3a, MuRF-1, and atrogin-1/MAFbx, key molecules in muscle atrophy. Our model is illustrated in Figure 8. The identification of nNOS as a regulator of unloading-induced muscle wasting suggests that pharmacological intervention targeting nNOS or its downstream or upstream pathways would prevent or diminish this debilitating process.
Figure 8. A model of nNOS involvement in tail suspension–induced muscle atrophy.
Under normal conditions nNOS is located at the sarcolemma as a peripheral member of the DGC. During tail suspension, nNOS dissociates from α1-sytrophin (syn) and dislocates into the cytoplasm, generating NO, which ultimately regulates Foxo transcription factors, and muscle-specific E3 ubiquitin ligases, MuRF-1, and atrogin-1/MAFbx, which promote muscle protein degradation by the ubiquitin-proteasome system.
Methods
Animals and tail suspension model.
Twelve-week-old female C57BL/6, nNOS-null and eNOS-null mice were purchased from the Jackson Laboratory. α1-Syntrophin-null mice were produced in our previous study (25). These mice were backcrossed to the C57BL/6 strain for more than 10 generations. The animals were allowed ad libitum access to food and drinking water. The Experimental Animal Care and Use Committee of the National Institute of Neuroscience approved all experimental protocols. The mice were randomly assigned to control or tail suspension groups. To induce muscle atrophy by disuse, mice were suspended so that their hind limbs were 1 mm off the cage floor for 14 days. After 14 days of tail suspension, some groups were allowed 7 days of reloading by normal weight bearing. Muscle weight was normalized to body weight and is presented as a percentage of control in each experiment.
Denervation model.
The left sciatic nerve of mice was excised for nearly the full length of the thigh (approximately 10 mm) from a small incision (approximately 4 mm) made in the mid-lateral thigh under general anesthesia under a surgical microscope (Olympus) (42). The mice were sacrificed 3 or 14 days after denervation by cervical dislocation under general anesthesia, and soleus and gastrocnemius muscles were excised for analysis. The right gastrocnemius muscle served as a control.
Reagents.
Lipopolysaccharide from E. coli (0.1 ml, 3 mg/kg; E. coli, serotype 055:B5; Sigma-Aldrich) was administered via intraperitoneal injection. 7NI (Dojindo) was dissolved in peanut oil (50 mg/kg). l-NAME was injected daily into the intraperitoneal cavity of mice (10 mg/kg body weight). Clenbuterol (1 mg/kg; Sigma-Aldrich), streptomycin (300 mg/kg; Sigma-Aldrich) and nifedipine (5 mg/kg; Wako) were dissolved in PBS. PBS was injected into control mice.
Tissue preparation.
Control and tail-suspended mice were sacrificed with cervical dislocation. Body and wet muscle were weighed. The gastrocnemius and soleus muscles were collected individually using standard dissection methods and cleaned of excess fat, connective tissue, and tendons. Several of the muscles were frozen in isopentane cooled by liquid nitrogen for histological and immunohistochemical analysis, and the other muscles were frozen directly in liquid nitrogen for RNA isolation or protein extraction and stored at –80°C.
Real time PCR.
Total RNA was isolated using TRIzol (GIBCO). For RT-PCR, first-strand cDNA was synthesized using oligo-dT primers. Expression levels of selected genes (nNOS, MuRF-1, atrogin-1/MAFbx, and 18S-rRNA) were analyzed using Applied Biosystems SYBR Green gene expression assays on ABI7700 Sequence Detection System (Applied Biosystems) following the manufacturer’s instructions.
H&E staining.
Ten-micrometer cryosections were cut in the middle part of the muscle belly to obtain the largest myofiber diameter, placed on poly-L-lysine–coated slides, air dried, and stained with H&E. The sections were viewed and photographed using an HC-2500 digital camera system (Fuji Photo Film).
Immunohistochemistry.
Cryostat sections of muscle tissue (10 μm thick), were postfixed in acetone or 4% paraformaldehyde at –20°C and preincubated in PBS containing 5% goat serum and 1% bovine serum albumin for 30 minutes at room temperature. Polyclonal anti-nNOS (Zymed), anti-Foxo3a (Sigma-Aldrich), and anti–laminin-α2 (Alexis) were applied overnight at 4°C. Following incubations with appropriate secondary antibodies, mounted sections were observed by using a Leica confocal microscope. Muscle fiber diameters were determined on cross sections of soleus muscle using the greatest distance between the opposite sides of the narrowest aspect of the fiber. Total number of muscle fibers was also counted on cross sections.
