Abstract
We investigated expression levels of Nicotinamide N-Methyltransferase (NNMT), an enzyme involved in the biotransformation of many drugs and xenobiotic compounds, in oral squamous cell carcinoma (OSCC). Measurements were performed by semi-quantitative RT-PCR and quantitative real-time PCR in tumor and matched adjacent healthy tissue. Interestingly, NNMT was up-regulated in most of the favorable OSCCs, while no marked NNMT expression alterations between tumor and normal mucosa were detected in most of the unfavorable OSCCs. Western blot and immunohistochemical analyses also were performed and the relationship between tumor characteristics and NNMT levels in OSCC were studied to evaluate the effectiveness of NNMT as a prognostic marker in the squamous cell carcinoma of the oral cavity. In summary, the present study suggests that NNMT may have potential as a biomarker and a therapeutic target for OSCC.
INTRODUCTION
Squamous cell carcinoma is the most common cancer type of the oral cavity, representing 90 percent of all oral cancers. Treatment of oral squamous cell carcinoma (OSCC) usually is based on surgery or radiation treatment, with or without concomitant chemotherapy.
Despite the advances in surgical techniques and adjuvant therapies, the mortality rate of OSCC has shown little improvement over the last three decades, the overall five-year survival of these patients remains less than 50 percent, and the diagnostic delay seems to be responsible for the poor prognosis of patients with OSCC (1).
Because traditional clinical methods lack sensitivity to detect OSCC early and are insufficient to predict the aggressiveness of tumors and disease outcome, integrated diagnostic and prognostic systems should be developed, combining clinical-pathological features and new molecular markers defined by expression profiling. In fact, recent studies have shown that each case possesses its own biological characteristics, clinical behavior, and, by extension, unique sensitivity to therapy. Thus, gene expression analysis appears to be a powerful tool for studying OSCC pathogenesis, progression, and metastatic potential (2).
Currently, several genes relating to tumor proliferation, growth, angiogenesis, and loss of cell adhesion are being evaluated for their potential as prognostic factors and a great deal of attention has been devoted to the involvement of the apoptotic pathway in OSCC tumorigenesis (3–5). Moreover, the diagnostic potential of circulating RNA for oral cancers has been described recently (6).
We wished to focus on the expression of genes critical in the drug metabolism process, namely on Nicotinamide N-Methyltransferase (NNMT), an enzyme belonging to Phase II Metabolizing Enzymes and involved in the biotransformation and detoxification of many xenobiotics (7–9).
In fact, the metabolism of drugs, toxic chemicals, hormones, and micronutrients is an important topic in the fields of pharmacology and endocrinology, and it is often implicated in many diseases and pathophysiological processes, such as cancer and resistance to chemotherapy (10).
To explore the involvement of NNMT in OSCC, in the present study we analyzed the enzyme expression in paired tumor (T) and non-tumor (NT) tissues obtained at surgery by Semiquantitative RT-PCR, Real-Time PCR, and Western blot analysis.
Immunohistochemical analyses also were performed and the relationship between tumor clinicopathological findings and the level of NNMT tumor expression was analyzed to evaluate the effectiveness of NNMT as a prognostic marker in the oral squamous cell carcinoma.
MATERIALS AND METHODS
Patients and Surgical Specimens
A total of 22 patients who underwent surgical treatment for OSCC with curative intention at our institution between April 2002 and May 2005 were analyzed in this study. Patients included 12 men and 10 women, 48 to 91 years old (mean age 68 years).
Tumors were classified according to U.I.C.C. 2000 classification (UICC 2000). Lymph nodes metastases were observed in nine cases (N+, unfavorable OSCCs), while the remaining 13 did not show any evidence of regional lymph nodes infiltration (N0, favorable OSCCs). Table 1 lists the clinicopathological findings of the 22 patients.
Table 1.
