Abstract
MicroRNAs act as negative regulators of gene expression by inhibiting the translation or promoting the degradation of target mRNAs. Recent studies have revealed key roles of microRNAs as regulators of the growth, development, function, and stress responsiveness of the heart, providing glimpses of undiscovered regulatory mechanisms and potential therapeutic targets for the treatment of heart disease.
Life from early embryogenesis to adulthood depends on the second-to-second function of the heart. Even subtle perturbations in the delicate process of heart formation during embryogenesis or in the transmission of electrical impulses in the adult heart can have catastrophic consequences. The heart is also highly sensitive to stress, which results in pathological growth and remodeling, frequently leading to a loss in contractile efficiency, pump failure, and sudden death.
Despite a detailed understanding of the molecular mechanisms governing cardiac development and contractility, cardiac malformations are the most prevalent birth defect and the number one cause of noninfectious infant mortality (1), and adult heart disease remains the primary cause of morbidity and mortality in the industrialized world (2). Thus, there is a dire need for new therapeutic strategies to enhance cardiac function and repair in the settings of congenital and acquired heart disease.
Recent studies have uncovered profound and unexpected roles for a family of tiny regulatory RNAs, known as microRNAs (miRNAs), in the control of diverse aspects of cardiac function and dysfunction, including myocyte growth, integrity of the ventricular wall, contractility, gene expression, and maintenance of cardiac rhythm (3–6). Specific miRNAs are misexpressed in diseased hearts (7–11), and gain- and loss-of-function experiments in mice have shown these miRNAs to be necessary and sufficient for multiple forms of heart disease (3–6, 8, 12, 13). Here we review the biology of miRNAs and consider their myriad roles in the heart as well as possible therapeutic opportunities for exploiting miRNAs in the settings of heart disease.
miRNAs: history, biology, and function
miRNAs are approximately 22 nucleotides in length and inhibit translation by interacting with the 3′ untranslated regions of specific mRNA targets (14). Through forward genetic screening in the nematode Caenorhabditis elegans, lin-4 and let-7, the first miRNAs to be discovered, were shown to function as regulators of developmental timing (15, 16). Subsequent studies in vertebrates suggested that miRNAs, rather than functioning as decisive regulatory on-off switches, more commonly function to modulate or fine-tune cellular phenotypes by repressing expression of proteins that are inappropriate for a particular cell type or by adjusting protein dosage (17). miRNAs have also been proposed to provide robustness to cellular phenotypes by eliminating extreme fluctuations in gene expression (18).
Numerous miRNAs are upregulated in response to cellular stress (19). Moreover, stress signaling modulates the efficiency of translational repression by miRNAs by regulating their association with mRNA targets in cytoplasmic structures known as stress granules (20). These findings point to the potential importance of miRNAs in the modulation of pathological processes, as has already been shown in cancer (21) and heart disease (as discussed below).
Based on sequence conservation and the ability to fold into a hairpin structure, the human genome is predicted to encode as many as a thousand miRNAs, which are estimated to regulate as many as 30% of mRNA transcripts (22). To date, the in vivo functions of only a handful of miRNAs have been determined in mammals (4–6, 8, 12, 23, 24), but their powerful effects on cellular phenotypes, effect on such a substantial fraction of the genome, and evolutionary conservation across divergent species underscore their importance as key regulators of physiological and pathological processes that are only beginning to be appreciated.
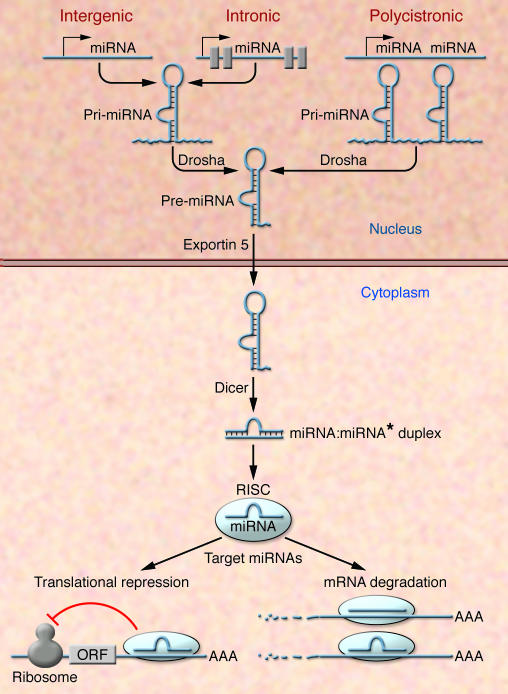
miRNAs are transcribed by RNA polymerase II and can be derived from individual miRNA genes, from introns of protein-coding genes, or from polycistronic transcripts that often encode multiple, closely related miRNAs (Figure 1). Pri-miRNAs, generally several thousand bases long, are processed in the nucleus by the RNase Drosha into 70–100 nucleotide, hairpin-shaped precursors, called pre-miRNAs. Following transport to the cytoplasm, the pre-miRNA is further processed by the RNA endonuclease Dicer to produce a double-stranded miRNA. The fully processed miRNA duplex is then incorporated into a multicomponent protein complex known as RNA-induced silencing complex (RISC) (25–27). During this process, one strand of the miRNA duplex is selected as a mature miRNA while the other strand, known as miRNA*, is in general rapidly removed and degraded. This selection process is primarily determined by the strength of base pairing at the end of the miRNA:miRNA* duplex (25–27). As part of the RISC, miRNAs negatively regulate gene expression through two major mechanisms, translational repression and mRNA cleavage, which depend on the extent of complementarity between the miRNA and its mRNA target and other criteria that are still being defined. Although it was originally thought that perfect base pairing was a requisite for mRNA cleavage, it has now become clear that even imperfect base pairing can lead to a decrease in target mRNA abundance (28, 29) (Figure 1).
