Abstract
Ataxia-telangiectasia mutated (ATM) kinase orchestrates nuclear DNA damage responses but is proposed to be involved in other important and clinically relevant functions. Here, we provide evidence for what we believe are 2 novel and intertwined roles for ATM: the regulation of ribonucleotide reductase (RR), the rate-limiting enzyme in the de novo synthesis of deoxyribonucleoside triphosphates, and control of mitochondrial homeostasis. Ataxia-telangiectasia (A-T) patient fibroblasts, wild-type fibroblasts treated with the ATM inhibitor KU-55933, and cells in which RR is inhibited pharmacologically or by RNA interference (RNAi) each lead to mitochondrial DNA (mtDNA) depletion under normal growth conditions. Disruption of ATM signaling in primary A-T fibroblasts also leads to global dysregulation of the R1, R2, and p53R2 subunits of RR, abrogation of RR-dependent upregulation of mtDNA in response to ionizing radiation, high mitochondrial transcription factor A (mtTFA)/mtDNA ratios, and increased resistance to inhibitors of mitochondrial respiration and translation. Finally, there are reduced expression of the R1 subunit of RR and tissue-specific alterations of mtDNA copy number in ATM null mouse tissues, the latter being recapitulated in tissues from human A-T patients. Based on these results, we propose that disruption of RR and mitochondrial homeostasis contributes to the complex pathology of A-T and that RR genes are candidate disease loci in mtDNA-depletion syndromes.
Introduction
The ataxia-telangiectasia mutated (ATM) kinase initiates a well-characterized response to DNA damage, resulting in cell-cycle arrest, DNA repair, or apoptosis (1–4). Mutations in the ATM gene, though tolerated, result in the fatal childhood disorder ataxia-telangiectasia (A-T), characterized by symptoms including predisposition to cancer, ataxia due to progressive cerebellar degeneration, immunodeficiency, and telangiectasias (spider veins) (1–4). ATM signaling is required to sense and initiate repair of DNA double-strand breaks. Therefore, nuclear genomic instability resulting from loss of this function is regarded as a major mechanism underlying the pathology of A-T (2–5). However, this disease presents with a wide-array of symptoms, not all of which are readily explained by nuclear genomic instability, and study of cell and animal models of A-T has led to much speculation about additional pathogenic mechanisms (6). One such mechanism for which there is substantial experimental evidence is oxidative stress (7–11). How ATM is involved in oxidative stress management remains unclear as do other potential roles in cellular homeostasis in the absence of DNA damage.
It has been proposed that a key downstream target of ATM signaling in mammalian cells is ribonucleotide reductase (RR), the rate-limiting enzyme in the de novo synthesis of deoxyribonucleoside triphosphates (dNTPs). Mammalian RR is a multimeric enzyme comprising the large R1 subunit and either of 2 small subunits, R2 or p53R2 (12–14). Expression of each RR subunit is cell-cycle regulated, with p53R2 induction occurring at the G1-S transition while R1 and R2 are S-phase specific (13, 15–17). However, R1 and p53R2 are present at a basal level throughout the cell cycle and in resting cells, suggesting that RR activity can contribute to dNTP synthesis even in nondividing cells (13, 18). Furthermore, differential induction of all RR subunits has been observed in response to different types of DNA damage (12–14, 19). The primary role postulated for ATM in this regard is the p53-dependent induction of p53R2 (14, 20). Whether ATM is involved in other aspects of RR regulation or in mitochondrial DNA (mtDNA) maintenance has not been investigated.
Human mtDNA encodes 13 essential subunits of the ATP-producing oxidative phosphorylation (OXPHOS) system and is essential for normal cellular energy metabolism (21). Mutations and rearrangements in mtDNA cause numerous human diseases and have been implicated in aging, age-related pathology, and diseases such as Parkinson’s disease and diabetes (22–24). In addition to reduced energy metabolism, a common underlying factor in the pathology of these diseases is the likely involvement of ROS that promote oxidative stress and associated cellular damage. Mitochondria are a major source of ROS (25–27), the production of which can be elevated by loss of normal mtDNA expression (28) and other defects in mitochondrial function (24). Thus, deciphering the pathways that control mtDNA expression and maintenance is important to fully understanding the increasingly recognized roles of mitochondrial dysfunction in human health.
In stark contrast to nuclear DNA, mtDNA is small (~16.5 kb) and circular, present at 102–104 copies/cell in most tissues, replicated at all stages of the cell cycle, and propagated continuously even in nondividing cells (29). Furthermore, unlike the many well-characterized DNA damage and checkpoint-signaling pathways that regulate nuclear DNA, those that control mtDNA replication and stability in mammalian cells are not well defined.
As the essential building blocks of DNA, dNTPs are required not only for replication and repair of nuclear DNA but also mtDNA. Thus, pathways that regulate the mitochondrial dNTP pool are almost certainly important for mtDNA maintenance, which is underscored by the existence of mtDNA-depletion syndromes caused by mutations in deoxynucleotide salvage pathways (30–34). This, coupled with our recent documentation of a role for the Mec1p checkpoint–signaling pathway (orthologous to the ATM pathway) and its downstream target RR in the regulation of mtDNA copy number in budding yeast (35, 36), suggests a dynamic interplay between the salvage and de novo pathways for dNTP synthesis in mtDNA replication and repair.
Another key factor required for mtDNA maintenance is human mitochondrial transcription factor A (h-mtTFA or TFAM), a DNA-binding protein of the high-mobility group box family (37). This protein binds upstream of mtDNA promoters and interacts with 2 other mitochondrial transcription factors to facilitate initiation by mitochondrial RNA polymerase (38). Since mitochondrial transcription also generates the primers for initiation of leading-strand DNA synthesis, h-mtTFA, like the rest of the transcription machinery, has a critical role in mtDNA replication (39). Because of this primer-generating function and its additional postulated role in packaging mtDNA, h-mtTFA has been implicated as a key factor in the regulation of mtDNA copy number in mammalian cells (21, 40–45). A similar role in mtDNA packaging and copy number regulation has been ascribed to the yeast homolog of h-mtTFA, Abf2p (46, 47). While there is substantial data supporting this role in mtDNA maintenance, including a strong correlation between mtTFA levels and mtDNA copy number in cultured cells purposely depleted of mtDNA (42–45) and in patient tissues with severe mtDNA depletion (42, 44), a function for mtTFA in mtDNA copy number regulation per se has not been unequivocally established (39). Furthermore, the involvement of other pathways that work independently of h-mtTFA to control mtDNA replication and stability in mammals remains possible.
Here, we present our results that describe 2 novel and interrelated functions for ATM that are potentially relevant to the disease pathology of A-T: the overall regulation of RR subunit expression/stability and proper mtDNA copy number dynamics and expression in the presence and absence of induced DNA damage.
Results
RR is required to maintain normal mtDNA levels in actively dividing cells.
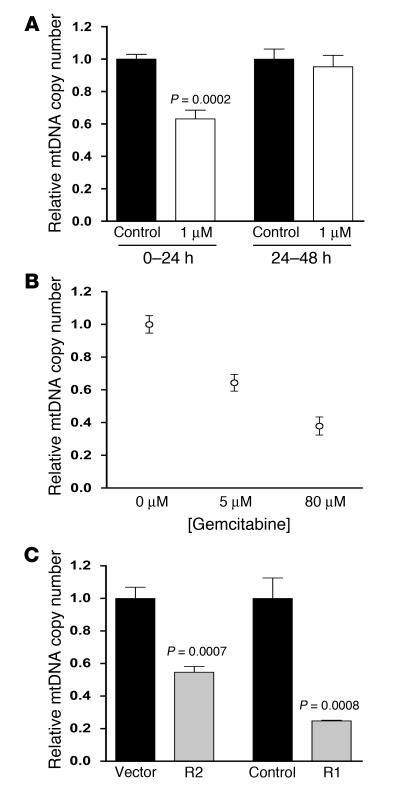
We previously reported that activation of RR increases mtDNA copy number and stability in Saccharomyces cerevisiae (35, 36). To determine whether mtDNA copy number regulation via RR was conserved in mammals, we inhibited the RR enzyme either pharmacologically with Triapine or gemcitabine or genetically via RNA interference (RNAi) of the R1 and R2 subunits. Exposure of proliferating primary human fibroblasts to Triapine, a specific RR inhibitor that prevents reconstitution of the tyrosyl radical in the small subunits required for nucleotide reduction (48, 49), resulted in mtDNA depletion (Figure 1A). Likewise, inhibition of the R1 large subunit of RR via gemcitabine, a cytidine analog that is resistant to reduction, resulted in significant mtDNA depletion (Figure 1B). Furthermore, reduction of the R2 or R1 subunits of RR in HeLa cells by RNAi as described (50, 51) resulted in a 50% or 80% depletion of mtDNA, respectively (Figure 1C). Interestingly, the effect of Triapine on mtDNA levels was only observed in actively proliferating cultures; that is, when Triapine was added to cells that had reached a higher level of confluence, no mtDNA depletion was observed (Figure 1A). These data suggest that other mechanisms of nucleotide production (e.g., salvage pathways) or mtDNA maintenance can compensate for lack of RR activity under these conditions. Altogether, these results clearly implicate RR in the replication and/or stability of mtDNA, but show that this effect is conditional.
Figure 1. Inhibition of RR causes mtDNA depletion during proliferation.