Patient and clinicopathological findings
| Cases | 22 |
| Age, years | |
| Mean | 68 |
| Range | 48–91 |
| Sex, male/female | 12/10 |
| Tumor size, cm | |
| Mean | 2.5 |
| Range | 0.7–5 |
| Grading | |
| G1 | 3 |
| G2 | 13 |
| G3 | 6 |
| Staging | |
| I | 6 |
| II | 5 |
| III | 3 |
| IV | 8 |
Fresh oral tissue was obtained at surgery. Tumor material and corresponding normal mucosa were collected, snap frozen in liquid nitrogen, and stored at −80°C prior to use. All the samples were collected as per the institutional committee guidelines.
RNA Extraction
An aliquot of the frozen tissue (20–40 mg) was homogenized in a lysis buffer, and total RNA was then extracted through the SV Total RNA Isolation System (Promega, Madison, WI, USA). RNA samples were tested by ultraviolet absorption at 260 nm to determine the concentration. The quality and concentration of the RNA samples were further confirmed by electrophoresis on denaturated one percent agarose gels.
Semiquantitative RT-PCR
One microgram of RNA was reverse-transcribed in a total volume of 20 μL for 60 min at 37°C with the First Strand cDNA Synthesis Kit II (Bio Basic Inc, Markham Ontario, Canada) using random nonamers. Of the reaction mixture, 0.5 μL was subjected to PCR in a total of 25 μL with Taq Polymerase and buffers purchased from GeneCraft (Ludinghausen, Germany). Gene-specific PCR primers for human NNMT and β-actin were designed, based on the DNA sequences in the National Center for Biotechnology Information databases. The sequences of these primers were: 5′-TGGCTTCTGGAGGAAAGAGA-3′ (forward) and 5′-AATCAGCAGGTCTC CCTTCA-3′ (reverse) for NNMT and 5′-CCAACCGCGAGAAGATGAC-3′ and 5′-GAGGCGTACAGGGATAGCACA-3′ for β-actin. Optimal PCR amplification conditions were determined, and PCR products were electrophoresed in two percent agarose gel. The gel was stained with ethidium bromide to visualize DNA through an ultraviolet illuminator. The intensities of the gene-specific PCR products were evaluated from images obtained using Quantity One 1-D Analysis Software (Bio-Rad, Hercules, CA, USA).
Real-time Quantitative PCR
To examine NNMT gene expression more quantitatively, we performed a real-time PCR assay using Rotor-Gene 6000 Real-time rotary analyzer (Corbett Life Science, Cambridge, UK) The cDNA, generated for semiquantitative RT-PCR, was used for real-time quantitative PCR. To avoid false-positive results attributable to the amplification of contaminating genomic DNA in the cDNA preparation, all primers were selected to flank an intron, and PCR efficiencies were tested for both primer pairs and found to be close to one. The following primers were used, 5′-ATTACAAGTTTGGTTCTAGGCACT-3′ (forward) and 5′-GGCCAGAGCCGAT GTCAAT-3′ (reverse) for NNMT, and 5′-CCAACCGCGAGAAGATGAC-3′ (forward) and 5′-GAGGCGTACAGGGATAGCACA-3′ (reverse) for β-actin.
Both genes were run in duplicates (45 cycles of 94°C for 15 seconds and 54°C for 30 seconds) using SYBRGreen chemistry. All samples were tested in triplicate with the reference gene β-actin for normalization of data to correct for variations in RNA quality and quantity. Direct detection of PCR products was monitored by measuring the fluorescence produced by SYBR Green I dye binding to double-stranded DNA after every cycle. These measurements were then plotted against cycle numbers. The parameter Ct (threshold cycle) was defined as the cycle number at which the first detectable increase above the threshold in fluorescence was observed. Fold changes in relative gene expression were calculated by 2−Δ(ΔCt) where ΔCt = Ct (NNMT) − Ct (β-actin) and Δ(ΔCt) = ΔCt (tumor tissue) − ΔCt (normal tissue).
Western Blot Analysis
Oral tissue extracts were prepared with lysis buffer (Phosphate-Buffered Saline (PBS), containing one percent Nonidet P40, 0.1 percent SDS, 1 mM phenylmethylsulfonylfluoride, 2 μg/mL aprotinin). Samples containing 50 μg protein were subjected to 15 percent sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes. After regular blocking and washing, the membranes were incubated with rabbit polyclonal antibody (1:1000 dilution) against NNMT (Richard Weinshilboum, Mayo Clinic, Rochester, MN, USA) for two hours followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit IgG (Pierce, Rockford, IL USA; 1:2000 dilution) for two hours. NNMT protein was visualized using an enhanced chemiluminescent substrate (SuperSignal West Femto Maximum Sensitivity Substrate, Pierce).