Western blotting.
Total skeletal muscle protein was extracted from mouse hindlimb muscle for western blot analysis. We used the Bradford method and Coomassie Brilliant Blue G-250 (Bio-Rad) to determine the protein concentrations. Then protein fractions were extracted with a reducing sample buffer containing 10% SDS, 70 mM Tris-HCl, 5% β-mercaptoethanol, and Complete inhibitor cocktail (Roche). Protein (15 or 30 μg per lane) was separated on an SDS-polyacrylamide gel. The resulting gel was subsequently transferred to a polyvinylidene difluoride membrane (Millipore) using 242 mA for 1 hour. The blot was later incubated with primary antibodies. The signals were detected using the enhanced chemiluminescence method (GE Amersham). Relative quantities of proteins in western blots were determined by scanning densitometry (Alpha Innotec) and expressed in arbitrary units. The following antibodies were used for immunoblotting: anti–α1-syntrophin (Biogenesis), anti–laminin-α2 (Alexis), anti-dystrophin (dys2), anti-utrophin, anti–β-dystroglycan, anti-dystrobrevin, anti-dysferlin, anti–caveolin-3 (Novocastra), anti-hsp90 (Stressgen), anti-nNOS, anti-IKKβ, anti-Akt, anti–p-Akt, anti–p-mTOR, anti-p50 (Transduction Laboratories), anti-Foxo3a, anti–p-Foxo3a, anti-Foxo1/4, anti–Na/K-ATPase (Upstate Biotechnology), anti-Hsp70, and anti-iNOS antibody (Santa Cruz Biotechnology Inc.). Anti–α-sarcoglycan antibody was kindly provided by Michihiro Imamura (National Institute of Neuroscience, National Center of Neurology and Psychiatry).
Nuclear and cytosolic protein extraction.
Nuclear extracts were prepared from mouse skeletal muscle according to the method of Hunter et al. (20). Briefly, the cytosolic extract was obtained from the first supernatant of the nuclear extract preparation. The supernatant was placed in Millipore Ultrafree-4 centrifugal columns that had been pre-wetted with 0.5 ml of dilution buffer (20 mM HEPES, 40 mM KCl, 10% glycerol, 0.2 mM EDTA, 1 mM DTT), and centrifuged (7,500 g) at 4°C for 30 minutes. Dilution buffer (0.8 ml) was added to the column, and the 30-minute spin was repeated. Protein concentrations were determined using the Bradford protein assay (Bio-Rad).
Immunoprecipitation.
Skeletal muscle samples were homogenized in 0.15 M NaCl, 10 mM HEPES (pH 7.5) and Complete inhibitor cocktail (Roche), with or without 1% digitonin (Wako). nNOS and caveolin-3 immunoprecipitation was performed with anti-nNOS goat polyclonal antibody (Santa Cruz Biotechnology Inc.) and anti–caveolin-3 antibody (Transduction Laboratories). The samples were incubated with protein G gel (GE Amersham) overnight at 4°C. After the gel was washed with the equilibrating buffer, the bound fraction was eluted with 1% SDS, 1 mM tris-(2-carboxyethyl) phosphine, and 14 mM Tris-HCl (pH 6.7) and concentrated. Coimmunoprecipitates were resolved by SDS-PAGE and analyzed by western blotting. α-Tubulin was used as internal control for total protein inputted.
Two-dimensional PAGE.
Muscle extracts from ground control and tail suspension mice were resolved on 2-dimensional PAGE and analyzed by western blotting as described in ref. 38.
EMSA.
EMSA was performed according to the manufacturer’s instructions (Panomics). Briefly, probes were end-labeled with biotin. Binding reactions were performed for 30 minutes in a volume of 10 ml. Specificity of DNA binding was determined by addition of a 5-fold molar excess of unlabeled competitor DNA to the binding reactions. The binding reactions were loaded onto 6% non-denaturing polyacrylamide gels and electrophoretically resolved in 0.5× tris-borate EDTA (TBE) buffer.
NOS catalytic assays.