(A) Relative mtDNA copy number for wild-type primary fibroblasts treated with Triapine during high proliferation (0–24 hours) or reduced proliferation (24–48 hours) conditions. The ratio of the amount of mtDNA to nuclear DNA is plotted with the ratio in control cells arbitrarily set to 1.0. The mean ± SEM is plotted with significant statistical differences via Student’s t test indicated. (B) Relative mtDNA copy number of gemcitabine-treated wild-type primary fibroblasts. Cultures were maintained in the concentrations indicated for 48 hours with untreated (0 μM) arbitrarily set to 1.0. The mean ± SEM is shown. (C) Relative mtDNA copy number (plotted as in A) of HeLa cells with R2 or R1 expression knocked down compared with vector or short hairpin RNA control cells, respectively. R2 was reduced 50% and R1 was undetectable via Western blot. The mean ± SEM is plotted with significant statistical differences via Student’s t test indicated.
ATM and RR are required to upregulate mtDNA copy number and biogenesis in response to ionizing radiation.
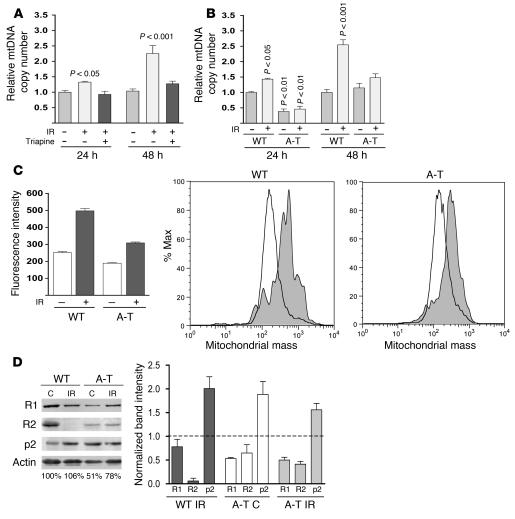
In yeast, dNTP synthesis is controlled by the Mec1p checkpoint pathway. The DNA damage–sensing kinase ATM is a mammalian ortholog of yeast Mec1p, which led us to ask whether there is a conserved role for the ATM-signaling pathway in the regulation of RR and mtDNA in human cells. We exposed wild-type primary human fibroblasts to ionizing radiation (IR), a DNA-damaging stress known to induce ATM signaling. Compared with nonirradiated control fibroblasts, there was a significant increase in the amount of mtDNA at 24 hours after irradiation that reached approximately 2.5-fold at 48 hours (Figure 2A). Similar results were obtained in IR-treated human Jurkat T cells (data not shown). To determine whether the observed increase in mtDNA copy number in response to IR required RR, as our previous data suggested, we incubated cells with Triapine. RR inhibition via Triapine completely prevented the IR-induced increase in mtDNA in these cells (Figure 2A).
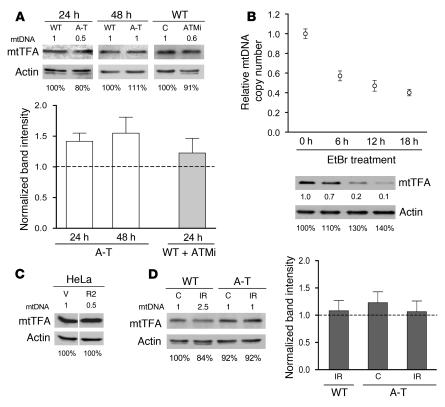
Figure 2. RR and ATM are required for increased mtDNA copy number and biogenesis in response to IR and proper regulation of RR subunit expression.
(A) Relative mtDNA levels (plotted as in Figure 1) in primary wild-type fibroblasts 24 and 48 hours after exposure to 17.5 Gy of IR with all values compared with nonirradiated, untreated wild-type cells at 24 hours. Cells were exposed to 1 μM Triapine for 24 hours either immediately after IR (24 hours) or after a 24-hour recovery (48 hours) as indicated. The mean ± SEM is plotted. A 1-way ANOVA was used to determine statistical significance as indicated. (B) Relative mtDNA copy number of wild-type and A-T primary fibroblasts 24 and 48 hours after exposure (+) to 17.5 Gy of IR is plotted as in Figure 1. (C) Mitochondrial mass of wild-type and A-T patient fibroblasts 48 hours after IR. Median MitoTracker Green FM fluorescence intensity (mean ± SEM) is plotted. FACS histograms from 1 representative replicate are shown in the right panels. The percentage of cells collected (% max; y axis) with the indicated amount of fluorescence on the x axis (log scale) is shown. (D) Western blot analysis of R1, R2, and p53R2 (p2) from nonirradiated A-T cells (C) or WT and A-T cells 48 hours after 17.5 Gy (IR) from a representative experiment. Actin was probed as a control (C) for protein loading (the relative amount of actin in each lane is shown below the actin panel; wild-type nonirradiated was set to 100%). Actin-normalized signals from 3 independent experiments are depicted graphically on the right of the panels with the wild-type nonirradiated control protein levels set to 1 as indicated by the dotted line.
To confirm that ATM was required for the observed mtDNA increase after IR, we next exposed A-T (ATM–/–) patient–derived fibroblasts to IR and quantified mtDNA. While the wild-type (ATM+/+) control fibroblasts again exhibited an IR-dependent increase in mtDNA at 24 and 48 hours after irradiation, A-T fibroblasts were completely nonresponsive (Figure 2B). Altogether, these results revealed a general mitochondrial response to IR that involves RR- and ATM-dependent upregulation of mtDNA levels.
It is well documented that mtDNA copy number and overall mitochondrial biogenesis are usually coordinately regulated. This was confirmed in our experiments with wild-type fibroblasts, where an approximately 2-fold increase in mitochondrial mass was observed 48 hours after irradiation (Figure 2C), closely matching the observed increase in mtDNA copy number (Figure 2, A and B). However, in A-T fibroblasts, this increase in mitochondrial mass, though present, was significantly blunted (Figure 2C). These results indicate that ATM is required for a full biogenesis response and to coordinate an increase in mtDNA with increases in mitochondrial mass under these conditions.
Altered steady-state levels and damage-induced dynamics of all 3 RR subunits in ATM null cells.
Given that RR subunits are a likely target of ATM signaling (14, 20) and considering our observation that the mtDNA copy number increase in response to IR was RR dependent, we examined the relative expression of the R1, R2, and p53R2 subunits of RR after IR by Western blotting. As reported by others (18, 20, 51–53), we observed an increase in p53R2 and a marked reduction of R2 in wild-type fibroblasts exposed to IR (Figure 2D). In addition, there was a slight reduction in the amount of the R1 subunit. However, a much different RR subunit expression profile was observed in A-T fibroblasts. First, compared with wild-type cells, there was a reduction in the steady-state levels of both R1 and R2 and a 2-fold increase in p53R2 in A-T cells in the absence of radiation (Figure 2D). Second, unlike wild-type cells, A-T cells exhibited minimal changes in RR steady-state profiles in response to irradiation (Figure 2D). These data demonstrate that the expression dynamics of all 3 RR subunits are abnormal (and affected differentially) by the loss of ATM both in irradiated and nonirradiated cells.
ATM is required to maintain normal mtDNA levels in actively dividing cells and for proper regulation of RR subunit abundance.
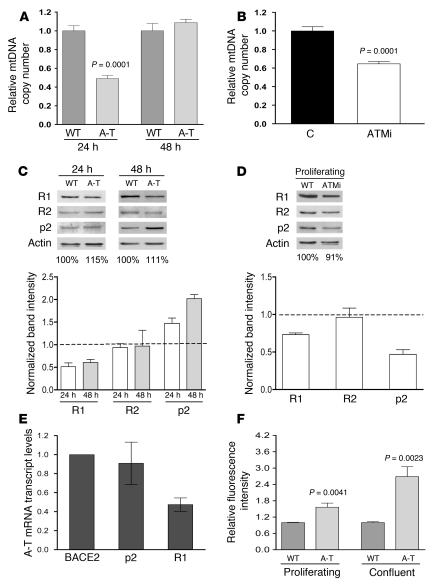
In the course of the IR experiments described above, we noticed that nonirradiated primary A-T fibroblasts showed a 50% reduction in mtDNA copy number at the 24-hour time point (~60% confluent) compared with wild-type fibroblasts (Figure 2B). This led us to consider that ATM may influence mtDNA copy number even in the absence of induced DNA damage. To address this, we first determined whether ATM contributes to the maintenance of mtDNA during normal growth in the absence of DNA damage. Consistent with this hypothesis, A-T patient fibroblasts proliferating under normal growth conditions exhibited a 50% depletion of mtDNA (Figure 3A). To verify that the ATM effect on mtDNA copy number was not specific to patient-derived fibroblasts that are chronically ATM null, we also inhibited ATM in wild-type primary fibroblasts with KU-55933, a specific competitive inhibitor of ATP binding to ATM (54). KU-55933 reduced mtDNA levels in wild-type fibroblasts, effectively reproducing the mtDNA depletion observed in proliferating A-T cells under the same conditions (Figure 3B). Furthermore, as was the case with IR exposure, A-T fibroblasts also exhibited anomalous expression of RR subunits, again including reduced expression of the R1 subunit, and upregulation of p53R2 (Figure 3C, compare to Figure 2D). We also observed aberrant regulation of RR subunit expression in wild-type fibroblasts treated with KU-55933. However, this resulted in a downregulation of both R1 and p53R2 (Figure 3D). These data demonstrate that short-term inhibition of ATM kinase activity, like chronic loss of ATM protein, leads to reduction of R1 but does not lead to elevated p53R2. Altogether, these data define 2 novel roles of ATM, regulation of RR subunit expression and maintenance of normal mtDNA levels.
