Abstract
Many eukaryotic mRNAs contain sequences that resemble segments of 28S and 18S rRNAs, and these rRNA-like sequences are present in both the sense and antisense orientations. Some are similar to highly conserved regions of the rRNAs, whereas others have sequence similarities to expansion segments. In particular, four 18S rRNA-like sequences are found in several hundred different genes, and the location of these four sequences within the various genes is not random. One of these rRNA-like sequences is preferentially located within protein coding regions immediately upstream of the termination codon of a number of genes. Northern blot analysis of poly(A)+ RNA from different vertebrates (chicken, cattle, rat, mouse, and human) revealed that a large number of discrete RNA molecules hybridize at high stringency to cloned probes prepared from the 28S or 18S rRNA sequences that were found to match those in mRNAs. Inhibition of polymerase II activity, which prevents the synthesis of most mRNAs, abolished most of the hybridization to the rRNA probes. We consider the hypotheses that rRNA-like sequences may have spread throughout eukaryotic genomes and that their presence in primary transcripts may differentially affect gene expression.
Our previous studies of gene expression revealed that a number of genes were differentially up- or down-regulated as a consequence of cell–cell or cell–substrate interactions (1, 2). In attempting to categorize these sequences through data base searches, we found a large number of mRNAs for ribosomal proteins (2) and rRNAs (unpublished observations). These initial observations led to a computer analysis of the GenBank and EMBL nucleic acid data bases. The results of these searches were striking: a large number of eukaryotic mRNAs contained sequences resembling those in different segments of the 28S and 18S rRNAs. In this paper, we examine these sequence similarities and describe the location within various mRNAs of the most abundant of the rRNA-like sequences. The results of the data base searches were confirmed by Northern hybridization analyses of poly(A)+ and total RNA using probes corresponding to those regions of rRNAs identified in the data base searches. We consider the possibility that the rRNA-like sequences in primary transcripts may function in the regulation of gene expression inasmuch as the observed sequences may interact with rRNAs in the antisense orientation or with ribosomal proteins in both orientations.
MATERIALS AND METHODS
Sequence Analysis.
The fasta program (3; Genetics Computer Group), version 7.2—UNIX was used to search the GenBank (release no. 95) and EMBL (release no. 47) data bases, or subsections of these data bases. The searches were done using the following mouse rRNA sequences: 28S rRNA (accession no. X00525X00525), 18S rRNA (accession no. X00686X00686), 5.8S rRNA (accession no. K01367K01367), and 5S rRNA (accession no. X71804X71804). In each case, the top 1000 sequence matches were retrieved. It should be noted that at present, sequence similarities located in intergenic regions would likely be underrepresented in these data base searches.
rRNA Probes.
Twenty-two regions of the rRNA sequences and two genomic regions flanking the 5S rRNA gene (see Fig. 1) were amplified from mouse genomic DNA using PCR, cloned into the TA vector (Invitrogen), and confirmed by sequence analysis. Insert fragments were excised from these vectors using EcoRI. Probe 28S-9 consisted of two double-stranded oligonucleotides. DNA fragments were used as probes after radiolabeling by random oligonucleotide priming (Boehringer Mannheim).
Figure 1.