Figure 1. miRNA biogenesis and function.
The primary transcripts of miRNAs, called pri-miRNAs, are transcribed as individual miRNA genes, from introns of protein-coding genes, or from polycistronic transcripts. The RNase Drosha further processes the pri-miRNA into 70–100 nucleotide, hairpin-shaped precursors, called pre-miRNA, which are exported from the nucleus by exportin 5. In the cytoplasm, the pre-miRNA is cleaved by Dicer into an miRNA:miRNA* duplex. Assembled into the RISC, the mature miRNA negatively regulates gene expression by either translational repression or mRNA degradation, which is dependent on sequence complementarity between the miRNA and the target mRNA. ORF, open reading frame.
The 5′ portion of an miRNA spanning bases two–eight, termed the seed region, is especially important for target recognition (30). The sequence of the seed, together with phylogenetic conservation of the target sequence, forms the basis for many current target prediction models. Although increasingly sophisticated computational approaches to predict miRNAs and their targets are becoming available, target prediction remains a major challenge and requires experimental validation. Ascribing the functions of miRNAs to the regulation of specific mRNA targets is further complicated by the ability of individual miRNAs to base pair with hundreds of potential high- and low-affinity mRNA targets and by the targeting of multiple miRNAs to individual mRNAs.
Blockade to miRNA processing by Dicer loss of function
The importance of miRNAs in the normal development of vertebrates is evidenced by Dicer loss of function in both zebrafish and mice (31, 32). Zebrafish lacking Dicer and thereby the capacity to synthesize miRNAs undergo relatively normal morphogenesis and organ development but die two weeks after fertilization due to a general growth arrest (32). Dicer-deficient mice die at gastrulation and lack multipotent stem cells (31). A caveat to the interpretation of these phenotypes, with respect to the requirement of miRNAs, is that Dicer may perform functions in addition to miRNA processing.
The roles of miRNAs in tissue development have also begun to be explored through conditional deletion of Dicer in specific cell lines and tissue types (33–36). To study the requirement of miRNAs in the mouse heart, a conditional Dicer allele was removed by expressing Cre recombinase under control of Nkx2.5 regulatory elements, which are expressed in the heart from day E8.5 onwards (37). Cardiac deletion of Dicer did not perturb the expression of cardiac markers of differentiation or patterning but resulted in death from cardiac failure by E12.5 due to a poorly developed ventricular myocardium (4). Since these experiments did not allow for deletion of Dicer during the earliest steps of cardiogenesis, it remains to be determined whether miRNAs play a role in specification of the cardiac lineage.
Cardiac hypertrophy and pathological remodeling
The heart responds to chronic and acute injury by hypertrophic growth. Cardiomyocyte hypertrophy is the dominant cellular response to virtually all forms of hemodynamic overload, endocrine disorders, myocardial injury, or inherited mutations in a variety of structural and contractile proteins (2, 38). Numerous signaling pathways, especially those involving aberrant calcium signaling, drive cardiac hypertrophy and pathological remodeling (39). Hypertrophic growth in response to stress involves different signaling pathways and gene expression patterns than physiological hypertrophy as occurs in response to exercise. Stress-mediated myocardial hypertrophy is a complex phenomenon associated with numerous adverse consequences with distinct molecular and histological characteristics causing the heart to dilate and decompensate which, through myocyte degeneration and death, often culminates in heart failure. As such, there has been intense interest in deciphering the underlying molecular mechanisms and in discovering novel therapeutic targets for suppressing adverse cardiac growth.