Figure 3. ATM is required for mtDNA maintenance and normal expression of RR subunits in the absence of DNA damage.
(A) Relative mtDNA copy number (plotted as in Figure 1) of wild-type and A-T primary fibroblasts at 24 or 48 hours. The mean ± SEM is plotted with significant statistical differences via Student’s t test indicated. (B) Relative mtDNA copy number of wild-type primary fibroblasts untreated (C) or treated for 24 hours with indicated concentration of ATM inhibitor KU-55933, after a 48-hour preincubation. The mean ± SEM is plotted with significant statistical differences via Student’s t test indicated. (C) Western blot analysis of R1, R2, and p53R2 (p2) of wild-type and A-T primary fibroblasts at 24 and 48 hours from a representative blot corresponding to conditions in A as depicted in Figure 2D. (D) Western blot analysis of R1, R2, and p53R2 of wild-type primary fibroblasts treated with 10 μM ATM inhibitor KU-55933 (ATMi) as described in B and depicted as in Figure 2D. (E) Relative levels of BACE2-normalized R1 and p53R2 mRNA transcript levels in A-T patient–derived fibroblasts. The mean ± SEM from 3 independent experiments is plotted. (F) Relative fluorescence intensity of dihydroethidium, a dye that detects cellular ROS, in wild-type and A-T primary fibroblasts at 24 hours (proliferating) or 6 days (confluent) with wild-type fluorescence arbitrarily set to 1.0 at each time point. The mean ± SEM of 1 representative experiment is plotted with significant statistical differences via Student’s t test indicated.
To begin to determine the mechanism of reduced RR subunit abundance in A-T cells, we utilized quantitative real-time RT-PCR to quantify the steady-state levels of the corresponding transcripts compared with a control transcript BACE2, which is unchanged in response to ATM expression status (55). In A-T fibroblasts, we observed an approximately 50% reduction in the steady-state level of R1 transcript (Figure 3E) that corresponded well with the observed reduction in R1 protein under these conditions (Figure 3C). These data suggest that the reduction of R1 protein in ATM null cells is due, at least in part, to decreased steady-state levels of its mRNA. Conversely, the upregulation of p53R2 in the A-T cell was not accompanied by a corresponding increase in its mRNA (Figure 3E).
ATM null cells have increasingly elevated ROS levels.
Through further investigation, we found that in contrast to actively dividing cultures (24-hour time points, ~60% confluent), less actively dividing cultures (48-hour time points, ~80% confluence) as well as confluent cultures of A-T fibroblasts exhibited normal mtDNA copy number (Figure 2B, Figure 3A, and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI31604DS1), an effect reminiscent of that observed when the RR inhibitor Triapine is added to growing versus more confluent cultures (compare Figure 3A with Figure 1A). One condition that has been reported to lead to unscheduled mitochondrial biogenesis (including coordinate increases in mtDNA) is oxidative stress (56). We therefore considered the possibility that the “rescue” of mtDNA depletion in less actively dividing A-T cells was due to oxidative stress, a well-documented phenotype of A-T (7–11). As a marker of oxidative stress, we measured cellular ROS levels in proliferating and confluent A-T cells using dihydroethidium staining and FACS analysis. While there was a moderately elevated level (~40%) of ROS in proliferating A-T cells (24 hour) compared with wild-type cells, this difference was significantly more pronounced in the more confluent (6 day) cells (~2.5-fold higher in A-T cells) (Figure 3F). Altogether, these results (Figure 3) strongly implicate ATM and RR in mtDNA maintenance in actively dividing cells and suggest that oxidative stress influences RR expression or activity in confluent cells or that RR-independent pathways (e.g., nucleotide salvage pathways) are able to maintain mtDNA under these conditions in the absence of ATM.
Altered mtDNA levels and RR subunit abundance in ATM null cells is not due to differences in cell-cycle progression.
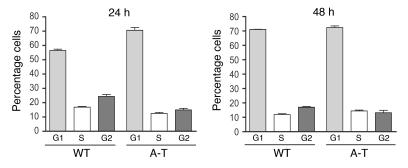
It is documented that A-T cells have defects in cell-cycle progression (2, 57). Due to the growth state–dependent nature of the mtDNA depletion in A-T cells and the fact that RR is regulated in a cell-cycle–dependent fashion, we examined whether the observed differences in A-T mtDNA copy number and RR subunit expression were due to defects in cell-cycle progression. To address this, we analyzed the cell-cycle distribution of primary wild-type and A-T patient–derived fibroblasts at the 24-hour and 48-hour time points where we previously detected changes in mtDNA copy number and RR subunits. At the 24-hour point, there were slight differences between the wild-type and A-T fibroblasts, with approximately 15% of the cells shifting from the S and G2 phases to G1 in the A-T cells compared with the wild-type profile (Figure 4). However, no significant differences were observed between the 2 cell lines at the 48-hour time point (Figure 4). Perhaps most noteworthy, the cell-cycle profile of the A-T fibroblasts at the 24-hour time point (where mtDNA depletion is observed; Figure 3A) was virtually identical to that of wild-type and A-T fibroblasts at the 48-hour time point (Figure 4), where no differences in mtDNA copy number were observed (Figure 3A). These data are most consistent with a cell-cycle–independent effect of loss of ATM on mtDNA copy number and RR subunit abundance. With regard to the RR subunit regulation, this conclusion is bolstered by an observed reduction in R1 and an elevation in p53R2 in 7-day confluent cultures (Supplemental Figure 1), conditions under which the cells were synchronized in the G0 stage of the cell cycle.
Figure 4. A-T–specific defects in RR and mtDNA levels are independent of cell cycle.
Cell-cycle profiles for wild-type and A-T patient fibroblasts, determined by propidium iodide staining and FACS analysis, are plotted. The y axes indicate the percentage of cells in each cell-cycle phase (G1, S, or G2) on the x axis at either 24 or 48 hours. The mean ± SEM of 1 representative experiment is plotted.
ATM- and RR-mediated changes in mtDNA copy number occur independently of h-mtTFA, a factor previously implicated in mtDNA maintenance and copy number regulation.
The high-mobility group–box transcription factor h-mtTFA is required for mtDNA replication and maintenance and has been postulated to be a key regulator of mtDNA copy number in human cells (21, 40–45). The latter function is based on a generally sound correlation between its steady-state levels and mtDNA copy number (42–45). For example, in transformed cell lines treated with ethidium bromide to deplete mtDNA (a standard technique in the field), h-mtTFA levels fall in parallel (45, 58). Given that we have identified both ATM and RR as factors involved in mtDNA maintenance, we sought to determine the extent to which h-mtTFA might be involved in their effects on mtDNA copy number. First, we examined the levels of h-mtTFA in A-T patient fibroblasts (from a 24-hour growth point) and wild-type fibroblasts treated with the ATM inhibitor KU-55933, both of which exhibit approximately 50% mtDNA depletion, as well as A-T cells from a 48-hour growth point, where mtDNA levels are normal (Figure 3, A and B). In none of these cases was a significant alteration of mtTFA levels observed (Figure 5A). If anything, there was a modest trend toward upregulation of h-mtTFA in the A-T fibroblasts. This was somewhat surprising given that we predicted mtTFA to mirror mtDNA levels based on published results in the field. To ensure that we were indeed capable of detecting a decrease in mtTFA levels in response to drug-induced mtDNA depletion in primary cell lines, we treated primary wild-type fibroblasts with ethidium bromide. In these cells, we observed a typical reduction in mtDNA and the previously documented parallel decrease in h-mtTFA abundance (Figure 5B). We next examined HeLa cells that were depleted of mtDNA due to stable RNAi suppression of the R2 subunit of RR. Again, similar to ATM null cells, there was no reduction in the amounts of h-mtTFA (Figure 5C). The steady-state amount of h-mtTFA was also unperturbed in IR-treated wild-type cells (Figure 5D) that had significantly increased mtDNA (~2.5-fold; Figure 2A). These data clearly demonstrate that alterations in mtDNA copy number can occur without corresponding changes in h-mtTFA and strongly suggest that mtDNA copy number changes mediated by ATM and RR are likewise independent of h-mtTFA.
Figure 5. ATM/RR-dependent alterations in mtDNA copy number occur independently of h-mtTFA.
(A, C, and D) Western blot analyses of h-mtTFA with the relative mtDNA copy number summarized in numerical form at the top of each panel based on the data presented in Figures 1, 2, and 3. Quantification of h-mtTFA levels relative to actin are plotted in A and D as described in Figure 2D. (A) Wild-type and A-T primary fibroblasts at 24 or 48 hours and wild-type primary fibroblasts untreated (C) or treated with 10 μM KU-55933 for 24 hours after a 48-hour preincubation. (B) Relative mtDNA copy number (as plotted in Figure 1) of ethidium bromide–treated (EtBr-treated) wild-type primary fibroblasts with concurrent Western blot analysis of h-mtTFA. Hours in EtBr are indicated, and numbers below actin and h-mtTFA panels indicate protein normalized to actin. (C) HeLa cells with expression of R2 stably knocked down 50% by RNAi (R2) or with empty vector (V). (D) Wild-type and A-T primary fibroblasts nonirradiated (C) or 48 hours after irradiation (IR).