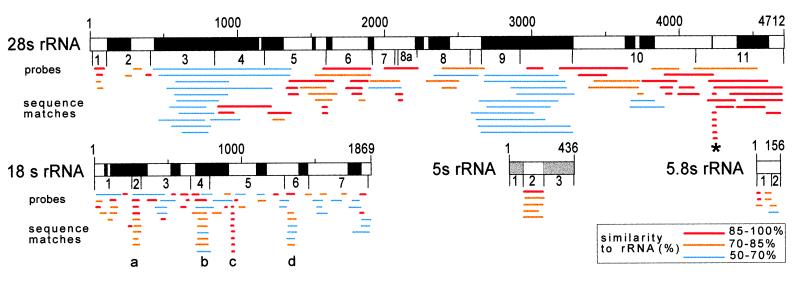
Sequence matches to mouse rRNAs based on fasta searches of nucleic acid data bases. Similarities of mRNAs and gene sequences to the mouse rRNA sequence are indicated as lines below the corresponding regions of rRNA. The extent of the homologies is indicated in color. The numbers above the bar representing the rRNA indicate the size of the rRNA in nucleotides. Locations of the 24 probes used for the hybridization analyses (see Fig. 4) are indicated below the bars. The black bars (28S and 18S rRNA) correspond to expansion segments. Note that the 18S rRNA sequence matches are from mouse data base entries only. The shaded bars (5S-1 and -3) are genomic sequences. Hundreds of sequences had similarity to the expansion segment at 28S nt 2654–3279. The region of the α-sarcin domain is indicated by the symbol ∗. The segments that form columns of sequence matches at nt 250–300 (18S-a), nt 679–768 (18S-b), nt 918–930 (18S-c), and nt 1286–1334 (18S-d) were found in more than 1000 mouse data base entries. The extent of these sequence similarities over approximately 1000 matches for 18S-a ranged from 83% over 29 nt to 75% over 24 nt; for 18S-b, they ranged from 80% over 35 nt to 72% over 33 nt; for 18S-c, they ranged from 100% over 13 nt to 100% over 8 nt; and for 18S-d, they ranged from 94% over 49 nt to 60% over 40 nt. In each of these ranges, the shorter match was to a subset of the longer nucleotide sequence.
Preparation of RNA and Northern Blot Analysis.
Total RNA was prepared from BALB/cByJ mouse embryos (embryonic day 17–18) and from the mouse C6 glioma cell line, using the guanidium thiocyanate extraction method (4). Poly(A)+ RNA was selected using Oligotex oligo(dT) beads (Qiagen). Poly(A)+ RNA from adult human, bovine, and chicken brains; from Drosophila embryos; and from yeast was obtained commercially, as was a Northern blot containing poly(A)+ RNA from various adult mouse tissues (CLONTECH). RNA was resolved on 1.2% agarose/formaldehyde gels and transferred to Hybond-N+ (Amersham). Filters were prehybridized and hybridized in Rapid-hyb buffer (Amersham) at 65°C, using a probe concentration of less than 2 ng/ml buffer. Filters were hybridized for a minimum of 4 h and were washed extensively at 65°C in 0.1× saline sodium citrate (SSC; 15 mM NaCl/1.5 mM sodium citrate)/0.5% SDS. Washed filters were exposed to XOMAT-AR film (Kodak) without an intensifying screen.
α-Amanitin Treatment.
C6 glioma cells were incubated with 50 μg/ml α-amanitin for 48 h, a treatment shown to be sufficient to inhibit luciferase reporter gene activity in cells transiently transfected with the pGL2-control plasmid (Promega; unpublished observations). After treatment with α-amanitin, the cells were rinsed once with PBS, and poly(A)+ RNA was prepared from 120 μg of total RNA and subjected to Northern blot analysis as described above.
RESULTS
rRNA-Like Sequences in Genes.
Data base searches revealed a diverse array of genes and mRNAs that contained sequences resembling those in the rRNAs of the mouse. These sequence similarities occurred in both the sense and antisense orientations and did not appear to be tags for any particular class of sequences. A representative sample of these data is presented diagramatically in Fig. 1. The 28S rRNA-like sequences that were present in different genes were often several hundred base pairs in length, with sequence similarities ranging from 50% to almost identical matches. The 18S rRNA-like sequences, on the other hand, tended to be much shorter, generally less than 60 bp, with sequence similarities ranging from approximately 65% to 100%. In contrast to the 28S- and 18S-like sequences which are found in thousands of genes, only a small number of genes contained sequences similar to the 5.8S and 5S rRNAs. Some specific examples of the various types of genes in which rRNA-like sequence similarities are found are listed in Table 1. The sequences were not random segments of the rRNA, but rather, were from particular regions of the rRNAs which were preferentially represented in the various genes. These rRNA-like segments can be seen forming columns of sequence matches aligned below particular portions of the 28S and 18S rRNA sequences themselves (shown diagramatically in Fig. 1).