Immunohistochemical Analyses
Serial sections (4-μm) from formalin-fixed, paraffin-embedded blocks of tumor representative areas were cut for each case. Only sections containing sufficient neoplastic and normal epithelium to assess the antibody reactivity with 1000 cells were considered for this study.
Immunohistochemistry was then performed on the remaining sections mounted on poly-L-lysine-coated glass slides. Deparaffined and rehydrated sections were incubated for 30 min in three percent H2O2/methanol to quench endogenous peroxidase activity, and then rinsed for 20 min with phosphate-buffered saline (Bio-Optica M107, Milan, Italy). Non-specific protein binding was attenuated by incubation for 30 min with five percent horse serum in PBS. Specimens were incubated overnight with polyclonal rabbit antibody anti-human NNMT (Richard Weinshilboum, Mayo Clinic) at a dilution of 1:500. The specificity of this antibody has been described in the literature (11).
The antibody was applied directly to the section and the slides were incubated overnight (4° C) in a “humidified chamber.” The sections were washed three times with PBS at room temperature. Immune complexes were subsequently treated with the secondary byotinilated antibody and then detected by streptavidin peroxidase, both incubated for 30 min at room temperature (Vectastain ABC kit, Vector Laboratories, Burlingame, CA, USA). After rinsing with three changes of PBS, the immunoreactivity was visualized by development for two minutes with 0.1 percent 3.3′-diaminobenzidine and 0.02 percent hydrogen peroxide (DAB substrate kit, Vector Laboratories Burlingame, CA, USA). Sections were counterstained with Mayer’s haematoxylin, mounted with permanent mounting medium and examined by light microscopy.
Positive controls consisted of tissue specimen sections of human kidney tumor with known antigenic reactivity. A negative control was performed in all cases by substituting the primary antibody for normal mouse serum. Negative controls in all instances resulted in a negative immunoreactivity for NNMT.
Statistical Analysis
Data were analyzed using GraphPad Prism software version 4.00 for Windows (GraphPad Software, San Diego, CA, USA). Significant differences (P < 0.05) between groups were determined using one-way analysis of variance and the Mean-Whitney test. Correlation between variables was assessed by Spearman test. Survival curves were analyzed according to the method of Kaplan-Meier and for differences between curves the P value was calculated by the log-rank test. A P value of less than 0.05 was accepted as statistically significant.
RESULTS
Semiquantitative RT-PCR and Real-time Quantitative PCR
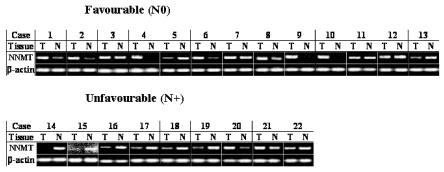
We first examined NNMT mRNA expression in paired tumor (T) and non-tumor (NT) surgical specimens of 22 patients who underwent surgery for OSCC by semiquantitative RT-PCR, as described under Materials. A transcript of 238 bp corresponding to NNMT mRNA was detected in all samples as expected (Figure 1). Correct identification of NNMT band was first verified by DNA cloning and sequencing (results not shown). Amplified fragments from tumor and non-tumor tissues were compared after being normalized by the intensity of β-actin gene, used as internal control. Compared with normal mucosa, OSCC exhibited significantly increased expression of NNMT in 11 of 22 (50 percent) examined patients (see Figure 1). Interestingly, NNMT was upregulated in most of the favorable OSCCs (N0), while no marked NNMT expression alterations between tumor and normal mucosa were detected in most of the unfavorable OSCCs (N+).
Figure 1.
Semiquantitative RT-PCR. NNMT expression was analyzed in set of paired tumor (T) and non tumor (N) tissues from favorable (N0, cases 1 to 13 ) and unfavorable (N+, cases 14 to 22) OSCCs. β-actin served as internal control.