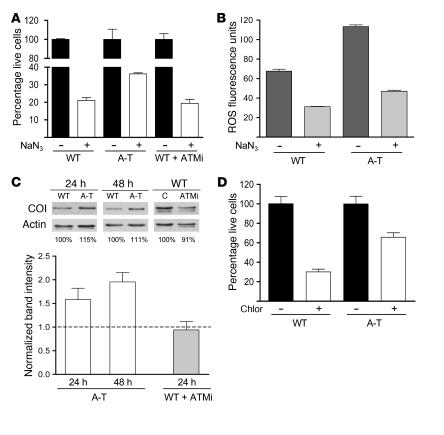
ATM null cells are resistant to inhibitors of mitochondrial respiration and translation.
The data presented thus far indicate that mitochondrial homeostasis is dysregulated in the absence of normal ATM signaling. It has also been reported previously that mitochondrial respiration is aberrantly upregulated in ATM null mouse cerebellum (59). To examine further the degree to which mitochondrial homeostasis is disrupted in the absence of ATM signaling, we determined the sensitivity of A-T patient fibroblasts and ATM-inhibited wild-type fibroblasts to the mitochondrial respiration poison sodium azide, an inhibitor of cytochrome oxidase (complex IV). Consistent with increased activity or abundance of complex IV, A-T fibroblasts were less sensitive to azide-induced growth inhibition than wild-type fibroblasts (Figure 6A). Interestingly, this effect was not observed in wild-type fibroblasts in which ATM was pharmacologically inhibited (Figure 6A). Since ROS production or detoxification capability between wild-type and A-T fibroblasts could account for the differential sensitivity to azide, we also analyzed ROS levels in azide-treated cells and found a similar reduction in ROS in both cell types. These data suggest that increased resistance to azide in the A-T fibroblasts is most likely due to less inhibition of respiration as opposed to differences in ROS production or detoxification capacity.
Figure 6. A-T patient–derived fibroblasts exhibit aberrant resistance to mitochondrial inhibitors and abnormal mtDNA-encoded CO I levels.
(A) Relative cell number with respect to untreated controls of wild-type, A-T, or ATM-inhibited wild-type cells (ATMi), treated with 8 mM sodium azide (NaN3) for 4 days. The percentage of live cells compared with untreated controls is plotted on the y axis with genotype and treatment on the x axis. The mean ± SEM of 1 representative experiment is shown. (B) Relative fluorescence intensity of dihydroethidium in wild-type and A-T primary fibroblasts at 40 hours of 8 mM NaN3 treatment. The mean ± SEM of 1 representative experiment is plotted. (C) Western blot analysis of mtDNA-encoded CO I for wild-type, A-T, and ATM-inhibited wild-type fibroblasts as described in Figure 2D. (D) Relative cell number with respect to untreated controls of wild-type and A-T cells treated with 240 μg/ml of mitochondrial translation inhibitor chloramphenicol (Chlor) for 4 days. The percentage of live cells compared with untreated controls is plotted on the y axis with genotype and treatment on the x axis. The mean ± SEM of 2 independent experiments is shown.
One possible mechanism of azide resistance in A-T cells is an increased amount of OXPHOS complex IV. Therefore, we examined the levels of cytochrome oxidase I (CO I), an mtDNA-encoded subunit of complex IV, in both wild-type and A-T patient–derived fibroblasts. Mirroring the results of the azide sensitivity assays, there was a significant increase in CO I levels in A-T cells but not in ATM-inhibited wild-type cells (Figure 7C). These data suggested to us that chronic loss of ATM results in an upregulation of cytochrome oxidase (and perhaps other mitochondrial OXPHOS complexes) as an attempt to compensate for the disruption in mitochondrial function in these cells. To determine whether one level of CO I induction might be occurring via upregulation of mitochondrial gene expression, we also determined the sensitivity of A-T patient–derived fibroblasts to the mitochondrial translation inhibitor chloramphenicol. As was the case with azide, A-T fibroblasts displayed increased resistance to chloramphenicol (Figure 6D).
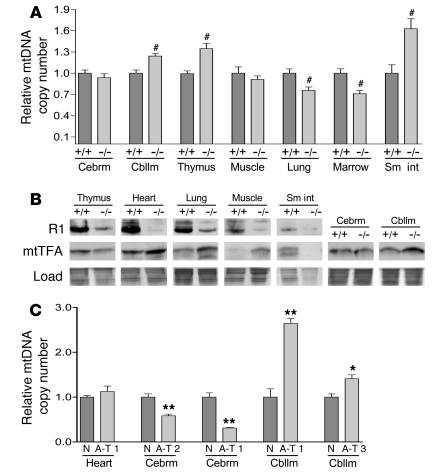
Figure 7. ATM null tissues exhibit loss of mtDNA homeostasis and depleted levels of RR subunit R1.
(A) Relative mtDNA copy number of wild-type (+/+) and ATM null (–/–) mouse tissues plotted as in Figure 1, with the mean of each wild-type tissue arbitrarily set to 1.0. The mean ± SEM is plotted with significant statistical differences via Student’s t test indicated (#P < 0.03). (B) Western blot of R1 and mtTFA in wild-type (+/+) and ATM null (–/–) mouse tissues. Simply Blue gel staining demonstrates loading. (C) Relative mtDNA copy number of normal or A-T patient tissues plotted as in Figure 1, with the mean of the normal control tissue (N) arbitrarily set to 1.0. The mean ± SEM is plotted with significant statistical differences via Student’s t test indicated (*P < 0.01, **P < 0.001). Cebrm, cerebrum; Cbllm, cerebellum; Sm int, small intestine.
ATM-deficient tissues have decreased steady-state levels of the R1 subunit of RR and altered mtDNA homeostasis.
Based on the cell culture results presented thus far, we concluded that ATM has 2 previously unidentified roles, one in regulating expression and/or stability of RR subunits and another in maintaining mitochondrial homeostasis. We next set out to find evidence of how loss of these functions might be manifested in vivo by examining tissues of ATM null mice and human A-T patients. First, we isolated various tissues from ATM null mice and probed them for alterations in mtDNA copy number and mtTFA (as measures of mtDNA-related mitochondrial homeostasis) and for expression of RR subunits. Here, we uncovered clear, tissue-specific alterations in mtDNA and mtTFA abundance (Figure 7). First, significant mtDNA depletion was detected in lung and bone marrow while cerebellum, thymus, and small intestine demonstrated significant increases in mtDNA. No significant differences were found in cerebrum and skeletal muscle (Figure 7A). Second, the levels of mtTFA were also misregulated, being elevated in cerebellum, muscle, heart, and lung, reduced in thymus and small intestine, and unperturbed in cerebrum (Figure 7B). Furthermore, as was the case in the cultured A-T cells, there was no correspondence between the steady-state levels of mtTFA and the observed alterations in mtDNA copy number (compare Figure 7A with Figure 7B). Third, there was a universal reduction in the steady-state level of the R1 subunit of RR in all ATM null mouse tissues analyzed (Figure 7B), consistent with our results in cultured A-T fibroblasts (Figure 2D and Figure 3C). We were unable to detect R1 in either wild-type or ATM null cerebellum or cerebrum. In addition, we also probed for the R2 and p53R2 subunits of RR but were unable to detect these in any of the wild-type or ATM null mouse tissues analyzed, thus precluding us from making conclusions regarding their relative abundance. Finally, we obtained a few rare samples of A-T patient tissue, analyzed these for mtDNA copy number, and found a similar spectrum of mtDNA alterations observed in the mouse tissue panel, including increased copy number in cerebella, normal copy number in heart, and reduced copy number in cerebra (Figure 7C). Based on the results from the A-T mouse model and human patients, we concluded that ATM is intimately involved in the regulation of the R1 subunit of RR and in the proper maintenance of mtDNA-related functions in tissues, with the precise effects of the latter being tissue specific.
Discussion
ATM kinase, mutations in which result in the disease A-T, is required for nuclear genome stability due to a critical role in sensing double-strand breaks. In this study, we have defined what we believe are 2 additional novel and interrelated roles for ATM in the regulation of the expression/stability of RR and in the maintenance of mitochondrial homeostasis, especially with regard to the dynamics and expression of mtDNA. The rationale for these conclusions will be discussed in detail below as will the potential relevance of our results to the pathology of A-T and mitochondrial disorders.
The first main conclusion we draw from our results is that ATM and RR are required to maintain and properly modulate mtDNA copy number in mammalian cells in the presence and absence of induced DNA damage. This is based on multiple lines of evidence reported here. First, reductions in mtDNA copy number are observed in response to pharmacologic inhibition of RR activity or a reduction in the amount of the R1 or R2 subunits of RR by RNAi (Figure 1). Second, increases in mtDNA copy number normally observed in response to IR are eliminated in wild-type fibroblasts treated with the RR inhibitor Triapine or in A-T patient fibroblasts (Figure 2, A and B). Third, consistent with wild-type fibroblasts in which RR activity is inhibited pharmacologically or via RNAi (Figure 1), actively proliferating A-T fibroblasts or wild-type fibroblasts treated with the ATM inhibitor KU-55933 exhibit approximately 50% depletion of mtDNA even in the absence of IR (Figure 3, A and B). These results clearly show that ATM and RR are required not only for mtDNA amplification in response to IR, but also for mtDNA maintenance during active cell proliferation. This conclusion is bolstered by the recent demonstration that mutations in p53R2 subunit of RR cause a subset of mtDNA depletion syndromes in humans as well as mtDNA depletion in mouse tissues (60). Therefore, at least one potential mechanism to explain the effects of loss of ATM function on RR subunit expression and mtDNA copy number could be the aberrant regulation of p53 in the absence of ATM signaling.