Table 1.
A Sample of transcription units with sequence similarity to rRNA
| Sequence | Similarity to rRNA | Accession code | Location in mRNA |
|---|---|---|---|
| Rat insulin-like growth factor binding protein | 98% in 63 nt of 28S (nucleotides 26–88) | S46785S46785 | 3′ UTR |
| Human pax-2 mRNA | 55% in 197 nt of 28S (nucleotides 813–997) | M89470M89470 | 3′ UTR |
| Mouse testin-2 mRNA | 98% in 354 nt of 28S (nucleotides 860–1213) | X78990X78990 | 5′ UTR/coding† |
| Human liver glycine methyltransferase mRNA | 89% in 111 nt of 28S (nucleotides 1772–1882) | X62250X62250 | 5′ UTR |
| Mouse brain β-spectrin (Spn-2) mRNA | 94% in 318 nt of 28S (nucleotides 1898–1582*) | musspna | 5′ UTR |
| Mouse pmuAUF1-3 RNA binding protein mRNA | 77% in 182 nt of 28S (nucleotides 1911–2102) | U11274U11274 | 5′ UTR/coding‡ |
| Human protein kinase C delta cDNA 5′ UTR | 68% in 289 nt of 28S (nucleotides 2964–3248) | Z22521Z22521 | 5′ UTR |
| Mouse heat shock 86 protein mRNA | 100% in 229 nt of 28S (nucleotides 4129–4357) | J04633J04633 | 5′ UTR |
| Mouse homeobox gene (H2.9) mRNA | 88% in 458 nt of 28S (nucleotides 4261–4712) | X59474X59474 | 5′ UTR |
| Mouse erythropoietin gene | 100% in 20 nt of 18S (nucleotides 257–276) | M12930M12930 | 3′ UTR |
| Mouse engrailed protein gene (En-1) | 80% in 25 nt of 18S (nucleotides 266–290) | L12702L12702 | 5′ UTR |
| Mouse CENP-B, centromere associated protein | 80% in 35 bp of 18S (nucleotides 708–742) | X55038X55038 | |
| Mouse c-mos (TTAAGAGGGACGG) | 100% in 13 nt of 18S (nucleotides 918–930) | M26092M26092 | LTR |
| Mouse IgE-binding factor mRNA | 100% in 13 nt of 18S (nucleotides 918–930) | muslfige | 3′ UTR |
| Mouse tumor necrosis factor (MuTNF) mRNA | 94% in 49 nt of 18S (nucleotides 1334–1286*) | M11731M11731 | Coding |
| Mouse nucleolin gene | 96% in 23 nt of 18S (nucleotides 1821–1799*) | X07699X07699 | Intron |
| Human serum amyloid A gene | 82% in 119 nt of 5S (nucleotides 112–229) | humsaa | Intron |
| Human FMR-1 gene | 80% in 120 nt of 5S (nucleotides 227–109*) | humfmrls | 5′ Genomic |
UTR, untranslated region; LTR, long terminal repeat.
These sequence matches were in the antisense orientation.
5′ UTR (248 nt)/coding (106 nt).
5′ UTR (130 nt)/coding (52 nt).
18S rRNA-Like Sequences.
Data base searches revealed that four short sequences within the 18S rRNA were closely similar to sequences found in hundreds to thousands of different genes. Within the 18S rRNA structure (Fig. 1), the sequence at nucleotides 250–300 (denoted 18S-a) is part of a stem loop; the sequence at nucleotides 679–768 (denoted 18S-b) forms one stem loop and is part of a second; the sequence at nucleotides 918–930 (denoted 18S-c) is situated within a stem; and the fourth sequence at nucleotides 1286–1334 (denoted 18S-d) forms one stem loop (5, 6). These same four sequence similarities were also identified in the 18S rRNAs and mRNAs from a variety of different organisms, including human, Xenopus laevis, and Drosophila melanogaster.
