Abstract
It was previously appreciated that the determination of skeletal muscle fiber type (fast or slow) could be regulated by class II histone deacetylases (HDACs), which function by inhibiting the transcription factor myocyte enhancer factor 2 (MEF2). In a report by Potthoff et al. in this issue of the JCI, it is further shown that HDACs are degraded via the ubiquitin/proteasome pathway, opening up a search for the putative E3 ligase that mediates the proteolysis of the responsible HDACs (see the related article beginning on page 2459). In a second report, by Suzuki et al., a new convergence between the biology of muscular dystrophy and muscle atrophy is elucidated (see the related study beginning on page 2468). It had previously been known that NO signaling is dysregulated during muscular dystrophy due to the disruption of the dystrophin glycoprotein complex (DGC), which anchors neuronal NOS (nNOS). Here it is shown that nNOS is similarly perturbed in a setting of skeletal muscle atrophy. Both of these studies suggest new avenues for the treatment of skeletal muscle disease.
Ubiquitination of class II HDACs determines slow– versus fast–muscle fiber type
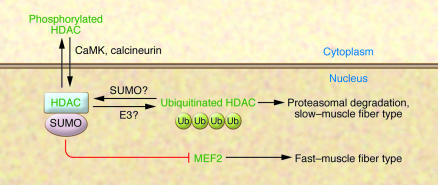
To the uninitiated, muscle may appear to be homogeneous, but in fact it is composed of distinct fiber types, referred to as slow and fast, defined by the myosin isotype expressed in the particular fiber (slow muscle expresses type I myosin; fast fibers can express types IIa, IIb, and IIx) and by the oxidative enzymes that are coexpressed in slow muscle. The variety in fiber type enables the animal to perform different types of work. It had previously been shown that fiber type could be perturbed by class II histone deacetylases (HDACs), which act by repressing the transcription factor myocyte enhancer factor 2 (MEF2), which in turn is required for the transcription of the oxidative genes found in slow fibers (Figure 1) (1). In a study published in this issue, Potthoff et al. (2) demonstrate that class II HDACs are regulated posttranscriptionally, apparently by the ubiquitin/proteasome pathway; despite being transcribed in soleus muscle, HDAC4 and HDAC5 protein levels were not observed there. However, when mice were treated with an inhibitor of the proteasome — the pharmacologic agent MG132 — an accumulation of HDACs in the soleus muscle was observed. Further, in these experiments on mice, ubiquitinated HDAC species could be isolated from the nuclear fraction of soleus muscle. Thus, in slow but not fast muscle fibers, there is a dearth of HDAC4 and -5 proteins, allowing MEF2 to activate the slow-muscle program (Figure 1).
Figure 1. Ubiquitination of class II HDACs causes their degradation via the proteasome, allowing MEF2 to induce a slow–muscle fiber phenotype.
One potential mechanism for regulation of HDAC sensitivity to ubiquitination is illustrated — SUMOylation. One model for MEF2 inactivation is that SUMOylated HDAC, which is associated with the SUMO E2, Ubc9, can associate with MEF2, leading to MEF2 SUMOylation and inactivation. In this issue of the JCI, Potthoff et al. (2) demonstrate that HDACs are degraded via ubiquitination in slow muscle fibers, resulting in active MEF2, which is necessary and sufficient to induce the expression of genes required for the slow–muscle fiber–type transcriptional program. Phosphorylation of the HDACs by CaMK (or dephosphorylation by calcineurin) does not seem, by itself, to perturb the sensitivity of HDAC to ubiquitination, but these proteins regulate HDAC localization to the nucleus; ubiquitinated HDAC proteins are found primarily in the nucleus, perhaps implicating a nuclear-localized E3 ubiquitin ligase. Ub, ubiquitin.
In the Potthoff et al. study, it was not possible to identify a particular class II HDAC as being necessary and sufficient to regulate MEF2 (2). Studies with knockout animals revealed that HDAC4, -5, and -9 were able to compensate for each other’s absence. It was only when double- and triple-knockout animals were employed that a clear fiber-type switch, from fast to slow, was observed (though an HDAC4, -5, -9 triple-knockout animal was not able to improve on the 80% slow-fiber composition observed in HDAC4, -5 and HDAC5, -9 double knockouts, perhaps indicating that in some fibers there may be additional mechanisms that are dominant over MEF2, keeping muscle fibers fast).