Our second main conclusion is that ATM plays a critical role in the overall regulation of RR subunit expression and/or stability. This is based on multiple lines of evidence as well. First, cultured A-T patient fibroblasts have a lower steady-state level of the large R1 subunit of RR and a higher level of the p53R2 subunit than wild-type fibroblasts under virtually all conditions in the absence of DNA damage (Figure 2D, Figure 3C, and Supplemental Figure 1). Second, all 3 RR subunits (R1, R2, and p53R2) exhibited uncharacteristic responses to IR, including no downregulate of R2 and loss of damage inducibility of p53R2, the latter having been postulated by others (14, 20). These aberrant responses were likely influenced by the fact that in A-T cells, R2 was already downregulated and p53R2 was already upregulated in the absence of IR (Figure 2D). Third, and perhaps most intriguing, is that R1 expression was downregulated in all tissues from ATM null mice that we analyzed (Figure 7B), indicating a clear role for ATM in RR regulation in vivo. The ramifications of reduced RR expression or activity in ATM null cells in the absence of an induced DNA damage insult is very likely relevant to the disease pathology of A-T. For example, this could exacerbate the already aberrant DNA damage response in A-T cells by limiting dNTPs needed for repair, altering normal cell-cycle timing and progression, and as we will discuss based on the results herein, affecting mitochondrial homeostasis that likely has its own pathogenic consequences.
Our results strongly suggest that in the absence of ATM, the observed downregulation of R1 is due to a direct effect on the transcription or stability of its mRNA transcript (Figure 3E). However, the aberrant upregulation of p53R2 appears to have occurred through a posttranscriptional mechanism since no significant elevation of its transcript was observed in A-T cells (Figure 3E). An R1 mRNA–binding activity important for R1 transcript stability has been described (61–63); however, the protein responsible has not been identified. This activity was found to be part of the PKC pathway. Given the known crosstalk between PKC and ATM signaling (64–66), it is tempting to speculate that disrupted PKC signaling in the absence of ATM may be responsible for the observed reduced R1 mRNA stability, resulting in reduced R1 protein. However, other forms of regulation are also possible.
Several important manifestations of loss of ATM or RR activity with regard to mitochondrial function follow from our results. The first is that mtDNA copy number and mitochondrial biogenesis are largely uncoupled in the absence of ATM (or RR activity). This conclusion is based on our observation that A-T cells maintain a mitochondrial biogenesis response in response to IR (albeit significantly blunted compared with wild-type cells; Figure 2C), even in the complete absence of mtDNA amplification under these conditions (Figure 2B). In addition, in cells depleted of mtDNA via loss of ATM or RR function, there is not a corresponding decrease in mitochondrial mass (data not shown). The end result in both of these cases is an imbalance in the usually strictly maintained cellular mtDNA/mitochondrial mass ratio. Second, mtDNA copy number alterations due to ATM or RR manipulations occur independently of corresponding changes in the steady-state levels of h-mtTFA (Figures 5 and 7). This result was not expected since the literature is replete with examples of how mtTFA abundance normally follows that of mtDNA (21, 41, 44, 45, 58, 67). Our results indicate that either the signaling required for h-mtTFA to properly track mtDNA is abrogated by inhibition of ATM or RR activity or that h-mtTFA is not as intimately involved in mtDNA copy number regulation as previously thought. Finally, our results demonstrating increased resistance of A-T cells to the mitochondrial inhibitors azide and chloramphenicol that correspond with increased levels of the mtDNA-encoded CO 1 protein (Figure 6) strongly suggest that ATM null cells aberrantly upregulate mitochondrial gene expression and/or OXPHOS activity in order to compensate for some loss in mitochondrial function. While the precise biological and pathological outcomes of this particular type of disruption in mitochondrial homeostasis remain to be investigated further, likely possibilities include altered energy metabolism, increased ROS production, altered susceptibility to apoptosis, or any of the multiple downstream consequences of mitochondrial defects that are prevalent in mitochondrial diseases.
While our results point to what we believe is a novel function for ATM in mtDNA maintenance, its impact on mtDNA regulation in the absence of DNA damage is conditional; that is, the mtDNA depletion observed in actively dividing A-T fibroblasts is not manifest in less actively dividing or nondividing cultures (Figure 3A and Supplemental Figure 1). The reasons for this are presently unknown, but there are several possible explanations. First, mtDNA replication occurs essentially independently of the cell cycle and continues even in nondividing cells (29). Under such conditions, it is generally accepted that salvage pathways for dNTP synthesis maintain mtDNA stability and replication (30–32, 34). Since RR and ATM specifically influence the de novo pathway for dNTP synthesis that is primarily operative in dividing cells (or in response to DNA damage), their effects are reduced or eliminated in confluent (or otherwise nondividing) conditions, where salvage pathways can compensate. A second possibility, which doesn’t necessarily exclude the first, is that mtDNA is being amplified in response to increased oxidative stress, as reported by others (56). Consistent with this possibility is our finding that, compared with those in proliferating cultures, A-T cells have higher amounts of ROS than wild-type cells, a difference that is enhanced when cell cultures are confluent (Figure 3F and Figure 6B). Therefore, we conclude that ATM and RR are major contributors to mtDNA maintenance under specific conditions, such as during proliferation and in response to DNA damage, but that mtDNA levels are also subject to additional layers of control (e.g., via salvage pathways and oxidative stress). These observations suggest that tissues with populations of actively dividing cells might be more prone to ATM-related mtDNA depletion than those in a postmitotic tissues. This likely influences the tissue-specificity of how loss of ATM has an impact on mtDNA copy number in vivo (Figure 7, A and C). Finally, while our results do not strongly implicate cell-cycle effects as a major contributor to the observed phenotypes, we cannot fully discount the possibility that some subset of the effects observed are a secondary consequence of the unique cell-cycle defects in the A-T cells. However, even if this were the case, the concept that RR and mitochondrial misregulation is a pertinent downstream consequence of loss of ATM signaling remains valid and of potential relevance to the pathology of A-T.
Our conclusions regarding the novel roles of ATM in the regulation of RR and mitochondrial homeostasis were confirmed in our analysis of tissues from ATM null mice, which provide a model for several important aspects of the human disease. In addition to the universal reduction in R1 levels in ATM null tissues already mentioned (Figure 7B), 2 mtDNA-related parameters, mtDNA copy number (Figure 7A) and expression of mtTFA (Figure 7B), were altered in a tissue-specific manner. Similar mtDNA copy number defects were observed in the human A-T patient tissues we analyzed (Figure 7B), indicating that disruption of mitochondrial homeostasis may also play a role in the human condition. Furthermore, as was observed in our cultured cell experiments (Figure 5), there was no correspondence between the steady-state levels of h-mtTFA and mtDNA in the ATM null mice tissues (Figure 7B), pointing to a general misregulation of normal mtDNA homeostasis in the absence of ATM signaling. The variable tissue-specific alterations in mtDNA in mouse and human tissues (both increases and decreases) are most likely a consequence of multiple pathways that can have an impact on this parameter in cells. For example, tissues with populations of dividing cells (e.g., bone marrow; Figure 5A) might be more prone to ATM-related mtDNA depletion due to their heightened dependence on the de novo pathway of dNTP synthesis. In addition, the documented oxidative stress in ATM null mouse cerebellum as well as patient cerebellum (7, 11) is consistent with the increased mtDNA copy number we observed in this tissue (Figure 7, A and C). In fact, it is probably a unique combination of all of these factors, and likely others yet to be defined, that determines the mtDNA copy number normally present in tissues as well as the unique tissue specificity of mtDNA copy number–related defects due to disruption of ATM and RR. We speculate that aberrant regulation of mitochondrial OXPHOS and/or ROS production resulting from inappropriate fluctuations in mtDNA copy number or mtDNA/h-mtTFA ratios in specific tissues may play a causative role in the pathogenesis of A-T, a disease with many symptoms in common with mitochondrial disorders, including neurodegeneration, ataxia, immunodeficiency, and premature aging (24). Future characterization of mitochondrial function in A-T cells and tissues and in ATM null mice will be critical in determining how mitochondrial dysfunction contributes to A-T–associated pathology and importantly to what extent this potential contribution is p53 dependent. Furthermore, the fact that loss of ATM protein and inhibition of its kinase activity by KU-55933 do not have completely overlapping effects on RR subunit regulation (Figures 3 and 5) and mitochondrial parameters (Figure 6), speaks to the existence of additional roles for the ATM protein (beyond its kinase activity) that are worthy of continued investigation. However, the fact that KU-55933 does cause mtDNA depletion in proliferating wild-type fibroblasts (Figure 3B), similar to that observed in A-T cells (Figure 3A), does implicate the kinase activity of ATM in this function.
In summary, this study has illuminated what we believe are novel roles for ATM signaling in the regulation of RR expression and normal mtDNA dynamics during proliferation and in response to genotoxic stress. Loss of either or both of these functions in A-T patients could certainly be expected to contribute significantly to the pathology of this disease in a manner that further compounds the loss of nuclear genomic stability associated with lack of DNA damage sensing by ATM. Furthermore, the requirement for proper amounts of mtDNA is underscored by the existence of inherited mtDNA depletion syndromes in humans, a fatal class of mitochondrial disease characterized by significant reductions of mtDNA in specific tissues (31–34). Relevant to the current study, several of these diseases are caused by nuclear gene mutations in enzymes involved in nucleotide metabolism, including those involved in salvage pathways of dNTP synthesis (30–34). However, mutations in the nucleotide salvage pathway enzymes represent only a subset of documented mtDNA depletion cases, and thus the genetic defects causing most mtDNA depletion syndromes have not been identified. Our results, which clearly implicate the de novo pathway of dNTP synthesis via RR in the maintenance of mtDNA, suggest that mutations in RR subunit genes may also represent a previously unrecognized cause of mtDNA depletion syndromes or other mtDNA-related disorders.