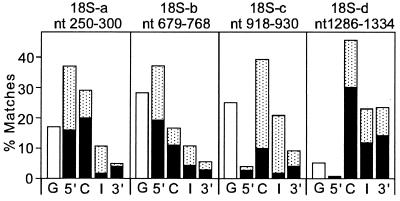
The distribution of these four sequences both within and outside of transcription units was determined, and the results are shown in Fig. 2. Two regions of 18S rRNA, 18S-a and 18S-b, were predominantly found in the 5′ UTR and in coding regions of mouse mRNAs. The third region, 18S-c, was found in coding regions and in introns, with little representation in 5′ UTRs. The fourth sequence, 18S-d, was predominantly found in coding regions but was represented significantly both in introns and in 3′ UTRs; this sequence was virtually absent from 5′ UTRs. The distributions of these sequences in coding regions, introns, and 3′ UTRs were similar in humans, Xenopus, and Drosophila. For example, the distribution of 18S-d in human sequences was almost identical to that in the mouse; 42% of the matches were in the coding region (72% in the reverse orientation), 20% were in introns, 12% were in 3′ UTRs, but this sequence was virtually absent from 5′ UTRs (4%). For two of the sequences (18S-a and 18S-b), however, the distribution in 5′ UTRs was lower in humans, Xenopus, and Drosophila.
Figure 2.
Location of mouse 18S rRNA-like sequences within and outside of transcription units. One hundred fasta matches to the 18S rRNA sequences indicated were analyzed for the location of the sequence similarity. The total number of matches in 5′ UTRs (5′), coding sequences (C), introns (I), and 3′ UTRs (3′) is indicated by the length of the filled/stippled bar. The relative number of matches identified either in the sense or reverse complementary orientation is indicated by the length in each bar of the stippled or filled regions, respectively. The total number of matches identified in genomic DNA sequences that were not in transcription units (G) is shown by the open bars.
The density of matches (number of matches per 10,000 nt of sequence) was also determined for each region. The results of this analysis indicated that the biases observed between the four sets of data base matches were the result of an increase or decrease in the density of matches. For example, sequence matches to 18S-a in 5′ UTRs occur 21 times in 10,000 nt of sequence. In 3′ UTRs, this same sequence match occurs only 1.2 times in 10,000 nt of sequence.
As shown in Fig. 2, the orientation of some of these sequences within the mRNAs of the mouse also appears to be nonrandom. Approximately 70% of the RNAs that have sequence similarities to 18S-b within their coding regions are reverse complementary matches. In contrast, 74% of the RNAs that are similar in their coding regions to 18S-c showed sequence matches in the sense orientation. Similar biases for the sense orientation are seen in introns that have sequence matches to 18S-a and 18S-c.
It is noteworthy that the fourth sequence (18S-d) that predominates in coding regions is sometimes found in a position immediately 5′ to the termination codon (Fig. 3). For example, myosin light chain 2 mRNA from human, mouse, and Xenopus contained this rRNA-like sequence near the stop codon. While the orientations of the rRNA-like sequences in these regions of mRNAs from different genes were complementary to the 18S rRNA sequence, they were present in all three reading frames (Fig. 3), resulting in very different amino acid sequences for the different proteins. This suggests that it is the RNA sequence or secondary structure rather than the amino acids encoded by this sequence that is significant.
Figure 3.