Real-time quantitative PCR was used to further evaluate NNMT expression in OSCC using the same cohort of 22 patients. Differential gene expression measurements (tumor versus normal tissue) confirmed the upregulation (50 percent of subjects, nine with favorable OSCC and two with unfavorable) in the tumor tissue with fold change values ranging between 1.1 and 38 (Table 2). Mean NNMT expression was 7.5-fold (median 3.8-fold) higher in cancerous specimen with respect to the accompanying normal mucosa. In the 11 patients with higher NNMT expression in the cancerous part, overexpression was up to threefold in five, between three- and ten-fold in four, and more than ten-fold in two patients.
Table 2.
NNMT expression levels in OSCCs in relation to lymph node metastases
| Case | Lymph Node | Tumor/Normal Tissue (-fold)a |
|---|---|---|
| 1 | N0 | 5.4 |
| 2 | N0 | 1.4 |
| 3 | N0 | 1.14 |
| 4 | N0 | 6.8 |
| 5 | N0 | 0.69 |
| 6 | N0 | 3.8 |
| 7 | N0 | 1.1 |
| 8 | N0 | 3.8 |
| 9 | N0 | 36 |
| 10 | N0 | 18 |
| 11 | N0 | 0.36 |
| 12 | N0 | 0.18 |
| 13 | N0 | 0.33 |
| 14 | N2b | 0.27 |
| 15 | N2b | 0.8 |
| 16 | N1 | 0.16 |
| 17 | N2c | 0.24 |
| 18 | N2b | 0.63 |
| 19 | N2b | 0.33 |
| 20 | N1 | 2.7 |
| 21 | N2b | 2.2 |
| 22 | N1 | 0.4 |
Differential NNMT expression measurements were performed by Real-Time PCR as described under Materials and Methods.
In the remaining 11 subjects (4 with favorable OSCC and 7 with unfavorable OSCC) NNMT mRNA expression did not increase in the tumor, even appearing slightly downregulated.
NNMT Gene Expression and Clinicopathological Parameters
We analyzed the relationship between clinicopathological parameters and the level of NNMT tumor expression, compared with the accompanying normal mucosa, as determined by real-time quantitative PCR analyses. The clinicopathological parameters investigated in this study were age, gender, tumor size (pT), histological grading, nodal metastasis, and pathological stage.
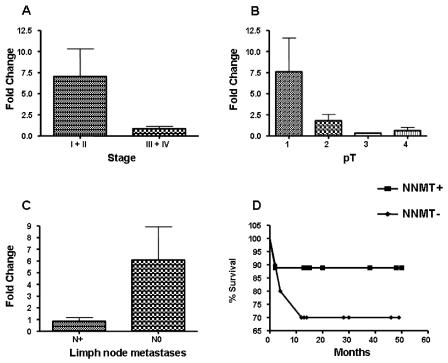
No statistical correlation was found between NNMT expression levels and age, gender, and histological grading of OSCC cases examined. Both pT and pathological staging showed an inverse correlation with NNMT mRNA levels, suggesting the possibility of this enzyme as a prognostic factor (Figure 2A,B). These correlations were statistically significant (pT, Rho = −0.480, P = 0.0238; stage, Rho = −0.5324, P = 0.0107 ). Mann Whitney test confirmed the inverse correlation between NNMT expression and stage (stage I + II versus III + IV; P = 0.0138).
Figure 2.
NNMT expression and clinicopathological parameters. The Mean-Whitney test was performed to evaluate any correlation between NNMT expression levels and stage (panel A), pT (panel B), and lymph node metastases (panel C). Panel D shows Kaplan-Meier survival curves of patients with OSCC (black square, patients bearing tumours with high NNMT expression levels; black diamond, patients bearing tumours with low NNMT expression levels).
Interestingly, NNMT mRNA levels inversely correlated with lymph node metastasis. A significant negative association of the amount of NNMT expressed by tumor tissue compared with the adjacent normal mucosa was in fact found with metastasis (P = 0.0417). The low NNMT expression detected in patients with metastasis supports the hypothesis that NNMT plays a role in tumor expansion, and tumors that downregulate it may be able to evade immunosurveillance and grow (Figure 2C).