Methods
Cell culture and transfections.
GM07532 wild-type and GM02052 A-T primary fibroblasts (R35X/R35X) were purchased from The Coriell Institute for Medical Research and grown in modified Eagle’s medium with Earle’s salts with 15% FCS (HyClone) and 2 mM glutamine (Invitrogen). Experiments were carried out between passages 12 and 20 for wild-type fibroblasts and 13 and 19 for A-T fibroblasts. HeLa cells were grown in DMEM with 10% Fetal Clone FIII FCS substitute (HyClone) and 2 mM glutamine.
Wild-type and A-T primary fibroblasts from asynchronously growing cultures were seeded to 1 × 105 cells in T25 flasks (in triplicate) and grown for 24 hours (proliferating, ~60% confluent), 48 hours (more slowly proliferating, ~80% confluent), or 6 days (confluent). Triapine was used at 1 μM final concentration. Gemcitabine was used at 5 or 80 μM as indicated. ATM inhibitor KU-55933 (KuDOS Pharmaceuticals) was used at 10 μM final concentration as indicated with a preincubation of 48 hours prior to experiment initiation. Ethidium bromide (Sigma-Aldrich) was used at 50 ng/ml for times indicated, and medium was supplemented with 100 μg/ml pyruvate (Invitrogen) and 50 μg/ml uridine (Sigma-Aldrich).
For mitochondrial inhibition studies, wild-type, A-T, and KU-55933–treated wild-type cells from asynchronously growing cultures were seeded in triplicate at 2.5 × 104 cells in 6-well dishes and grown for 4 days in the presence of 8 mM sodium azide (Fisher Scientific) or 240 μg/ml chloramphenicol (Cellgro; Mediatech Inc.). Medium was changed every 24 hours in sodium azide experiments due to inclusion of KU-55933–treated cultures. No media replacement occurred during chloramphenicol treatment. At experiment completion, cells were trypsinized and counted on a hemacytometer.
Stably transfected R2 knockdown HeLa cells were seeded to 2.5 × 105 cells in T25 flasks (in triplicate) and grown for 48 hours. Transiently transfected R1 knockdown HeLa cells were grown for 72 hours after transfection.
HeLa cells were stably transfected with 1 μg of either pZZeo mu6 or pZZeo mU6 R2 siRNA generated as previously described (51) using Effectene Transfection Reagent (QIAGEN) according to manufacturer’s recommendations. Cells transfected with empty vector were continually maintained in 200 μg/ml Zeocin (Invitrogen), and the total transfected population was used as vector control in copy number experiments after 10 days. Cells transfected with the R2 siRNA vector were selected by culturing in 200 μg/ml zeocin until colonies formed. R2 knockdown clones were expanded and continually maintained in 200 μg/ml zeocin, and R2 knockdown was confirmed in selected clones via Western blot. Two clones with 50% knockdown were used in parallel for mtDNA copy number analysis.
HeLa cells were transiently transfected with human R1 short hairpin RNA as described (50). R1 knockdown was confirmed via Western blot with maximal knockdown at 72 hours. Cell pellets were frozen at –80°C prior to DNA extraction and PCR analysis.
Irradiation protocol.
Asynchronously proliferating cultures of wild-type and A-T primary fibroblasts (1 × 105 cells) were irradiated with 17.5 Gy of radiation using a 137Cs source or an X-Rad 320 x-ray irradiator (Precision X-Ray Inc.). Experiments were done in triplicate, and samples were housed in 15 ml falcon tubes in 10–15 ml of 50% growth medium/50% PBS during irradiation. After irradiation, samples were centrifuged at 453 g for 5 minutes, seeded in fresh medium in T25 flasks, and allowed to recover for 24 or 48 hours prior to harvesting for mtDNA copy number and FACS analysis. Nonirradiated controls were treated identically to irradiated samples. Triapine, where indicated, was added at 1 μM for a 24-hour incubation period either immediately after irradiation or after 24 hours of recovery.
DNA extraction and real-time quantitative PCR.
Total cellular DNA was extracted using a standard SDS lysis protocol as described (68). DNA pellets were resuspended in 50–100 μl of TE (1 mM EDTA, 10 mM Tris, pH 8.0) and either incubated for 1 hour at 65°C or at room temperature overnight prior to PCR analysis. A Bio-Rad iCycler iQ was used for real-time PCR analysis, essentially as described (69). In brief, a ratio of mtDNA to the nuclear 18S gene was generated for each sample. Human mtDNA primer sequences and the 18S primer sequences used for both human and mouse amplifications are shown below and were taken from Bai et al. (69). Mouse mtDNA primer sequences are from Brown, and Clayton (70): 18S 1546, forward, 5′-TAGAGGGACAAGTGGCGTTC-3′; 18S 1650, reverse, 5′-CGCTGAGCCAGTCAGTGT-3′; human mt3212, forward, 5′-CACCCAAGAACAGGGTTTGT-3′; human mt3319, reverse, 5′-TGGCCATGGGTATGTTGTTAA-3′; mouse COI, forward, 5′-GCCCCAGATATAGCATTCCC-3′; mouse COI, reverse, 5′-GTTCATCCTGTTCCTGCTCC-3′.
Two independent reactions for mitochondrial and nuclear primer sets were run for each sample. The 25 μl reaction volumes contained 12.5 μl of Bio-Rad iQ SYBR Green Supermix, 0.5 μl of a 25 μM stock of each primer, 1.5 μl of water, and 10 μl of template genomic DNA. DNA samples were diluted 20- to 480-fold, and a 2-fold dilution series was run for each sample to insure accurate sample profiling and linearity. The PCR protocol consisted of 50°C for 2 minutes and 95°C for 10 minutes, then 40 cycles of 95°C for 15 seconds and 60°C annealing/extension for 1 minute, with real-time data collection as described (69). Melt curves from 55°C to 95°C with 0.5°C increments every 10 seconds were included to confirm single PCR products, which were verified in early reactions by running products on 2% agarose gels.
Western blotting.
All Western blotting samples were collected in tandem with mtDNA copy number samples. Wild-type and A-T primary fibroblasts and stably transfected R2 knockdown HeLa cells were grown or treated with IR as described above. For primary cells, at least 3 T25 flasks were combined prior to protein extraction and Western blotting. Cell pellets were harvested, lysed in lysis buffer (1% SDS, 10 mM Tris, p.H. 7.4), and passed through a QIAshredder column (QIAGEN). Protein determination was carried out using the Bio-Rad DC Protein Assay (Bio-Rad) according to manufacturer’s protocol with slight modifications. We ran 50 μg of total protein on a 12% bis-acrylamide gel and transferred it to a 0.2 μm nitrocellulose membrane (Pall). For mouse and human tissue, frozen pulverized tissue (see Tissue DNA extraction) was digested in CelLytic MT Mammalian Tissue Lysis/Extraction Reagent (Sigma-Aldrich) and boiled for 5 minutes. Boiled tissue samples were mechanically disrupted and passed through a QIAshredder column; protein concentration was determined, and Western blotting was performed as described for cell culture, except 80 μg of total protein was run. Primary antibody was incubated overnight at 4°C. Anti-R2 (E-16 or H-300) and p53R2 (N-16 or C-18) antibodies were purchased from Santa Cruz Biotechnology Inc. The anti-R1 (AD203 or H-300) antibodies were from Santa Cruz Biotechnology Inc. or Chemicon International, and anti-actin (20–33) antibody was from Sigma-Aldrich. Human and mouse mtTFA antibodies were gifts from David Clayton. Films were quantified using a GE Healthcare densitometer and ImageQuaNT software. Only films for which linearity of the loading control standard could be demonstrated were analyzed. SimplyBlue SafeStain (Invitrogen) was used according to manufacturer’s recommendations to demonstrate loading for mouse tissue blots.
Determination of RR mRNA transcript levels.
Asynchronous cultures of wild-type and A-T primary fibroblasts were seeded at 1 × 105 cells in T150 flasks and grown for 4 days. Medium was replaced 1 day prior to RNA extraction. We harvested 5 × 105 cells and performed RNA extraction using an RNeasy Mini Kit (QIAGEN) according to manufacturer’s instructions. During the procedure, a QIAshredder (QIAGEN) was used to homogenize cells, and total cellular RNA was twice eluted in the same 50 μl of RNase-free water. Sample RNA preps were quantified using SYBR green fluorescence against a known standard. We converted 12 μg of total RNA per sample into gene-specific cDNA with reverse primers to BACE2, R1, and p53R2 (sequences below) using M-MuLV Reverse Transcriptase (New England BioLabs Inc.) and associated protocol according to manufacturer’s instructions. cDNA preps were then purified in approximate volume using a QIAquick PCR Purification Kit (QIAGEN) according to the manufacturer’s protocol to remove components that inhibited the real-time PCR reaction. Samples were then analyzed in triplicate for each primer set in the linear range (BACE2, R1, and p53R2) using real-time PCR in a Bio-Rad iCycler iQ as described above. The relative levels of R1 and p53R2 between wild-type and A-T samples were determined using the 2ΔCT equation after normalization to the BACE2 CT values. R1, p53R2, and BACE2 primers did not amplify from no reverse transcriptase control reactions. The primer sequences used for cDNA conversion and real-time PCR were as follows: R1, forward, 5′-GGCTACTGGCAGCTACATTGC-3′; R1, reverse, 5′-CGCTTGTTCCCACCTTGATCC-3′; p53R2, forward, 5′-GGGCTGGATCAGGATGAGAGA-3′; p53R2 reverse, 5′-TGGAAAGATGACAAACCGGCG-3′; BACE2, forward, 5′-GGCTACTACCTGGAGATGCTG-3′; BACE2, reverse, 5′-TATGTAGGAGTGCGGGGTTCC-3′.