Location of an 18S rRNA sequence upstream of termination codons. The bottom sequence (18S) is the reverse complement of mouse 18S rRNA, nucleotides 1286–1334. Above this sequence are aligned the sequences of Xenopus homeobox XgbX2 mRNA (XGB, accession no. U04867U04867), mouse myosin light chain 2 mRNA (MYO, accession no. M91602M91602), mouse ATP-dependent RNA helicase (HEL, accession no. U46690U46690), mouse lactate dehydrogenase-A4 mRNA (LDL, accession nos. M17516M17516 and M29170M29170), human epithelial membrane protein 2 (EMP, accession no. X94771X94771), and mouse pre-T cell receptor α-type chain precursor mRNA (PTR, accession no. U16958U16958). Matches are represented as vertical bars. The translation termination sequences are boxed. The number 1 in the HEL sequence represents the sequence CG.
Hybridization Analysis Using Mouse rRNA Probes.
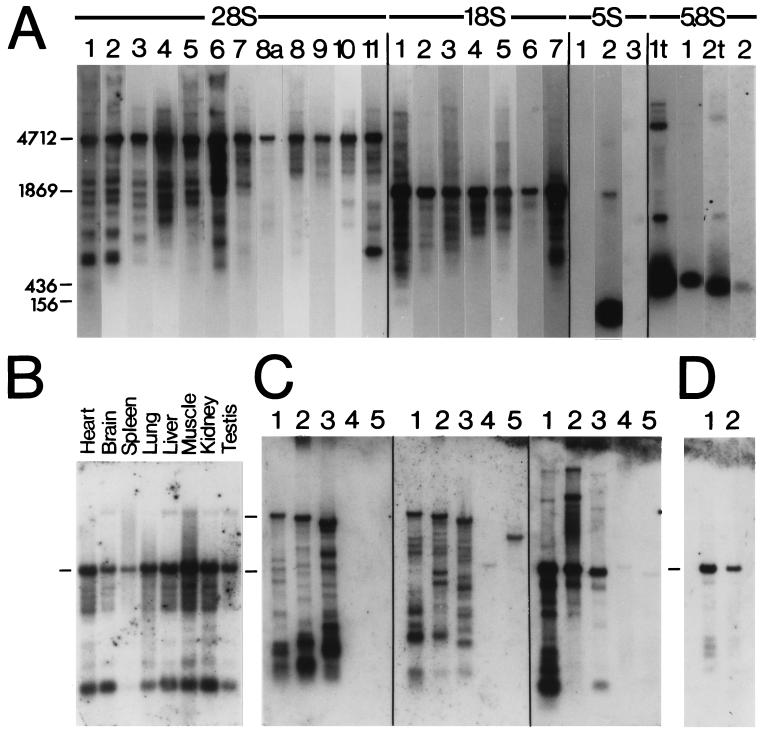
Northern blots of total and poly(A)+ RNA prepared from mouse embryos (Fig. 4A) were probed with cloned mouse rRNA fragments, and the results indicate that there are many mRNAs with sequence similarities to the segments in 28S and 18S rRNAs identified in the data base searches. The quality of the target RNA in the blots shown in Fig. 4A was comparable between probes since the same 5 Northern strips were used for all 23 hybridizations. For example, the same filter used for the 5S-2 hybridization was stripped and reprobed with the 18S-3 probe. In addition, a probe (28S-8a) based on a 28S rRNA sequence (nucleotides 2095–2219) that failed to identify any significant number of matches in the fasta analysis also failed to hybridize extensively to any bands other than that of the 28S rRNA. Moreover, there was no correlation between GC content and the hybridization results presented in Fig. 4A. For example, probe 28S-1 is 64 bp long, has a GC content of 43%, and hybridized to a large number of bands. Probe 5.8S-2 is similar in size (65 bp) and has a GC content of 74%, yet this probe failed to hybridize to any bands except for the 5.8S rRNA that was carried over into the poly(A)+ RNA. Probes 5S-1 and 5S-3 are genomic sequences that flank the 5S rRNA transcription unit. The GC content of these probes was 74% and 68%, respectively. They failed to hybridize to any bands on the Northern blots, although these same probes did hybridize to Southern blots of mouse genomic DNA (not shown). At lower stringency washing conditions (6× SSC/55°C), which should allow greater mismatch with the probe, the 5S rRNA probe did not hybridize to any additional bands. These findings do not exclude the possibility that a set of mRNAs contain sequence similarities to the 5S and 5.8S rRNAs, but they do indicate that such similarities were not detectable in RNA prepared from mouse embryos.