Cardiac myocytes are normally surrounded by a fine network of collagen fibers. In response to pathological stress, cardiac fibroblasts and extracellular matrix proteins accumulate disproportionately and excessively (40). Myocardial fibrosis, a characteristic of all forms of pathological hypertrophy, leads to mechanical stiffness, which contributes to contractile dysfunction (41). Another hallmark of pathological hypertrophy and heart failure is the reactivation of a set of fetal cardiac genes, including those encoding atrial natriuretic peptide, B-type natriuretic peptide, and fetal isoforms of contractile proteins, such as skeletal α-actin and β-myosin heavy chain (βMHC). These genes are typically repressed postnatally and replaced by the expression of a set of adult cardiac genes (42).
The consequences of fetal gene expression on cardiac function and remodeling (e.g., fibrosis) are not completely understood, but the upregulation of βMHC, a slow ATPase, and downregulation of αMHC, a fast-contracting ATPase, in response to stress has been implicated in the diminution of cardiac function (43). βMHC is the predominant myosin isoform expressed in the heart prenatally whereas αMHC is upregulated after birth. Thyroid hormone signaling soon after birth provides the stimulus for the β– to αMHC switch (44). There are differences in the relative abundance of α– and βMHC in rodent and human hearts; α represents more than 95% of MHC in the adult rodent heart whereas it constitutes only about 30% of MHC in the adult human heart (45). Nevertheless, downregulation of αΜΗC and upregulation of βMHC is a common response to cardiac injury irrespective of the species. Moreover, changes in the proportion of MHC isoforms correlate with the level of mechanical performance and efficiency of the heart. Relatively minor changes in the ratio of αMHC to βMHC have been shown to have profound effects on cardiac contractility in humans and rodents (43, 46–48). Thus, much attention has focused on understanding the mechanisms that regulate αMHC and βMHC switching and on potential approaches for therapeutically manipulating these mechanisms.
Regulation of cardiac hypertrophy and heart failure by stress-responsive miRNAs
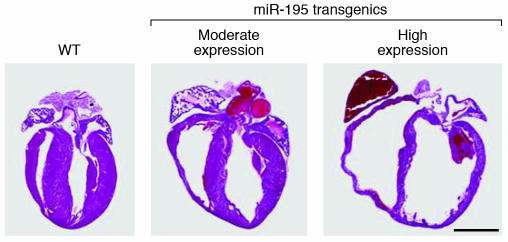
In several recent studies, microarray analyses were performed to determine whether miRNAs are dysregulated in hypertrophic and failing hearts. These studies point to a collection of miRNAs that are up- and downregulated during pathological cardiac remodeling in rodents and humans (7–11). In vitro experiments using either overexpression or knockdown of miRNAs in cultured cardiomyocytes indicate that a subset of these miRNAs are indeed actively involved in cardiomyocyte hypertrophy (7–11). Intriguingly, forced expression of certain stress-inducible miRNAs in primary cardiomyocytes or in the hearts of transgenic mice is sufficient to drive hypertrophic growth and myocyte disarray (7–11). Cardiac-specific overexpression of miRNA-195 (miR-195), which is consistently upregulated in rodent and human hypertrophic hearts, for example, results in dilated cardiomyopathy and heart failure in mice as early as two weeks of age (8) (Figures 2 and 3), implying that upregulation of miR-195 during cardiac hypertrophy actively contributes to the disease process. Based on target predictions for miR-195, we speculate that the cardiac phenotype of these transgenic mice results from the downregulation of multiple prosurvival proteins by this miRNA. Since miR-195 belongs to a small family of related miRNAs, it will be interesting to investigate the potential involvement of the other family members in cardiac disease.
Figure 2. Induction of cardiac hypertrophy and heart failure by miR-195.
H&E sections of 2-week-old wild-type and transgenic mice expressing miR-195 under control of the αMHC promoter. In transgenic as compared with wild-type mice, moderate levels of miR-195 expression (26-fold) cause cardiac hypertrophy, and higher levels of expression (29-fold) cause dilated cardiomyopathy with ventricular dilatation and wall thinning. Reproduced with permission from Proceedings of the National Academy of Sciences of the United States of America (8). Scale bar: 2 mm.
Figure 3. Roles of specific miRNAs in cardiac hypertrophy.
Stress signals drive a pathological remodeling response of the adult heart, which is characterized by hypertrophy, ventricular dilatation, and fibrosis. miR-208 is required for stress-dependent remodeling and miR-195, which is induced by cardiac stress, is sufficient to drive cardiac remodeling. miR-133 appears to play a negative role in cardiac hypertrophy (5, 6, 8).