Moreover, we addressed the question of whether the extent of NNMT expression is associated with patient prognosis. To determine associations, patients were stratified into two groups, according to the level of NNMT intratumor expression, compared with the adjacent normal mucosa. The survival rate was calculated using the Kaplan-Meier method and compared between the two groups. The curves revealed an improved overall survival rate for patients bearing tumors with NNMT expression levels higher than adjacent normal tissue, although without reaching statistical significance. These results seem to indicate a favorable prognosis in patients with tumors expressing high NNMT, whereas the absence of marked NNMT expression seems to constitute a hallmark of aggressive biological behavior (Figure 2D).
Western Blot Analysis
To confirm the above-reported results, NNMT expression was detected at protein level by Western blot analysis. The analysis showed a single immunoreactive band at approximately 30 kDa, which corresponds to the known molecular mass of NNMT. Consistent with the results of real-time PCR analyses, lanes loaded with equal protein amounts showed a markedly increased NNMT expression in tumor samples compared with the paired normal tissues in most of the favorable OSCC (N0) (Figure 3). Conversely, a marked NNMT downregulation was evident in two unfavorable cases (N+) (see Figure 3), while the remaining specimens tested (N+) showed a faint detectable band in both normal and cancerous tissue (not shown).
Figure 3.
Western blot analysis of NNMT. Tumor (T) and matched adjacent healthy (N) tissue lysates were prepared as described in Materials and Methods; 50 μg of protein from 15 percent SDS-PAGE were transferred to polyvinylidene fluoride membranes. Blot was probed with anti-NNMT antibody, and detected with enhanced chemiluminescence. (cases 1–11, favorable OSCCs; cases18 and 21, unfavorable OSCCs).
Immunohistochemical Analyses
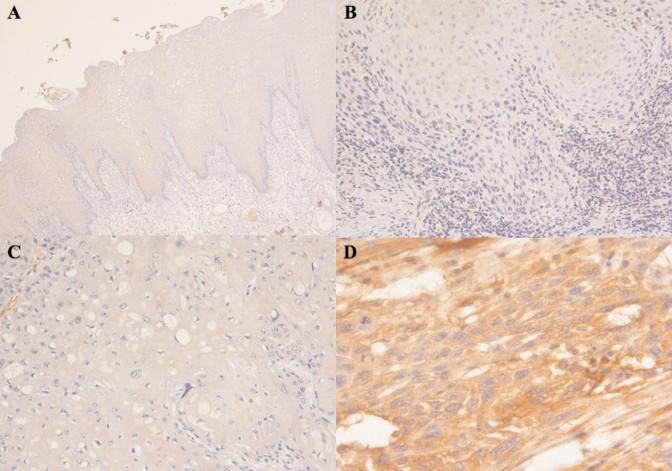
Next, we examined NNMT expression in OSCC tissues and their corresponding non-cancerous mucosa by immunohistochemistry with the same anti-NNMT antibody used in the Western blot analysis. Consistent with the results reported above, the normal mucosa displayed absent or very weak cytoplasmic NNMT immunoreactivity, with occasionally moderate nuclear staining. NNMT immunohistochemical staining of most of the favorable OSCCs was very strong and uniform in the cytoplasm, while the reaction was pale in the tumors with lymph node metastasis. A representative result for each group is shown in Figure 4.
Figure 4.
Immunohistochemistry. A, Normal oral mucosa displayed absent or very weak cytoplasmic NNMT immunoreactivity in all epithelial layers. (×100); B, Moderately differentiated squamous cell carcinoma with lymph nodes metastasis. Prevalently negativity of the nests of the tumor cells. (×160); C, Poorly differentiated squamous cell carcinoma with lymph nodes metastasis. Complete negativity of the tumor cells. (×400); D, Moderately differentiated squamous cell carcinoma with lymph nodes negative. Prevalently citoplasmatic positivity of the tumor cells. (×200).
Finally, in agreement with real-time PCR results, patients with absent or low NNMT immunoreactive tumors manifested poorer overall survival rates in comparison with patients with moderate or high NNMT immunoreactive tumors, but the difference was not statistically significant (not shown).