Figure 4.
Northern analysis with rRNA probes under high stringency conditions. (A) Mouse embryo RNA probed with 28S-1 to 28S-11, 18S-1 to 18S-7, 5S-1 to 5S-3, and 5.8S-1 to 5.8S-2 inserts (see Fig. 1). Lane numbering corresponds to probes. All lanes were loaded with 1 μg of poly(A)+ RNA, except for lanes 5.8S-1t and 5.8S-2t, which had 10 μg of total RNA. (B) Northern analysis of RNA from multiple adult mouse tissues with 28S.11 insert probe. Each lane was loaded with 2 μg of poly(A)+ RNA from heart, brain, spleen, lung, liver, skeletal muscle, kidney, and testis. (C) Northern hybridizations using 1 μg of poly(A)+ RNA from adult human (lane 1), cow (lane 2), and chicken brain (lane 3); Drosophila embryo (lane 4); and yeast (lane 5). The left, center, and right gel sets were probed with the 28S.2, the 28S-11, and the 18S-1 insert probes, respectively. (D) Northern hybridizations using poly(A)+ RNA prepared from cells that were cultured for 2 days in the absence (lane 1) or presence (lane 2) of 50 μg/ml α-amanitin. The positions of the 28S, 18S, 5.8S, and 5S rRNAs are indicated to the left in A. In B and D, the position of the 18S rRNA, and in panel C, the positions of the 28S and 18S rRNAs are shown at the left.
The Northern analysis in Fig. 4A was performed using RNA prepared from whole embryos, and the hybridization pattern is therefore a mixture of RNAs from all tissues. To investigate the hybridization pattern in individual tissues, a Northern blot containing poly(A)+ RNA from a variety of adult mouse tissues was probed with the 28S-11 probe (Fig. 4B), a probe that was found to hybridize to a large number of bands in whole embryo poly(A)+ RNA (Fig. 4A). The overall pattern in skeletal muscle seen with this probe was almost identical to that obtained using whole embryo poly(A)+ RNA. From tissue to tissue, the patterns obtained with this probe were similar but not identical. In contrast, the pattern in the spleen was less extensive and quite different from that observed in any of the other tissues tested. The results indicate that a variety of tissues contain RNAs that are capable of hybridizing to 18S and 28S rRNA probes. As discussed below, these RNAs failed to hybridize to probes prepared from 5S and 5.8S rRNAs.
rRNA-Like Sequences in Different Eukaryotes.
To investigate the occurrence of the rRNA-like sequences in the genes of other organisms, Northern blots were prepared using poly(A)+ RNA prepared from adult human, cattle, and chicken brains, D. melanogaster embryos, and Saccharomyces cerevisiae. These blots were probed with three of the mouse rRNA probes used in Fig. 4A. The results of these hybridizations indicate that RNAs from human, cattle, and chicken sources all contain sequences capable of hybridizing at high stringency with mouse rRNA probes (Fig. 4C). The lack of hybridization to the fly and yeast RNAs may reflect the stringent conditions adopted in the present studies as well as rRNA differences between these organisms. For example, the mouse 18S-1 probe is 98% identical to the human 18S rRNA over 250 nt but is only 76% and 78% identical to the fly and yeast 18S rRNAs, respectively.
α-Amanitin Perturbation of mRNA Populations.