Another miRNA consistently induced by cardiac stress, miR-21, appears to function as a regulator of cardiac growth and fetal gene activation in primary cardiomyocytes in vitro although some confusion remains regarding its exact role (7, 10). Cheng et al. (7) reported that knockdown of miR-21 using a 2′-O-methyl–miR-21 antisense oligonucleotide in cardiomyocytes in culture can suppress cardiomyocyte growth and fetal gene expression in response to the hypertrophic agonists angiotensin II and phenylephrine. However, Tatsuguchi et al. (10) showed inhibition of miR-21 in cardiomyocytes using locked nucleic acid–modified antisense oligonucleotides to induce myocyte hypertrophy while overexpression by an miR-21 duplex resulted in a decrease in cardiomyocyte cell size. The basis for these differences is currently unclear. It is notable in this regard that miR-21 has been reported to modulate cell growth both positively and negatively in multiple cancerous cell lines (49, 50) and to promote apoptosis in transformed cells (49, 51).
miRNAs that are downregulated in the hypertrophic and failing heart are also of interest, especially since forced expression of a subset of these prevents hypertrophic cell growth in neonatal rat myocytes (9, 10). It is tempting to speculate that these miRNAs function as negative regulators of cell growth or as regulators of prosurvival pathways such that their downregulation predisposes the heart to pathological remodeling. A major challenge for the future will be to identify the mRNA targets of the miRNAs that participate in cardiac remodeling and to understand the functions of their target mRNAs.
Requirement of miR-208 for stress-dependent cardiac growth and gene expression
Encoded within intron 27 of the gene encoding αMHC is miR-208 (5). Like αMHC, miR-208 is expressed specifically in the heart with trace expression in the lung (44). miR-208 is processed out of the αMHC pre-mRNA rather than being transcribed as a separate transcript (5). Intriguingly, however, miR-208 displays a remarkably long half-life of at least 14 days and can thereby exert functions even when αMHC mRNA expression has been downregulated. Although genetic deletion of miR-208 in mice failed to induce an overt phenotype, microarray analysis of hearts from wild-type and miR-208–/– animals at two months of age revealed loss of miR-208 to result in pronounced expression of numerous fast skeletal muscle contractile protein genes, which are normally not expressed in the heart. Thus, it is tempting to speculate that under normal conditions miR-208 is coexpressed with the sole cardiac-specific MHC gene to maintain the cardiomyocyte contractile phenotype by repressing the expression of skeletal muscle genes in the heart.
The most remarkable function of miR-208 was revealed by the aberrant response of miR-208–/– mice to cardiac stress (5). In response to pressure overload by thoracic aortic constriction or signaling by calcineurin (a calcium, calmodulin-dependent phosphatase that drives pathological remodeling of the heart), miR-208–/– mice showed virtually no hypertrophy of cardiomyocytes or fibrosis and were unable to upregulate βMHC expression (Figure 4). In contrast, other stress-responsive genes, such as those encoding atrial natriuretic peptide and B-type natriuretic peptide, were strongly induced in miR-208–/– animals, demonstrating that miR-208 is dedicated specifically to the control of βMHC expression, which can be uncoupled from other fetal cardiac and stress-responsive genes. Clearly, miR-208 is also essential for expression of the genes involved in cardiac fibrosis and hypertrophic growth (Figure 3).
Figure 4. Requirement of miR-208 for cardiomyocyte hypertrophy and fibrosis.
(A) Schematic diagram of a heart following thoracic aortic banding (TAB). (B) Sections of hearts of approximately 3-month-old wild-type and miR-208–/– mice are shown following sham operation or TAB for 21 days. High-magnification views of the ventricular wall are shown at the bottom. Trichrome staining identifies fibrosis in blue. Note that hypertrophy and fibrosis are diminished in miR208–/– mice compared with wild-type following TAB. Scale bars: 2 mm (top); 20 μm (bottom). Reproduced with permission from Science (5).
βMHC expression is repressed by thyroid hormone signaling and is upregulated in the hypothyroid state (52). miR-208–/– animals were also resistant to upregulation of βMHC expression following treatment with the thyroid T3 hormone inhibitor propylthiouracil, which induces hypothyroidism. Intriguingly, however, expression of βMHC before birth was normal in miR-208–/– mice, indicating that miR-208 is dedicated specifically to the postnatal regulation of βMHC expression, which coincides with the acquisition of thyroid hormone responsiveness of the gene encoding βMHC.
A clue to the mechanism of action of miR-208 comes from the resemblance of miR-208–/– hearts to hyperthyroid hearts, both of which display a block in βMHC expression, upregulation of stress-response genes, and protection against pathological hypertrophy and fibrosis (52). The upregulation of fast skeletal muscle genes in miR-208–/– hearts also mimics the induction of fast skeletal muscle fibers in the hyperthyroid state (53) and raises the question of whether a similar miRNA-mediated mechanism might govern fast-fiber gene expression in skeletal muscle.