NOS catalytic assays were carried out according to the method described by Brenman et al. (24). Muscles from wild-type and nNOS-null mice with or without tail suspension were homogenized in 10 volumes of buffer containing 25 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM EGTA, and 0.1 mM NaCl. The homogenate was centrifuged at 20,000 g to separate the soluble fraction. The pellet was extracted in the same buffer containing 0.5 M NaCl and centrifuged at 20,000 g to create a particulate fraction. Aliquots from these fractions were assayed in 125-μl reactions containing 1.8 × 105 cpm of [3H]-arginine (53.0 Ci/mmol; GE Amersham), 1 mM NADPH, 640 μM CaCl2, 1 μM calmodulin, and 3 μM each of tetrahydrobiopterin, FAD, and FMN. After incubation for 20 minutes at 37°C, the assays were terminated with 2 ml of 20 mM HEPES, pH 5.5, and 2 mM EDTA. The samples were then applied to AG50WX-8 columns (Na+ form; Dowex), which were centrifuged, and the supernatant was collected. [3H]-citrulline was quantified by liquid scintillation spectroscopy.
Subcellular fractionation.
The subcellular fractionation was performed according to the method described by Brenman et al. (24). The gastrocnemius muscle was homogenized in 10 volumes (w/v) of buffer A (25 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA). The nuclei of the muscle were pelleted by centrifugation at 1,000 g. The supernatant was then centrifuged at 20,000 g to yield the supernatant S1. The resulting heavy microsomal pellet was resuspended in buffer B (500 mM NaCl added to buffer A), incubated for 30 minutes at 4°C with agitation, and centrifuged at 15,000 g, yielding supernatant S2. The pellet from this last centrifugation was resuspended in buffer B containing 0.5% Triton X-100, incubated for 30 minutes at 4°C with agitation, and centrifuged at 15,000 g to create supernatant S3 and the final pellet. The fractions were resolved using the sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis. The proteins were transferred to a polyvinylidene difluoride membrane (Millipore), which was later incubated with anti-nNOS antibody. The bands were quantified in densitometry.
Direct measurement of NO by EPR spectrometry.
Concentrations of NO in the skeletal muscle of mice were measured using the NO-trapping technique combined with EPR spectroscopy (29). Spin traps react with unstable free radicals such as NO to form a relatively stable radical adduct. This long-lived adduct formation results in accumulation of a steady-state formation of these free radicals, and thus the resultant radical adduct can be detected readily by EPR spectroscopy. We used a Fe-MGD complex as the trapping agent to quantify NO levels in the skeletal muscle tissues of the mice. Solutions of FeSO4 (62 mg/kg; Wako) and MGD (348 mg/kg; Dojindo) were injected subcutaneously. The Fe-MGD complex formed had a high specificity for NO (27, 29), and the NO-Fe-MGD complex was detected by EPR spectroscopy. The amplitude of the signal measured from the peak-to-peak height of the lower field side signal in the 3-line spectrum is known to be proportional to the amount of NO (29). The level of NO-Fe-MGD complex was estimated by comparing it with the signal height of a standard solution of a chemically synthesized NO complex. The concentration in tissues was determined 30 minutes after injection of the NO-trapping reagent. Thirty minutes after administration of the reagent, the gastrocnemius and soleus muscle were removed and weighed (approximately 100–120 mg). The tissue was minced and subjected to immediate measurement of NO by EPR spectrometry. X-band EPR spectra were measured at room temperature with a TE-200 EPR spectrometer (JEOL). The homogenates were drawn into a capillary tube (75 mm in length, 46 μl in internal volume) that had been inserted first into an EPR quartz tube (outside diameter, 5 mm), then introduced into the cavity. The instrument settings were as follows: center field, 331 mT; field scan, 4 mT; sweep time, 2 min; time constant, 0.3 s; modulation amplitude, 0.32 mT; modulation frequency, 9.44 GHz; microwave power, 60 mW.
Statistics.
Statistical differences were determined by either 2-tailed unpaired Student’s t test or the Mann-Whitney test. All data are expressed as mean ± SEM. Statistical significance is defined as P < 0.05.
Acknowledgments
We thank S. Masuda, A. Fukase, and T. Harano for technical support; K. Asanuma and J. Yoshitake for EPR spectrometry; Y. Onodera and M. Tateyama for immunohistochemistry; and all members of the Department of Molecular Therapy, National Institute of Neuroscience, for technical assistance and useful discussion, especially M. Yoshida, M. Ikemoto, and Y. Mochizuki. We also thank K. Ono for correcting our English. This work was supported by Research on Nervous and Mental Disorders (grant 16B-2); Health Science Research Grants for research on the human genome and gene therapy (H16-genome-003) and for research on brain science (H15-kokoro-021 and H18-kokoro-019) from the Japanese Ministry of Health, Labor and Welfare; Grants-in-Aid for Scientific Research (14657158, 15390281, 16590333, 17590857, and 18590392) from the Japanese Ministry of Education, Culture, Sports, Science and Technology; and the Ground-based Research Program for Space Utilization, promoted by Japan Space Forum.