Mitochondrial mass, membrane potential, and ROS detection.
A BD FACSCalibur Flow Cytometer was used for cell sorting, and data were analyzed using FlowJo 3.1 software (Tree Star). Triplicate cultures of asynchronous wild-type and A-T primary fibroblasts (1 × 105 cells) were seeded in T25 flasks and grown for 24 hours (proliferating), 48 hours, or 6 days as indicated. Cells were stained with 80 μM dihydroethidium, 60 nM MitoTracker Green FM, or 80 nM MitoTracker Red (Invitrogen) in modified Eagle’s medium with Earle’s salts medium for 20 to 25 minutes at 37°C and 5% CO2 in the dark. Cells were maintained in PBS with 1% FCS on ice in the dark until FACS analysis was performed. Each experiment was calibrated with unstained cells, and 3,000–12,000 gated cells were analyzed in all experiments.
Cell-cycle analysis.
Wild-type and A-T primary fibroblasts (3 × 105 cells) were seeded from low density, asynchronous cultures and trypsinized at times indicated. Cell pellets were washed and resuspended in 300 μl washing buffer (5 mM EDTA in PBS). Cells were vortexed upon dropwise addition of 700 μl of ice-cold 100% ethanol and pelleted at 11,000 g. Cells were then resuspended in 500 μl of washing buffer with the addition of 50 μl of 10 mg/ml RNase A (Sigma-Aldrich) and incubated with rocking at 37°C for 20 minutes. Cells were pelleted at 11,000 g and resuspended in 200–300 μl of propidium iodide solution (Sigma-Aldrich) (100 μg/ml in PBS) and immediately analyzed by FACS on a BD FACSCalibur Flow Cytometer. Identical gates were used for both wild-type and A-T cells to evaluate the percentage of cells in G1, S, and G2-M phases. In all cases, less than 5% of cells presented as doublets or higher order aggregates and were excluded from analysis.
Tissue DNA extraction.
Breeding pairs of inbred 129SvEV mice heterozygous for a null allele of ATM (71) were obtained from Howard Mount (University of Toronto, Toronto, Ontario, Canada) and maintained in collaboration with the Yale Animal Resource Center according to Institutional Animal Care and Use Committee–approved protocols. Pups were genotyped as previously described (71) with primer sequences as follows: ATM, forward, 5′-CCGACTTCTGTCAGATGTTGC-3′; ATM, reverse, 5′-ATTTGCAGGAGTTGCTGAGCG-3′; ATM, neo reverse, 5′-GGGTGGGATTAGATAAATGCC-3′.
Tissues from 3 ATM–/– mice at 10 weeks of age were fast frozen on dry ice or liquid nitrogen and stored at –80°C until powdered by mortal and pestle on dry ice. The entire organ was taken for cerebrum, cerebellum, thymus, and lung. Skeletal muscle from the left quadriceps was indicated as muscle. Bone marrow was extracted from the right and left femur, and small intestine was taken from the jejunum region. Total cellular DNA was extracted in duplicate and processed independently using a DNeasy Tissue Kit (QIAGEN) according to manufacturer’s protocol, and samples were processed by real-time quantitative PCR as described above. Real-time relative mtDNA copy numbers reflect all mouse duplicate tissue extractions combined.
Normal and A-T human tissue samples matched by age, sex, and postmortem interval prior to tissue-harvesting were obtained from the National Institute of Child Health and Human Development (NICHD) Brain and Tissue Bank for Developmental Disorders (University of Maryland, Baltimore, Maryland, USA) and stored at –80°C. Tissue sections were grossly pulverized, and duplicate samples were powdered by mortal and pestle on dry ice and processed independently as described above. Samples of heart, cerebrum, and cerebellum were obtained from a single A-T patient (A-T 1), and 2 other A-T patients (A-T 2 and A-T 3) provided secondary cerebrum and cerebellum samples.
Statistics.
In all bar graphs, the mean ± SD from at least 2 triplicate experiments is plotted. The P values for individual comparisons based on unpaired, 2-tailed Student’s t tests are indicated in the figures. A P value of less than 0.05 was considered significant. For multiple comparisons 1-way ANOVA analysis was employed.
Acknowledgments
This work was supported by Program Project Grant P01 ES011163 and by a grant from the AT Children’s Project. The authors wish to thank Howard Mount for providing the ATM null mice, Robert Johnson at the Brain and Tissue Bank for careful oversight of normal and A-T tissue allocation, David Clayton for the human and mouse mtTFA antibodies, Sharen McKay for critical input into the project, and Susan Kaech for many helpful discussions.
Footnotes
Nonstandard abbreviations used: A-T, ataxia-telangiectasia; ATM, A-T–mutated; CO I, cytochrome oxidase I; dNTP, deoxyribonucleoside triphosphate; h-mtTFA, human mitochondrial transcription factor A; IR, ionizing radiation; mtDNA, mitochondrial DNA; mtTFA, mitochondrial transcription factor A; OXPHOS, oxidative phosphorylation; RNAi, RNA interference; RR, ribonucleotide reductase.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2723–2734 (2007). doi:10.1172/JCI31604
References
- 1.Abraham R.T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 2.Lavin M.F., Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu. Rev. Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 3.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 4.Shiloh Y., Kastan M.B. ATM: genome stability, neuronal development, and cancer cross paths. Adv. Cancer Res. 2001;83:209–254. doi: 10.1016/s0065-230x(01)83007-4. [DOI] [PubMed] [Google Scholar]
- 5.Kastan M.B., Lim D.S. The many substrates and functions of ATM. Nat. Rev. Mol. Cell Biol. 2000;1:179–186. doi: 10.1038/35043058. [DOI] [PubMed] [Google Scholar]
- 6.Shiloh Y. ATM (ataxia telangiectasia mutated): expanding roles in the DNA damage response and cellular homeostasis. Biochem. Soc. Trans. 2001;29:661–666. doi: 10.1042/0300-5127:0290661. [DOI] [PubMed] [Google Scholar]
- 7.Barlow C., et al. Loss of the ataxia-telangiectasia gene product causes oxidative damage in target organs. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9915–9919. doi: 10.1073/pnas.96.17.9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reichenbach J., et al. Elevated oxidative stress in patients with ataxia telangiectasia. Antioxid. Redox Signal. 2002;4:465–469. doi: 10.1089/15230860260196254. [DOI] [PubMed] [Google Scholar]
- 9.Reliene R., Fischer E., Schiestl R.H. Effect of N-acetyl cysteine on oxidative DNA damage and the frequency of DNA deletions in atm-deficient mice. Cancer Res. 2004;64:5148–5153. doi: 10.1158/0008-5472.CAN-04-0442. [DOI] [PubMed] [Google Scholar]
- 10.Schubert R., et al. Cancer chemoprevention by the antioxidant tempol in Atm-deficient mice. Hum. Mol. Genet. 2004;13:1793–1802. doi: 10.1093/hmg/ddh189. [DOI] [PubMed] [Google Scholar]
- 11.Kamsler A., et al. Increased oxidative stress in ataxia telangiectasia evidenced by alterations in redox state of brains from Atm-deficient mice. Cancer Res. 2001;61:1849–1854. [PubMed] [Google Scholar]
- 12.Guittet O., et al. Mammalian p53R2 protein forms an active ribonucleotide reductase in vitro with the R1 protein, which is expressed both in resting cells in response to DNA damage and in proliferating cells. J. Biol. Chem. 2001;276:40647–40651. doi: 10.1074/jbc.M106088200. [DOI] [PubMed] [Google Scholar]
- 13.Nordlund P., Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka H., et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 15.Bjorklund S., Skogman E., Thelander L. An S-phase specific release from a transcriptional block regulates the expression of mouse ribonucleotide reductase R2 subunit. EMBO J. 1992;11:4953–4959. doi: 10.1002/j.1460-2075.1992.tb05602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson E., Skogman E., Thelander L. The TATA-less promoter of mouse ribonucleotide reductase R1 gene contains a TFII-I binding initiator element essential for cell cycle-regulated transcription. J. Biol. Chem. 1995;270:30162–30167. doi: 10.1074/jbc.270.50.30162. [DOI] [PubMed] [Google Scholar]
- 17.Liu X., et al. The ribonucleotide reductase subunit M2B subcellular localization and functional importance for DNA replication in physiological growth of KB cells. Biochem. Pharmacol. 2005;70:1288–1297. doi: 10.1016/j.bcp.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Hakansson P., Hofer A., Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J. Biol. Chem. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 19.Filatov D., Bjorklund S., Johansson E., Thelander L. Induction of the mouse ribonucleotide reductase R1 and R2 genes in response to DNA damage by UV light. J. Biol. Chem. 1996;271:23698–23704. doi: 10.1074/jbc.271.39.23698. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi T., et al. p53R2-dependent pathway for DNA synthesis in a p53-regulated cell cycle checkpoint. Cancer Res. 2001;61:8256–8262. [PubMed] [Google Scholar]