DISCUSSION
The survival of patients with oral squamous cell carcinoma remains unaffected despite recent therapeutic advances.
In particular, the presence of lymph node metastases seems to be the most important prognostic factor in oral squamous cell carcinoma, and it is generally accepted that survival is poorer in patients with lymph node metastases than in those without. Therefore, the optimal management of cervical lymph node metastases is very important to improve survival, and the identification of reliable and clinically applicable markers, which allow their preoperative detection is crucial. In fact, the detection of predictive markers of cervical lymph node metastasis in the primary tumor would be helpful in deciding the optimal treatment for Stage I and II OSCCs, and in achieving a high survival rate (12).
In the present study we investigated the expression of the enzyme Nicotinamide-N-Methyltransferase in paired tumor and non-tumor tissues obtained from 22 patients with OSCC.
Compared with normal mucosa, we found a significantly increased NNMT expression in 11 of 22 (50 percent) examined OSCCs and, interestingly, this up-regulation was detected in most of the favorable cases, while no marked NNMT expression alterations between tumor and normal mucosa were detected in most of the metastatic tumors.
NNMT catalyses the N-methylation of nicotinamide, pyridines and other structural analogs, playing a pivotal role in the biotransformation and detoxification of many xenobiotics (7,8). In fact, N-methylation is one method by which drug and other xenobiotic compounds are metabolized by the liver and the enzyme NNMT is responsible for this activity which uses S-adenosyl methionine as the methyl donor. In the liver, where the enzyme is predominantly expressed, NNMT activity has a bimodal frequency distribution, an observation that raises the possibility that this enzyme activity may be regulated by a genetic polymorphism. Such a polymorphism could have functional implications for individual differences in metabolism and the therapeutic effect of drugs (8). However, its function also could lead to the production of toxic products, as shown in Parkinson’s disease, where enhanced NNMT activity seems to produce toxic N-methylpiridinium compounds, which have been advanced as possible neurotoxins, underlying nigrostriatal degeneration (13).
As reported above, NNMT is highly expressed in the liver, while a low expression has been detected in the kidney, lung, skeletal muscle, placenta, heart, and brain. In tumors, abnormal expression of NNMT has been reported in glioblastoma, stomach adenocarcinoma (14,15,16), papillary thyroid cancers (11,17), colon (18), and renal carcinoma (19,20), where its upregulation appears to be inversely related to tumor size, thus suggesting that the enzyme may be significant in an initial step of malignant conversion. In addition, RNA analysis of pancreatic juices from patients with pancreatic cancer contains increased levels of NNMT mRNA with respect to the non-cancer controls (21). However, the function of NNMT in cancer cell metabolism is yet unclear (11,17).
As already mentioned, the enzyme catalyzes the formation of N1-methylnicotinamide, thus being involved in the major route of nicotinamide catabolism (22). Indeed, N-methylation has been proposed as a metabolic pathway for nicotinamide excretion, and NNMT is the only enzyme known to utilize nicotinamide as a methyl acceptor substrate. Therefore, NNMT could participate in the regulation of nicotinamide intracellular levels, modulating its excretion after N-methylation.
In this regard, NNMT upregulation could affect all fundamental events, in which nicotinamide appears to be involved. Nicotinamide is part of the NAD molecule, which participates in a wide range of biological processes, including energy production, cellular resistance to stress or injury, and longevity (23). In addition, several enzymes, which use NAD as substrate, including ADP-ribosyltransferases, CD38, and sirtuins, each contain a nicotinamide-product site, and can be inhibited by this compound (24). Because of this type of product inhibition, the salvage and/or elimination of nicotinamide are crucial steps in NAD metabolism and the enzyme NNMT could play a fundamental role in the regulation of these cellular events.