Footnotes
Nonstandard abbreviations used: DGC, dystrophin glycoprotein complex; EPR, electron paramagnetic resonance; Foxo, forkhead box O; IKKβ, inhibitor of NF-κB kinase β; l-NAME, N-nitro- l-arginine methylester; MAFbx, muscle atrophy F-box protein; MGD, N-methyl-D-glucamine-dithiocarbamate; mTOR, mammalian target of rapamycin; MuRF-1, muscle-specific RING finger protein 1; 7NI, 7-nitroindazole; nNOS, neuronal NOS.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2468–2476 (2007). doi:10.1172/JCI30654
See the related Commentary beginning on page 2459.
References
- 1.Tidball J.G. Mechanical signal transduction in skeletal muscle growth and adaptation. J. Appl. Physiol. 2005;98:1900–1908. doi: 10.1152/japplphysiol.01178.2004. [DOI] [PubMed] [Google Scholar]
- 2.Ikemoto M., et al. Space shuttle flight (STS-90) enhances degradation of rat myosin heavy chain in association with activation of ubiquitin-proteasome pathway. FASEB J. 2001;15:1279–1281. doi: 10.1096/fj.00-0629fje. [DOI] [PubMed] [Google Scholar]
- 3.Bodine S.C., et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 4.Gomes M.D., Lecker S.H., Jagoe R.T., Navon A., Goldberg A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodine S.C., et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 6.Song Y.H., et al. Muscle-specific expression of IGF-1 blocks angiotensin II-induced skeletal muscle wasting. J. Clin. Invest. 2005;115:451–458. doi: 10.1172/JCI200522324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rommel C., et al. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 8.Latres E., et al. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J. Biol. Chem. 2005;280:2737–2744. doi: 10.1074/jbc.M407517200. [DOI] [PubMed] [Google Scholar]
- 9.Cai D., et al. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 10.Sandri M., et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y.P., Schwartz R.J., Waddell I.D., Holloway B.R., Reid M.B. Skeletal muscle myocytes undergo protein loss and reactive oxygen-mediated NF-kappaB activation in response to tumor necrosis factor alpha. FASEB J. 1998;12:871–880. doi: 10.1096/fasebj.12.10.971. [DOI] [PubMed] [Google Scholar]
- 12.Greer E.L., Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 13.Lehtinen M.K., et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto M., Han S., Kitamura T., Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J. Clin. Invest. 2006;116:2464–2472. doi: 10.1172/JCI27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berman J., Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 16.Lecker S.H., et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 17.Kamei Y., et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J. Biol. Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 18.Skurk C., et al. The FOXO3a transcription factor regulates cardiac myocyte size downstream of AKT signaling. J. Biol. Chem. 2005;280:20814–20823. doi: 10.1074/jbc.M500528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obsilova V., et al. 14-3-3 Protein interacts with nuclear localization sequence of forkhead transcription factor FoxO4. Biochemistry. 2005;44:11608–11617. doi: 10.1021/bi050618r. [DOI] [PubMed] [Google Scholar]
- 20.Hunter R.B., et al. Activation of an alternative NF-kappaB pathway in skeletal muscle during disuse atrophy. FASEB J. 2002;16:529–538. doi: 10.1096/fj.01-0866com. [DOI] [PubMed] [Google Scholar]
- 21.Hunter R.B., Kandarian S.C. Disruption of either the Nfkb1 or the Bcl3 gene inhibits skeletal muscle atrophy. J. Clin. Invest. 2004;114:1504–1511. doi: 10.1172/JCI200421696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acharyya S., et al. Dystrophin glycoprotein complex dysfunction: A regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell. 2005;8:421–432. doi: 10.1016/j.ccr.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Acharyya S., et al. Cancer cachexia is regulated by selective targeting of skeletal muscle gene products. J. Clin. Invest. 2004;114:370–378. doi: 10.1172/JCI200420174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brenman J.E., Chao D.S., Xia H., Aldape K., Bredt D.S. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- 25.Kameya S., et al. Alpha1-syntrophin gene disruption results in the absence of neuronal-type nitric-oxide synthase at the sarcolemma but does not induce muscle degeneration. J. Biol. Chem. 1999;274:2193–2200. doi: 10.1074/jbc.274.4.2193. [DOI] [PubMed] [Google Scholar]