- 21.Shadel G.S., Clayton D.A. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 22.Bender A., et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 23.Kraytsberg Y., et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 24.Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balaban R.S., Nemoto S., Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Kushnareva Y., Murphy A.N., Andreyev A. Complex I-mediated reactive oxygen species generation: modulation by cytochrome c and NAD(P)+ oxidation-reduction state. Biochem. J. 2002;368:545–553. doi: 10.1042/BJ20021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turrens J.F. Superoxide production by the mitochondrial respiratory chain. Biosci. Rep. 1997;17:3–8. doi: 10.1023/a:1027374931887. [DOI] [PubMed] [Google Scholar]
- 28.Bonawitz N.D., Rodeheffer M.S., Shadel G.S. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Mol. Cell. Biol. 2006;26:4818–4829. doi: 10.1128/MCB.02360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnusson J., Orth M., Lestienne P., Taanman J.W. Replication of mitochondrial DNA occurs throughout the mitochondria of cultured human cells. Exp. Cell Res. 2003;289:133–142. doi: 10.1016/s0014-4827(03)00249-0. [DOI] [PubMed] [Google Scholar]
- 30.Marti R., Nishigaki Y., Vila M.R., Hirano M. Alteration of nucleotide metabolism: a new mechanism for mitochondrial disorders. Clin. Chem. Lab. Med. 2003;41:845–851. doi: 10.1515/CCLM.2003.128. [DOI] [PubMed] [Google Scholar]
- 31.Elpeleg O., Mandel H., Saada A. Depletion of the other genome-mitochondrial DNA depletion syndromes in humans. J. Mol. Med. 2002;80:389–396. doi: 10.1007/s00109-002-0343-5. [DOI] [PubMed] [Google Scholar]
- 32.Mancuso M., et al. Mitochondrial DNA depletion: mutations in thymidine kinase gene with myopathy and SMA. Neurology. 2002;59:1197–1202. doi: 10.1212/01.wnl.0000028689.93049.9a. [DOI] [PubMed] [Google Scholar]
- 33.Saada A. Deoxyribonucleotides and disorders of mitochondrial DNA integrity. DNA Cell Biol. 2004;23:797–806. doi: 10.1089/dna.2004.23.797. [DOI] [PubMed] [Google Scholar]
- 34.Salviati L., et al. Mitochondrial DNA depletion and dGK gene mutations. Ann. Neurol. 2002;52:311–317. doi: 10.1002/ana.10284. [DOI] [PubMed] [Google Scholar]
- 35.O’Rourke T.W., et al. Differential involvement of the related DNA helicases Pif1p and Rrm3p in mtDNA point mutagenesis and stability. Gene. 2005;354:86–92. doi: 10.1016/j.gene.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 36.Taylor S.D., et al. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:3010–3018. doi: 10.1091/mbc.E05-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parisi M.A., Clayton D.A. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 38.McCulloch V., Shadel G.S. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell. Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonawitz N.D., Clayton D.A., Shadel G.S. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 40.Alam T.I., et al. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ekstrand M.I., et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 42.Larsson N.G., Oldfors A., Holme E., Clayton D.A. Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem. Biophys. Res. Commun. 1994;200:1374–1381. doi: 10.1006/bbrc.1994.1603. [DOI] [PubMed] [Google Scholar]
- 43.Larsson N.G., et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 44.Poulton J., et al. Deficiency of the human mitochondrial transcription factor h-mtTFA in infantile mitochondrial myopathy is associated with mtDNA depletion. Hum. Mol. Genet. 1994;3:1763–1769. doi: 10.1093/hmg/3.10.1763. [DOI] [PubMed] [Google Scholar]
- 45.Seidel-Rogol B.L., Shadel G.S. Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic Acids Res. 2002;30:1929–1934. doi: 10.1093/nar/30.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X.J., Wang X., Kaufman B.A., Butow R.A. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 2005;307:714–717. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- 47.Stigter D. Packaging of single DNA molecules by the yeast mitochondrial protein Abf2p: reinterpretation of recent single molecule experiments. Biophys. Chem. 2004;110:171–178. doi: 10.1016/j.bpc.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 48.Finch R.A., et al. Triapine (3-aminopyridine-2-carboxaldehyde- thiosemicarbazone): a potent inhibitor of ribonucleotide reductase activity with broad spectrum antitumor activity. Biochem. Pharmacol. 2000;59:983–991. doi: 10.1016/s0006-2952(99)00419-0. [DOI] [PubMed] [Google Scholar]
- 49.Finch R.A., Liu M.C., Cory A.H., Cory J.G., Sartorelli A.C. Triapine (3-aminopyridine-2-carboxaldehyde thiosemicarbazone; 3-AP): an inhibitor of ribonucleotide reductase with antineoplastic activity. Adv. Enzyme Regul. 1999;39:3–12. doi: 10.1016/s0065-2571(98)00017-x. [DOI] [PubMed] [Google Scholar]
- 50.Lin Z.P., et al. Excess ribonucleotide reductase R2 subunits coordinate the S phase checkpoint to facilitate DNA damage repair and recovery from replication stress. Biochem. Pharmacol. 2007;73:760–772. doi: 10.1016/j.bcp.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 51.Lin Z.P., Belcourt M.F., Cory J.G., Sartorelli A.C. Stable suppression of the R2 subunit of ribonucleotide reductase by R2-targeted short interference RNA sensitizes p53(–/–) HCT-116 colon cancer cells to DNA-damaging agents and ribonucleotide reductase inhibitors. J. Biol. Chem. 2004;279:27030–27038. doi: 10.1074/jbc.M402056200. [DOI] [PubMed] [Google Scholar]
- 52.Tsai M.H., et al. Transcriptional responses to ionizing radiation reveal that p53R2 protects against radiation-induced mutagenesis in human lymphoblastoid cells. Oncogene. 2006;25:622–632. doi: 10.1038/sj.onc.1209082. [DOI] [PubMed] [Google Scholar]
- 53.Xue L., et al. Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res. 2003;63:980–986. [PubMed] [Google Scholar]
- 54.Hickson I., et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 55.Chen S., Wang G., Makrigiorgos G.M., Price B.D. Stable siRNA-mediated silencing of ATM alters the transcriptional profile of HeLa cells. Biochem. Biophys. Res. Commun. 2004;317:1037–1044. doi: 10.1016/j.bbrc.2004.03.149. [DOI] [PubMed] [Google Scholar]
- 56.Lee H.C., et al. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem. J. 2000;348:425–432. [PMC free article] [PubMed] [Google Scholar]
- 57.Xu Y., Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 58.Piechota J., et al. Differential stability of mitochondrial mRNA in HeLa cells. Acta Biochim. Pol. 2006;53:157–168. [PubMed] [Google Scholar]
- 59.Stern N., et al. Accumulation of DNA damage and reduced levels of nicotine adenine dinucleotide in the brains of Atm-deficient mice. J. Biol. Chem. 2002;277:602–608. doi: 10.1074/jbc.M106798200. [DOI] [PubMed] [Google Scholar]
- 60.Bourdon A., et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 61.Chen F.Y., Amara F.M., Wright J.A. Defining a novel ribonucleotide reductase r1 mRNA cis element that binds to an unique cytoplasmic trans-acting protein. Nucleic Acids Res. 1994;22:4796–4797. doi: 10.1093/nar/22.22.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen F.Y., Amara F.M., Wright J.A. Posttranscriptional regulation of ribonucleotide reductase R1 gene expression is linked to a protein kinase C pathway in mammalian cells. Biochem. Cell Biol. 1994;72:251–256. doi: 10.1139/o94-036. [DOI] [PubMed] [Google Scholar]
- 63.Chen F.Y., Amara F.M., Wright J.A. Regulation of mammalian ribonucleotide reductase R1 mRNA stability is mediated by a ribonucleotide reductase R1 mRNA 3′-untranslated region cis-trans interaction through a protein kinase C-controlled pathway. Biochem. J. 1994;302:125–132. doi: 10.1042/bj3020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li B., et al. Distinct roles of c-Abl and Atm in oxidative stress response are mediated by protein kinase C delta. Genes Dev. 2004;18:1824–1837. doi: 10.1101/gad.1223504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakajima T. Signaling cascades in radiation-induced apoptosis: roles of protein kinase C in the apoptosis regulation. Med. Sci. Monit. 2006;12:RA220–RA224. [PubMed] [Google Scholar]
- 66.Nakajima T., et al. Regulation of radiation-induced protein kinase Cdelta activation in radiation-induced apoptosis differs between radiosensitive and radioresistant mouse thymic lymphoma cell lines. Mutat. Res. 2006;595:29–36. doi: 10.1016/j.mrfmmm.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 67.Moraes C.T. What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 2001;17:199–205. doi: 10.1016/s0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 68. Sambrook, J., and Russell, D.W. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 999 pp. [Google Scholar]
- 69.Bai R.K., Perng C.L., Hsu C.H., Wong L.J. Quantitative PCR analysis of mitochondrial DNA content in patients with mitochondrial disease. Ann. N. Y. Acad. Sci. 2004;1011:304–309. doi: 10.1007/978-3-662-41088-2_29. [DOI] [PubMed] [Google Scholar]
- 70.Brown T.A., Clayton D.A. Release of replication termination controls mitochondrial DNA copy number after depletion with 2′,3′-dideoxycytidine. Nucleic Acids Res. 2002;30:2004–2010. doi: 10.1093/nar/30.9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mount H.T., et al. Progressive sensorimotor impairment is not associated with reduced dopamine and high energy phosphate donors in a model of ataxia-telangiectasia. J. Neurochem. 2004;88:1449–1454.. doi: 10.1046/j.1471-4159.2003.02278.x. [DOI] [PubMed] [Google Scholar]