Speculation as to potential modulators of HDAC ubiquitination
How is HDAC turnover regulated in slow muscle? This is a classic chicken-and-egg problem, since one might at first presume that muscle fibers are homogenous until HDAC/MEF2-induced regulation is established. However, there must be some distinction that allows for fast– versus slow–muscle fiber patterning. There did seem to be some diminution in HDAC protein levels in soleus muscle from transgenic animals overexpressing either calcium/calmodulin-dependent protein kinase (CaMK) or calcineurin (2), and it has been established that CaMK phosphorylates HDACs, blocking their nuclear localization (3, 4). However, the majority of ubiquitinated protein was isolated from the nuclear fraction, where it is dephosphorylated. Also, in an in vitro ubiquitination assay, it was observed that the phosphorylation state of the HDACs did not affect their ability to be ubiquitinated (2). Therefore, the decrease in HDAC4 and -5 levels observed in the calcineurin and CaMK transgenic animals may not have been due to the ubiquitination mechanism that was the focus of the current study.
A more attractive potential mechanism for regulating the ubiquitination of class II HDACs is SUMOylation. Small ubiquitin-like modifier (SUMO) is a peptide that, in a manner similar to that of ubiquitin, can be conjugated on particular substrate lysines. However, unlike ubiquitination, SUMOylation does not lead to protein degradation — rather, it has been linked to changes in protein function, including perturbations in transcription factor activity (5). Class II HDACs and MEF2 itself have been shown to be SUMOylated, which enhances the repressive activity of the HDACs and decreases the activity of MEF2 (6, 7). If the SUMOylation of HDAC proteins protects them from being ubiquitinated, that would create a feed-forward mechanism, allowing them to then bind and repress MEF2. Whether SUMOylation is the mechanism for controlling HDAC ubiquitination has yet to be determined. What will also help to clarify the actual underlying mechanism is the identification of the E3 ubiquitin ligase responsible for class II HDAC turnover in the soleus and an understanding of whether the ligase is regulated by some additional posttranslational modification of the HDACs, MEF2, or itself. One would think that the E3 ligase cannot itself be restricted in expression via the MEF2 mechanism shown here since its activity is necessary before the slow/fast–muscle fiber–type delineation is determined and since the Potthoff et al. study (2) shows that MEF2 activity is not only necessary, but sufficient, to make muscle fibers slow; however, such deductive reasoning can only be confirmed or denied after the responsible ligase is identified. The clear implication though is that there are additional patterning mechanisms upstream of MEF2 activity.
Dysregulated nNOS induces muscle atrophy
Cachexias are wasting syndromes that feature a decrease in lean body mass due to skeletal muscle atrophy. The loss of skeletal muscle mass causes muscle weakness and frailty. In severe cachexias, a myopathy may be observed, which can compromise the diaphragm muscle (8).
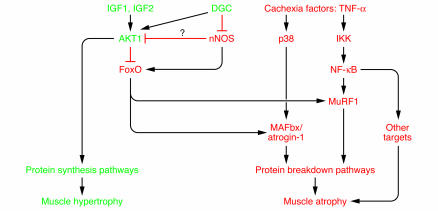
Loss of function in the diaphragm is also the eventual cause of death in a series of conditions that had until recently been thought to be distinct from atrophy syndromes: muscular dystrophies. Several of the dystrophies are caused by mutations in distinct components of the dystrophin glycoprotein complex (DGC), which helps to anchor the muscle cytoskeleton to the cell membrane via dystrophin and its binding partners. In dystrophic settings, there is an obvious loss of structural patency of proteins that were previously thought to be purely structural (9). However, a paper published recently in Cancer Cell demonstrated a convergence of atrophy and dystrophy signaling (10). In that study, it was shown that dystrophin was lost from the cell membrane under atrophy conditions, causing a loss of continuity between the cell membrane of the muscle fiber and the extracellular matrix. This resulted in a decrease in Akt signaling, which is required both to induce an increase in muscle fiber size, via protein synthesis, and to block muscle protein turnover (11); Akt accomplishes this last task in part by phosphorylating the forkhead box O (FoxO) family of transcription factors, thus keeping them out of the nucleus (12, 13). Transgenic overexpression of FoxO1 in skeletal muscle is sufficient to cause muscle atrophy (14). FoxO translocation to the nucleus mediates muscle atrophy in part by increasing the expression of the E3 ubiquitin ligases muscle-specific RING finger protein 1 (MuRF-1) and muscle atrophy F-box protein (MAFbx; also known as atrogin-1) (12, 13). These E3 ligases, MuRF-1 and MAFbx, are required for a significant portion of the protein turnover that is distinct to the atrophy condition since approximately a third of the muscle mass normally lost during atrophy is spared when either ligase is deleted, as in the production of MuRF-1– or MAFbx-null animals (15).