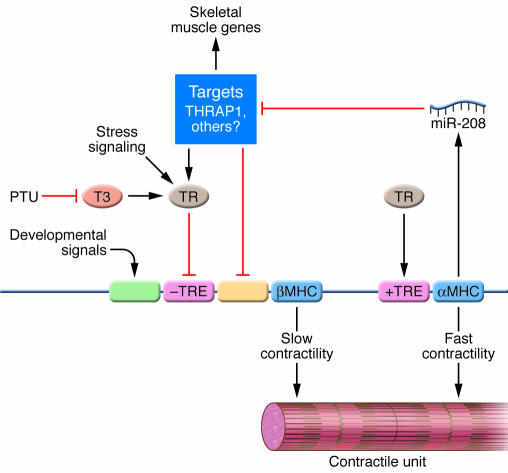
These findings suggest that miR-208 acts, at least in part, by repressing expression of a common component of stress-response and thyroid hormone–signaling pathways in the heart. Among the strongest predicted targets of miR-208 is the thyroid hormone receptor (TR) coregulator TR-associated protein 1 (THRAP1), which can exert positive and negative effects on transcription (54, 55). The TR acts through a negative TRE to repress βMHC expression in the adult heart (44). Thus, the increase in THRAP1 expression in the absence of miR-208 would be predicted to enhance the repressive activity of the TR toward βMHC expression, consistent with the blockade of βMHC expression in miR-208–/– hearts. However, although THRAP1 appears to be a bona fide target of miR-208, these data do not exclude the potential involvement of additional targets in the regulation of βMHC expression (Figure 5).
Figure 5. A model for the mechanism of action of miR-208.
αMHC and βMHC promote fast and slow contractility, respectively. TRs act through positive and negative TREs to activate and repress expression of the αMHC and βMHC genes, respectively, which are linked. Propylthiouracil (PTU) prevents T3 biosynthesis, resulting in hypothyroidism and upregulation of βMHC due to loss of the repressive action of the TR on the negative TRE. Stress signals also activate βMHC expression, at least in part through the TR. Developmental signals drive βMHC expression before birth through separate regulatory elements. miR-208, encoded by the αMHC gene, negatively regulates mRNA targets encoding THRAP1 and other negative regulators of βMHC expression. miR-208 also represses an activator of fast skeletal muscle genes. Modified with permission from Science (5).
Since even a subtle shift in the balance of αMHC versus βMHC expression toward βMHC reduces mechanical performance and efficiency of the adult heart (43), it might be of therapeutic value to exploit miR-208 regulation to prevent an increase in βMHC expression during cardiac disease. The cardiac specificity and importance of miR-208 to the cardiac stress response but not to normal cardiac development make miR-208 (and its downstream effectors) an attractive therapeutic target for manipulating βMHC levels.
Roles for miR-1 in cardiac development and contractility
miR-1 is a muscle-specific miRNA that forms a bicistronic cluster with miR-133 (56). The vertebrate genome contains 2 distinct loci for the miR-1/133 cluster with identical mature miRNA sequences, which is likely the result of an ancient genomic duplication. Cardiac expression of both miR-1-1 and miR-1-2 depend on the serum response factor transcription factor (12). While in vitro data suggest a possible role for miR-1 in myocyte hypertrophy (9), its primary function appears to be more related to cardiac development. Cardiac-specific overexpression of miR-1 in the embryonic heart inhibits cardiomyocyte proliferation and prevents expansion of the ventricular myocardium (12). These abnormalities were ascribed to the translational repression of the basic helix-loop-helix transcription factor Hand2, which is required for ventricular growth (57) although it seems likely that additional miR-1 mRNA targets contribute to the phenotype. On the other hand, targeted deletion of miR-1-2 in mice resulted in 50% lethality, largely due to ventricular-septal defects (4). A large proportion of the surviving mutant animals experienced electrophysiological defects and suffered sudden death, which was attributed to the upregulation of the mRNA encoding the transcription factor Irx5, a direct target of miR-1. Prior studies showed that Irx5 negatively regulates the Kv4.2 potassium channel and is thereby critical for maintaining the ventricular repolarization gradient (58). The surviving miR-1 mutant animals also displayed hyperplasia of the myocardium with persistent nuclear division after birth. The finding that loss of only one of the two miR-1 genes without any compensation by the remaining locus or changes in miR-133 expression induces such a severe spectrum of abnormalities underscores the importance of this specific miRNA and the sensitivity of the heart to relatively subtle changes in miRNA expression.