In the present study, both pT and pathological stage showed an inverse correlation with NNMT mRNA levels, suggesting the possibility of this enzyme as a prognostic factor, and suggesting a role of NNMT in tumor growth, in agreement with the behavior previously observed in renal carcinoma (20). Even though the effects of NNMT upregulation deserve further investigation, it is tempting to speculate that the tumor cells, through this aberrant enzyme expression, attempt to control cellular events related to nicotinamide concentration or regulate transmethylation reactions using S-adenosylmethionine. Moreover, owing to the broad substrate specificity exhibited by NNMT, its methyltransferase activity could alter the efficacy and/or adverse effect profile of standard doses of chemotherapeutic drugs.
Although the association of NNMT with a few different types of tumors has been described, as reported above, to the best of our knowledge this is the first study dealing with OSCC and, most importantly, showing some correlation between such enzyme and tumor metastases, with a possible prognostic meaning. In fact, a significant negative association of the amount of NNMT expressed by tumor tissue was found with metastasis, at both the mRNA and protein level, and an improved overall survival rate was shown for patients bearing tumors with NNMT expression levels higher than adjacent normal tissue, although without reaching statistical significance. Although the involvement of NNMT in cancer progression must be investigated, the observations reported above support the hypothesis that the enzyme plays a role in tumor expansion, and tumors that downregulate it may be able to evade immunosurveillance and grow. In this regard, knowing the status of NNMT expression would be helpful in predicting the prognosis for each patient.
In summary, the present study suggests that NNMT may have potential as a biomarker and a therapeutic target for OSCC. Moreover, for the first time, a correlation between NNMT upregulation and a favorable prognosis of OSCC patients has been described. It is our future aim to study NNMT expression further in a large cohort of matched and well-characterized OSCC samples, and the applicability of screening NNMT expression as a prognostic determinant as well as its inhibition as a possible molecular approach to the treatment of OSCC.
ACKNOWLEDGMENTS
We gratefully acknowledge Dr Richard Weinshilboum, Mayo Clinic, Rochester, MN, for providing NNMT antibody.
Footnotes
Online address: http://www.molmed.org
REFERENCES
- 1.Brinkman BM, Wong DT. Disease mechanism and biomarkers of oral squamous cell carcinoma. Curr Opin Oncol. 2006;18:228–33. doi: 10.1097/01.cco.0000219250.15041.f8. [DOI] [PubMed] [Google Scholar]
- 2.Carinci F, et al. Potential markers of tongue tumor progression selected by cDNA microarray. Int J Immunopathol Pharmacol. 2005;18:513–24. doi: 10.1177/039463200501800311. [DOI] [PubMed] [Google Scholar]
- 3.Tomioka H, Morita K, Hasegawa S, Omura K. Gene expression analysis by cDNA microarray in oral squamous cell carcinoma. J Oral Pathol Med. 2006;35:206–11. doi: 10.1111/j.1600-0714.2006.00410.x. [DOI] [PubMed] [Google Scholar]
- 4.Lo Muzio L, et al. Survivin as prognostic factor in squamous cell carcinoma of the oral cavity. Cancer Lett. 2005;225:27–33. doi: 10.1016/j.canlet.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 5.Lo Muzio L, et al. Genetic analysis of oral squamous cell carcinoma by cDNA microarrays focused apoptotic pathway. Int J Immunopathol Pharmacol. 2006;19:675–82. doi: 10.1177/039463200601900323. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, et al. Serum circulating human mRNA profiling and its utility for oral cancer detection. J Clin Oncol. 2006;24:1754–60. doi: 10.1200/JCO.2005.03.7598. [DOI] [PubMed] [Google Scholar]
- 7.Rini J, Szumlanski C, Guerciolini R, Weinshilboum RM. Human liver nicotinamide N-methyltransferase, ion-pairing radiochemical assay, biochemical properties and individual variation. Clin Chim Acta. 1989;186:359–74. doi: 10.1016/0009-8981(90)90322-j. [DOI] [PubMed] [Google Scholar]