In the study by Suzuki et al. (16), also published in this issue of the JCI, a second convergence between dystrophy and atrophy signaling is identified. It was previously shown that neuronal NOS (nNOS), normally bound to the DGC, is dysregulated during muscular dystrophy, meaning that its cellular localization is disturbed and it is no longer apparent at the sarcolemma (17, 18). Further, it was also previously demonstrated that maintenance of NO could ameliorate dystrophy symptoms (19, 20). Suzuki et al. (16) now report that nNOS is similarly dysregulated during atrophy conditions. When it is untethered from the DGC, upon unweighting of the skeletal muscle in an atrophy model called hind-limb suspension, the nNOS is free to enter the rest of the cellular compartment, where it is required to potentiate FoxO-mediated transcription and thus upregulate expression of the E3 ligases MAFbx and MuRF-1 (Figure 2). This requirement was demonstrated using nNOS–/– animals; in the nNOS–/– animals, hind-limb suspension surprisingly did not cause the upregulation of MuRF-1 and MAFbx as observed in wild-type animals.
Figure 2. Dysregulation of nNOS from the sarcolemma: a new overlap in dystrophy/atrophy signaling.
In the setting of muscle atrophy, nNOS activates FoxO, which is required for upregulation of the E3 ligases MuRF-1 and MAFbx. These E3s in part mediate the increase in protein turnover seen during muscle atrophy. IGF1 was previously shown to be able to block MuRF-1 and MAFbx upregulation by activating the Akt pathway. Akt phosphorylates the FoxO transcription factors, keeping them out of the nucleus. nNOS might act by inhibiting Akt signaling, thus allowing FoxO transcription factors to traffic to the nucleus. Alternatively, nNOS could act directly on FoxO proteins or via an undiscovered mechanism to activate the FoxO-induced muscle atrophy program. In this issue of the JCI, Suzuki et al. (16) demonstrate that nNOS is dysregulated, departing from DGC and resulting in the activation of FoxO transcription. IKK, inducible IκB kinase.
How does nNOS perturb FoxO-mediated transcription? A prior study reported that nitrosylation could inhibit the activity of Akt (21). Were such a thing to occur in the present setting, the inhibition of Akt would result in the observed nuclear localization and activation of FoxO. No perturbation in Akt phosphorylation was observed, and the measurements of downstream Akt signaling — phosphorylation of S6 kinase 1 and mammalian target of rapamycin (mTOR) — did not seem to be dramatically decreased (16). However, in this study, the phosphorylation differences were too subtle to make any definitive judgements. Also, Akt is inactivated by nitrosylation without its phosphorylation state being altered (21). Thus, Akt inactivation still represents a potential mechanism for the effects observed. Also, it is possible that direct nitrosylation of FoxO transcription factors helps increase their activity and may help to block their phosphorylation. Of course, still other mechanisms are possible, including alternative ways to phosphorylate or activate FoxO proteins.
Prospects for new treatments of muscle disease
Both the Potthoff et al. (2) and Suzuki et al. (16) studies suggest new avenues for the treatment of muscle weakness and atrophy. If a fast– to slow–muscle fiber switch is deemed desirable — and it would be, since slow muscle is relatively resistant to muscle atrophy — then a class II HDAC inhibitor could effect such a change. Also, HDAC inhibitors have been shown to be beneficial in the setting of muscular dystrophy (20, 22). The Suzuki et al. study opens up a new avenue for the treatment of muscle atrophy; for example, if nNOS could be stabilized on the sarcolemma under disease conditions by perturbing the mechanism for its release, then it would not be free to activate FoxO. Of course, this strategy depends on identifying the untethering mechanism. More immediately, a strategy to inhibit nNOS activity in skeletal muscle may be beneficial, as has been the case in muscular dystrophy.