Expression of miR-1 has also been reported to be upregulated in humans with coronary artery disease (3). In order to examine the potential involvement of miR-1 in cardiac pathogenesis, in vivo gene transfer was employed to either enhance or inhibit miR-1 expression in the infarcted myocardium. While injection of miR-1 into the infarcted myocardium exacerbated arrhythmogenesis, miR-1–specific knockdown resulted in suppressed arrhythmias. These data imply that miR-1 is involved in electrical remodeling and arrhythmias, effects that were attributed to the translational repression of the KCNJ2 and GJA1 channels. KCNJ2 encodes Kir2.1, the main K+ channel subunit responsible for regulating the cardiac resting membrane potential (59), while GJA1 encodes connexin 43, the main cardiac gap junction channel responsible for intercellular conductance in the ventricle (60).
The apparent importance of miR-1 in inhibiting myocyte proliferation suggests that strategies that diminish miR-1 expression could be efficacious in cardiac regeneration. However, the finding that increasing or decreasing miR-1 expression causes lethal cardiac arrhythmias highlights not only the exquisite sensitivity of the heart to the levels of expression of this miRNA but also poses significant hurdles to the possible therapeutic manipulation of miR-1 levels in the settings of cardiac conduction abnormalities or myocardial repair.
Roles for miR-133 in cardiac growth and cardiac conduction
The miR-133 family consists of 2 homologous miR-133a members, designated 133a-1 and 133a-2, and a nearly homologous miR-133b. While the two 133a miRNAs are expressed as part of the same bicistronic unit with miR-1, miR-133b is expressed as a separate transcript (56). In several recent studies (7, 8, 10) of the dysregulation of miRNAs during cardiac hypertrophy, variable patterns of miR-133 expression were reported; sequence homology among the family members made it difficult to determine the relative expression of individual members of this family.
A recent report found miR-133 to be downregulated in human heart disease as well as in three models of cardiac hypertrophy (6). Interestingly, miR-133 was dysregulated during both physiological and pathological hypertrophy, suggesting it participates in a general hypertrophic program. Overexpression in vitro by infecting both neonatal and adult mouse myocytes with an adenoviral vector expressing miR-133 reduced the hypertrophic response to agonist stimulation. Conversely, in vivo knockdown in mice of miR-133 by infusion of an antisense RNA oligonucleotide appeared sufficient to induce significant hypertrophic growth of the heart with induction of fetal gene expression compared with saline-treated mice (6). These results suggest an active role for miR-133 in the inhibition of cardiac hypertrophy (Figure 3).
While these studies imply that strategies to upregulate miR-133 levels in vivo might serve as a therapy for preventing pathological cardiac growth, the recent demonstration of a correlation between miR-133 overexpression and abnormalities in cardiac electrical activity raises concerns about such a therapeutic approach. Xiao et al. (13) reported miR-133 to play a role in cardiac conductance abnormalities during diabetes by lowering the protein levels of ether-a-go-go–related gene (ERG), which encodes a key K+ channel (IKr) responsible for rapid delayed rectifier K+ current in cardiac cells. In a rabbit model of diabetes, cardiac expression of miR-133 was dramatically increased, coinciding with a pronounced depression of ERG protein, but not mRNA. Exogenous delivery of miR-133 in myocytes also resulted in downregulation of ERG protein, and an antisense inhibitor of miR-133 abrogated the repressive effect of miR-133 on ERG, indicating the specificity of miR-133 action on ERG expression (13).
Therapeutic opportunities and challenges
The obvious importance of miRNAs for cardiac function and dysfunction suggests opportunities for therapeutically exploiting the biology of miRNAs in the settings of congenital and acquired heart disease, especially for cardiac arrhythmias and pathological cardiac remodeling. In considering miRNAs as therapeutic targets, it is important to bear in mind that the rationale for therapeutically targeting miRNAs differs from classical approaches for drug development in which the goal is to develop drugs that are highly specific for a single target (generally a receptor or an enzyme). Because single miRNAs have many targets, modulating the expression or activity of one pathogenic or beneficial miRNA can, in principle, influence an entire gene network and thereby modify complex disease phenotypes. Of course, the broad and as yet poorly understood consequences of modulating miRNA function also pose significant challenges with respect to specificity and possible off-target effects.
The use of chemically modified oligonucleotides to target either a specific miRNA or to disrupt the binding between an miRNA and a specific mRNA target in vivo represents a potentially effective means of inactivating pathological miRNAs. An especially promising approach in this regard is the use of single-stranded RNA oligonucleotides, called antagomirs, which efficiently inactivate miRNAs through complementary base pairing (61, 62). Chemical modification and conjugation to cholesterol have been shown to stabilize and facilitate delivery of antagomirs to tissues following i.v. administration (61).