- 26.Hosaka Y., et al. Alpha1-syntrophin-deficient skeletal muscle exhibits hypertrophy and aberrant formation of neuromuscular junctions during regeneration. J. Cell Biol. 2002;158:1097–1107. doi: 10.1083/jcb.200204076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lecour S., et al. Levels of nitric oxide in the heart after experimental myocardial ischemia. J. Cardiovasc. Pharmacol. 2001;37:55–63. doi: 10.1097/00005344-200101000-00007. [DOI] [PubMed] [Google Scholar]
- 28.Asanuma K., et al. Diffusion of cytotoxic concentrations of nitric oxide generated luminally at the gastro-oesophageal junction of rats. Gut. 2005;54:1072–1077. doi: 10.1136/gut.2004.063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagano T., Yoshimura T. Bioimaging of Nitric Oxide. Chem. Rev. 2002;102:1235–1270. doi: 10.1021/cr010152s. [DOI] [PubMed] [Google Scholar]
- 30.Kato N., Sato S., Yokoyama H., Kayama T., Yoshimura T. Sequential changes of nitric oxide levels in the temporal lobes of kainic acid-treated mice following application of nitric oxide synthase inhibitors and phenobarbital. Epilepsy Res. 2005;65:81–91. doi: 10.1016/j.eplepsyres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Moore P.K., Bland-Ward P.A. 7-nitroindazole: an inhibitor of nitric oxide synthase. Methods Enzymol. 1996;268:393–398. doi: 10.1016/s0076-6879(96)68041-0. [DOI] [PubMed] [Google Scholar]
- 32.Reynaert N.L., et al. Nitric oxide represses inhibitory kappaB kinase through S-nitrosylation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8945–8950. doi: 10.1073/pnas.0400588101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu M.C., et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 34.Segalat L., Grisoni K., Archer J., Vargas C., Bertrand A., Anderson J.E. CAPON expression in skeletal muscle is regulated by position, repair, NOS activity, and dystrophy. Exp. Cell Res. 2005;302:170–179. doi: 10.1016/j.yexcr.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Shiao T., et al. Defects in neuromuscular junction structure in dystrophic muscle are corrected by expression of a NOS transgene in dystrophin-deficient muscles, but not in muscles lacking alpha- and beta1-syntrophins. Hum. Mol. Genet. 2004;13:1873–1884. doi: 10.1093/hmg/ddh204. [DOI] [PubMed] [Google Scholar]
- 36.Kapur S., Bedard S., Marcotte B., Cote C.H., Marette A. Expression of nitric oxide synthase in skeletal muscle: a novel role for nitric oxide as a modulator of insulin action. Diabetes. 1997;46:1691–1700. doi: 10.2337/diab.46.11.1691. [DOI] [PubMed] [Google Scholar]
- 37.Stamler J.S., Meissner G. Physiology of nitric oxide in skeletal muscle. Physiol. Rev. 2001;81:209–237. doi: 10.1152/physrev.2001.81.1.209. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida M., et al. Dystrophin-associated protein A0 is a homologue of the Torpedo 87K protein. FEBS Lett. 1995;367:311–314. doi: 10.1016/0014-5793(95)00574-s. [DOI] [PubMed] [Google Scholar]
- 39.Yusuf I., Zhu X., Kharas M.G., Chen J., Fruman D.A. Optimal B-cell proliferation requires phosphoinositide 3-kinase-dependent inactivation of FOXO transcription factors. Blood. 2004;104:784–787. doi: 10.1182/blood-2003-09-3071. [DOI] [PubMed] [Google Scholar]
- 40.Huang H., Regan K.M., Lou Z., Chen J., Tindall D.J. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–297. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- 41.Kandarian S.C., Jackman R.W. Intracellular signaling during skeletal muscle atrophy. Muscle Nerve. 2006;33:155–165. doi: 10.1002/mus.20442. [DOI] [PubMed] [Google Scholar]
- 42.Mochizuki Y., et al. Participation of bone marrow-derived cells in fibrotic changes in denervated skeletal muscle. Am. J. Pathol. 2005;166:1721–1732. doi: 10.1016/S0002-9440(10)62482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]