As in all good science, the current reports (2, 16) not only offer exciting new answers, in this case as to how muscle homeostasis is maintained; they also help to identify important new questions, setting the basis for future study.
Footnotes
Nonstandard abbreviations used: CaMK, calcium/calmodulin-dependent protein kinase; DGC, dystrophin glycoprotein complex; FoxO, forkhead box O; HDAC, histone deacetylase; MAFbx, muscle atrophy F-box protein; MEF2, myocyte enhancer factor 2; MuRF-1, muscle-specific RING finger protein 1; nNOS, neuronal NOS; SUMO, small ubiquitin-like modifier.
Conflict of interest: D. J. Glass is an employee of Novartis Institutes for BioMedical Research.
Citation for this article: J. Clin. Invest. 117:2388–2391 (2007). doi:10.1172/JCI33379.
See the related article beginning on page 2468.
References
- 1.McKinsey T.A., Zhang C.L., Olson E.N. Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 2002;14:763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki N., et al. NO production results in suspension-induced muscle atrophy through dislocation of neuronal NOS. J. Clin. Invest. . 2007;117:2468–2476. doi: 10.1172/JCI30654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinsey T.A., Zhang C.L., Lu J., Olson E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. . Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y., Randall W.R., Schneider M.F. Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. J. Cell Biol. 2005;168:887–897. doi: 10.1083/jcb.200408128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirkin V., Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr. Opin. Cell Biol. . 2007;19:199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Zhao X., Sternsdorf T., Bolger T.A., Evans R.M., Yao T.-P. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol. Cell. Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregoire S., Yang X.-J. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell. Biol. 2005;25:2273–2287. doi: 10.1128/MCB.25.6.2273-2287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanson P., et al. Acute corticosteroid myopathy in intensive care patients. Muscle Nerve. 1997;20:1371–1380. doi: 10.1002/(sici)1097-4598(199711)20:11<1371::aid-mus4>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 9.Glass D.J. A signaling role for dystrophin: inhibiting skeletal muscle atrophy pathways. Cancer Cell. 2005;8:351–352. doi: 10.1016/j.ccr.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 10.Acharyya S., et al. Dystrophin glycoprotein complex dysfunction: a regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell. 2005;8:421–432. doi: 10.1016/j.ccr.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Glass D.J. Skeletal muscle hypertrophy and atrophy signaling pathways. Int. J. Biochem. Cell Biol. 2005;37:1974–1984. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 12.Sandri M., et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stitt T.N., et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 14.Kamei Y., et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. . J. Biol. Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 15.Bodine S.C., et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 16.Potthoff M.J., et al. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. . 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grozdanovic Z., Gosztonyi G., Gossrau R. Nitric oxide synthase I (NOS-I) is deficient in the sarcolemma of striated muscle fibers in patients with Duchenne muscular dystrophy, suggesting an association with dystrophin. Acta Histochem. 1996;98:61–69. doi: 10.1016/S0065-1281(96)80051-1. [DOI] [PubMed] [Google Scholar]
- 18.Crosbie R.H., Barresi R., Campbell K.P. Loss of sarcolemma nNOS in sarcoglycan-deficient muscle. FASEB J. . 2002;16:1786–1791. doi: 10.1096/fj.02-0519com. [DOI] [PubMed] [Google Scholar]
- 19.Wehling M., Spencer M.J., Tidball J.G. A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J. Cell Biol. . 2001;155:123–132. doi: 10.1083/jcb.200105110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Archer J.D., Vargas C.C., Anderson J.E. Persistent and improved functional gain in mdx dystrophic mice after treatment with L-arginine and deflazacort. FASEB J. . 2006;20:738–740. doi: 10.1096/fj.05-4821fje. [DOI] [PubMed] [Google Scholar]
- 21.Yasukawa T., et al. S-nitrosylation-dependent inactivation of Akt/protein kinase B in insulin resistance. J. Biol. Chem. 2005;280:7511–7518. doi: 10.1074/jbc.M411871200. [DOI] [PubMed] [Google Scholar]
- 22.Anderson I. Deacetylase inhibitors aid recovery in muscular dystrophy. Lancet Neurol. 2006;5:906. doi: 10.1016/s1474-4422(06)70593-2. [DOI] [PubMed] [Google Scholar]