Another challenge to be overcome in the targeted inhibition of miRNAs comes from the fact that many miRNAs belong to families of closely related or even identical miRNAs, which may be expressed in multiple cell types. Thus, the consequences of targeting an miRNA in tissues other than the diseased tissue need to be considered. Because miRNAs have a multitude of mRNA targets, it may also be important to selectively inactivate the effects of an miRNA on specific targets, without unwanted effects on closely related miRNAs. One potential means of circumventing this lack of gene specificity would be to block miRNA actions in a gene-specific manner by specifically disrupting the binding between an miRNA and its designated binding site. Gene-specific miRNA mimics and miRNA-masking antisense approaches to perturb miRNA-mRNA interactions in vitro have recently been described although the applicability of these approaches still needs to be demonstrated in vivo (63). The possibilities of manipulating miRNAs therapeutically are not limited to the inhibition of pathogenic miRNAs. One can also imagine creating artificial miRNAs with salutary effects by promoting the expression of beneficial gene products (e.g., tumor suppressor proteins).
Many of the challenges to be faced in this field are similar to those faced in gene therapy, such as modes of delivery, specificity, toxicity, reversibility, and regulation. It will be important to design strategies for delivery of antagomirs or inactivating oligonucleotides to specific tissues, perhaps by combining them with tissue-specific homing signals. Methods for regulating and reversing the effects of such synthetic anti-miRNAs will also need to be developed, which is an issue of particular relevance given the stability of antagomirs in vivo.
The biology of miRNAs also promises to provide new insights into the cellular basis of disease. Gene profiling to identify mRNAs that are upregulated in response to the downregulation of a specific miRNA may allow the identification of unexpected regulators of disease processes and potentially interesting therapeutic targets. In this regard, it will also be important to determine whether miRNAs that respond to cellular stress act through cell type–specific or widely expressed targets to evoke their responses. The latter issue becomes important when considering strategies to therapeutically inactivate specific miRNAs in diseased tissues without affecting more general cellular functions. The distinctive signature patterns of miRNA expression associated with various disease states will also allow for new diagnostics.
Looking to the future
The heart, more than most other organs, is especially sensitive to relatively subtle changes in gene dosage and to gene modifier effects, particularly on developmental pathways. Thus, the ability of miRNAs to fine-tune gene expression programs portends their importance in many facets of cardiac biology. Given the vast number of miRNAs and the profound effects on the heart of the few that have been examined so far (Table 1), it is a virtual certainty that many new and unanticipated roles of miRNAs in the control of normal and abnormal cardiac function are awaiting discovery. Lessons learned from the heart will undoubtedly provide insights into important and as yet unappreciated roles of miRNAs in diverse aspects of development and disease.
Table 1 .
In vivo cardiac phenotypes due to changes in miRNA expression
Acknowledgments
Work in Eric N. Olson’s laboratory was supported by grants from the NIH and the Donald W. Reynolds Cardiovascular Clinical Research Center. E. van Rooij was supported by a National Scientist Development Grant by the American Heart Association.
Footnotes
Nonstandard abbreviations used: ERG, ether-a-go-go–related gene; MHC, myosin heavy chain; miR-195, miRNA-195; miRNA, microRNA; RISC, RNA-induced silencing complex; THRAP1, TR-associated protein 1; TR, thyroid hormone receptor; TRE, thyroid hormone response element.
Conflict of interest: The authors are cofounders of Miragen Therapeutics, a company focused on the development of microRNA-based therapies for heart failure and cardiac repair.
Citation for this article: J. Clin. Invest. 117:2369–2376 (2007). doi:10.1172/JCI33099.