- 8.Aksoy S, Szumlanski CL, Weinshilboum RM. Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J Biol Chem. 1994;269:14835–40. [PubMed] [Google Scholar]
- 9.Aksoy S, Brandriff BF, Ward A, Little PF, Weinshilboum RM. Human nicotinamide N-methyltransferase gene, molecular cloning, structural characterization and chromosomal localization. Genomics. 1995;29:555–61. doi: 10.1006/geno.1995.9966. [DOI] [PubMed] [Google Scholar]
- 10.Szakacs G, et al. Predicting drug sensitivity and resistance, profiling ABC transporter genes in cancer cells. Cancer Cell. 2004;6:129–37. doi: 10.1016/j.ccr.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 11.Xu J, et al. Enhanced expression of nicotinamide N-methyltransferase in human papillary thyroid carcinoma cells. J Clin Endocrinol Metab. 2003;88:4990–6. doi: 10.1210/jc.2002-021843. [DOI] [PubMed] [Google Scholar]
- 12.Myo K, et al. Cyclin D1 gene numerical aberration is a predictive marker for occult cervical lymph node metastasis in TNM Stage I and II squamous cell carcinoma of the oral cavity. Cancer. 2005;104:2709–16. doi: 10.1002/cncr.21491. [DOI] [PubMed] [Google Scholar]
- 13.Williams AC, Ramsden DB. Autotoxicity, methylation and a road to the prevention of Parkinson’s disease. J Clin Neurosci. 2005;12:6–11. doi: 10.1016/j.jocn.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Markert JM, et al. Differential gene expression profiling in human brain tumors. Physiol Genomics. 2001;5:21–33. doi: 10.1152/physiolgenomics.2001.5.1.21. [DOI] [PubMed] [Google Scholar]
- 15.Jang JS, Cho HY, Lee YJ, Ha WS, Kim HW. The differential proteome profile of stomach cancer, identification of the biomarker candidates. Oncol Res. 2004;14:491–9. doi: 10.3727/0965040042380441. [DOI] [PubMed] [Google Scholar]
- 16.Lim BH, Cho BI, Kim YN, Kim JW, Park ST, Lee CW. Overexpression of nicotinamide N-methyltransferase in gastric cancer tissues and its potential post-translational modification. Exp Mol Med. 2006;38:455–65. doi: 10.1038/emm.2006.54. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Capezzone M, Xu X, Hershman JM. Activation of nicotinamide N-methyltransferase gene promoter by hepatocyte nuclear factor-1beta in human papillary thyroid cancer cells. Mol Endocrinol. 2005;19:527–39. doi: 10.1210/me.2004-0215. [DOI] [PubMed] [Google Scholar]
- 18.Roessler M, et al. Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer. Clin Cancer Res. 2005;11:6550–7. doi: 10.1158/1078-0432.CCR-05-0983. [DOI] [PubMed] [Google Scholar]
- 19.Yao M, et al. Gene expression analysis of renal carcinoma, adipose differentiation-related protein as a potential diagnostic and prognostic biomarker for clear-cell renal carcinoma. J Pathol. 2005;205:377–87. doi: 10.1002/path.1693. [DOI] [PubMed] [Google Scholar]
- 20.Sartini D, Muzzonigro G, Milanese G, Pierella F, Rossi V, Emanuelli M. Identification of nicotinamide N-methyltransferase as a novel tumor marker for renal clear cell carcinoma. J Urol. 2006;176:2248–54. doi: 10.1016/j.juro.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 21.Rogers CD, et al. Differentiating pancreatic lesions by microarray and QPCR analysis of pancreatic juice RNAs. Cancer Biol Ther. 2006;5:1383–9. doi: 10.4161/cbt.5.10.3323. [DOI] [PubMed] [Google Scholar]
- 22.Seifert R, Hoshino J, Kroger H. Nicotinamide methylation. Tissue distribution, developmental and neoplastic changes. Biochim Biophys Acta. 1984;801:259–64. doi: 10.1016/0304-4165(84)90075-8. [DOI] [PubMed] [Google Scholar]
- 23.Williams AC, Ramsden DB. Nicotinamide homeostasis, a xenobiotic pathway that is key to development and degenerative diseases. Med Hypotheses. 2005;65:353–62. doi: 10.1016/j.mehy.2005.01.042. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J. Are poly(ADP-ribosyl)ation by PARP-1 and deacetylation by Sir2 linked? Bioassays. 2003;25:808–14. doi: 10.1002/bies.10317. [DOI] [PubMed] [Google Scholar]