References
- 1.Hoffman J.I. Incidence of congenital heart disease: II. Prenatal incidence. Pediatr. Cardiol. 1995;16:155–165. doi: 10.1007/BF00794186. [DOI] [PubMed] [Google Scholar]
- 2.Rosamond W., et al. Heart disease and stroke statistics--2007 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2007;115:e69–e171. doi: 10.1161/CIRCULATIONAHA.106.179918. [DOI] [PubMed] [Google Scholar]
- 3.Yang B., et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y., et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 5.van Rooij E., et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 6.Care A., et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 7.Cheng Y., et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am. J. Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Rooij E., et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayed D., Hong C., Chen I.Y., Lypowy J., Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ. Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 10.Tatsuguchi M., et al. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J. Mol. Cell. Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thum T., et al. MicroRNAs in the human heart. A clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y., Samal E., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. . Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 13.Xiao J., et al. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J. Biol. Chem. 2007;282:12363–12367. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- 14.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Reinhart B.J., et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 16.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 17.Lee C.T., Risom T., Strauss W.M. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 2007;26:209–218. doi: 10.1089/dna.2006.0545. [DOI] [PubMed] [Google Scholar]
- 18.Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 19.Marsit C.J., Eddy K., Kelsey K.T. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 20.Leung A.K., Calabrese J.M., Sharp P.A. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. . Proc. Natl. Acad. Sci. U. S. A. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond S.M. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Berezikov E., et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 23.Thai T.H., et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez A., et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 26.Gregory R.I., et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 27.Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 28.Bagga S., et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 29.Lim L.P., et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 30.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein E., et al. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 32.Wienholds E., Koudijs M.J., van Eeden F.J., Cuppen E., Plasterk R.H. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 33.Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harfe B.D., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanellopoulou C., et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cobb B.S., et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moses K.A., DeMayo F., Braun R.M., Reecy J.L., Schwartz R.J. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad F., Seidman J.G., Seidman C.E. The genetic basis for cardiac remodeling. Annu. Rev. Genomics Hum. Genet. 2005;6:185–216. doi: 10.1146/annurev.genom.6.080604.162132. [DOI] [PubMed] [Google Scholar]
- 39.Heineke J., Molkentin J.D. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 40.Brower G.L., et al. The relationship between myocardial extracellular matrix remodeling and ventricular function. Eur. J. Cardiothorac. Surg. 2006;30:604–610. doi: 10.1016/j.ejcts.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Berk B.C., Fujiwara K., Lehoux S. ECM remodeling in hypertensive heart disease. J. Clin. Invest. 2007;117:568–575. doi: 10.1172/JCI31044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinsey T.A., Olson E.N. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J. Clin. Invest. 2005;115:538–546. doi: 10.1172/JCI200524144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abraham W.T., et al. Coordinate changes in Myosin heavy chain isoform gene expression are selectively associated with alterations in dilated cardiomyopathy phenotype. Mol. Med. 2002;8:750–760. [PMC free article] [PubMed] [Google Scholar]
- 44.Morkin E. Control of cardiac myosin heavy chain gene expression. Microsc. Res. Tech. 2000;50:522–531. doi: 10.1002/1097-0029(20000915)50:6<522::AID-JEMT9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 45.Nakao K., Minobe W., Roden R., Bristow M.R., Leinwand L.A. Myosin heavy chain gene expression in human heart failure. J. Clin. Invest. 1997;100:2362–2370. doi: 10.1172/JCI119776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyata S., Minobe W., Bristow M.R., Leinwand L.A. Myosin heavy chain isoform expression in the failing and nonfailing human heart. Circ. Res. 2000;86:386–390. doi: 10.1161/01.res.86.4.386. [DOI] [PubMed] [Google Scholar]
- 47.Krenz M., Robbins J. Impact of beta-myosin heavy chain expression on cardiac function during stress. J. Am. Coll. Cardiol. 2004;44:2390–2397. doi: 10.1016/j.jacc.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 48.Herron T.J., McDonald K.S. Small amounts of alpha-myosin heavy chain isoform expression significantly increase power output of rat cardiac myocyte fragments. Circ. Res. 2002;90:1150–1152. doi: 10.1161/01.res.0000022879.57270.11. [DOI] [PubMed] [Google Scholar]
- 49.Si M.L., et al. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 50.Cheng A.M., Byrom M.W., Shelton J., Ford L.P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan J.A., Krichevsky A.M., Kosik K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 52.Klein I. Thyroid hormone and cardiac contractility. Am. J. Cardiol. 2003;91:1331–1332. doi: 10.1016/s0002-9149(03)00433-8. [DOI] [PubMed] [Google Scholar]
- 53.Vadaszova A., Zacharova G., Machacova K., Jirmanova I., Soukup T. Influence of thyroid status on the differentiation of slow and fast muscle phenotypes. Physiol. Res. 2004;53(Suppl. 1):S57–S61. [PubMed] [Google Scholar]
- 54.Park S.W., et al. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol. Cell. 2005;19:643–653. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 55.Pavri R., et al. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 56.Chen J.F., et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srivastava D., et al. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 58.Costantini D.L., et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–358. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diaz R.J., et al. Selective inhibition of inward rectifier K+ channels (Kir2.1 or Kir2.2) abolishes protection by ischemic preconditioning in rabbit ventricular cardiomyocytes. Circ. Res. 2004;95:325–332. doi: 10.1161/01.RES.0000137727.34938.35. [DOI] [PubMed] [Google Scholar]
- 60.Jongsma H.J., Wilders R. Gap junctions in cardiovascular disease. Circ. Res. 2000;86:1193–1197. doi: 10.1161/01.res.86.12.1193. [DOI] [PubMed] [Google Scholar]
- 61.Krutzfeldt J., et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krutzfeldt J., et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 63.Xiao J., et al. Novel approaches for gene-specific interference via manipulating actions of microRNAs: examination on the pacemaker channel genes HCN2 and HCN4. J. Cell. Physiol. 2007;212:285–292. doi: 10.1002/jcp.21062. [DOI] [PubMed] [Google Scholar]