To determine whether rRNA-like sequences in different genes were present mainly in mRNAs, α-amanitin was used to selectively inhibit the transcription of mRNAs. By incubating rat C6 glioma cells and mouse LMTK− fibroblast cells (not shown) for 2 days in α-amanitin, it was expected that many preexisting mRNAs would be degraded over this time period while rRNA synthesis would continue. The results of the hybridizations indicated that this was indeed the case (Fig. 4D). In the presence of α-amanitin, the rRNAs were still present at approximately the same levels in total RNA, but in poly(A)+ RNA, many other bands were missing or present at reduced levels. The remaining common bands may represent long-lived mRNAs or may be transcripts of RNA polymerases I or III. The fact that α-amanitin eliminated or reduced the level of many of the bands suggests that a population of RNA polymerase II transcripts contains sequences with a significant similarity to the 28S and 18S rRNAs.
DISCUSSION
The rRNA-like sequences that we identified within transcription units were of several different types and were not random with respect to their positions within the rRNA sequences themselves. Although a large number of sequence matches to the 28S and 18S rRNAs were identified, only a few examples have been noted in the literature (e.g., see refs. 7–10). In some of these cases, probes to the genes under study cross-hybridized to the rRNAs on Northern blots (8–10). Specific examples of genes known to have sequences similar to the rRNAs include those for the small nucleolar RNAs. These small RNAs, which are 67–280 nt long in metazoans (11), contain short sequences complementary to the rRNAs (up to 21 nt) and appear to be involved in rRNA processing and methylation (11–13). These small nucleolar RNA complementarities do not correspond to the major sequence matches identified in Fig. 1.
Sequence Similarities to Expansion Segments of 28S rRNA.
In addition to genes containing sequences nearly identical to segments of rRNA, a number of genes that contained sequences that were more divergent were also identified. It is noteworthy that within the 28S rRNA sequence, two regions (nucleotides 438-1124 and nucleotides 2654–3279) corresponding to expansion segments show this type of sequence match (Fig. 1). Expansion segments are sequences present in eukaryotic rRNAs that are not present in Escherichia coli rRNAs (refs. 14 and 15; reviewed in ref. 16). Although the location of these expansion segments is at fixed positions within the molecule in different evolutionary lineages, their sizes and sequences vary considerably between different organisms, and it is thought that they may have once been mobile elements that have lost the ability to transpose (reviewed in ref. 16).
Sequence similarities to the 28S rRNA expansion segments were observed in the coding regions of a number of genes. Although these regions are GC-rich, it was surprising to find that both of these expansion segments in the mouse contained six open reading frames between nucleotides 480–780 and nucleotides 2640–3020. In addition, nucleotides 480-1130 have three open reading frames in the sense orientation and nucleotides 2640–3250 continue to be open in five reading frames. There is no obvious codon bias in any of these reading frames, and it seems unlikely that regions of the 28S rRNA are translated. Translation of these regions, however, remains a formal possibility, and it is perhaps pertinent that in E. coli, 23S rRNA can encode a pentapeptide that confers resistance to the antibiotic erythromycin (17).
Dispersed Middle Repetitive DNA Sequences Derived from rRNA Genes.
The identification of a large number of genes containing sequences similar to those in rRNAs suggests that the sequences may comprise a new group of middle repetitive sequences. These rRNA-like sequences appear to be dispersed throughout the genome, and the number of copies of each particular sequence, as identified by data base searches, varies greatly.
The origin of rRNA-like sequences in transcribed and nontranscribed parts of any genome is not known, nor is it apparent why they appear to be more prevalent in multicellular organisms than in yeast, for example. Although the shorter rRNA sequence similarities in mRNA may have evolved by convergent evolution, other mechanisms must be considered for the longer sequences. Mechanisms involving staggered chromosomal breaks and reverse transcription, such as those that occur in the generation of pseudogenes, may account for retroposition of rRNA fragments (18). The large amount of rRNA within the cell may also drive forward processes that are otherwise rare events, such as trans-splicing (19, 20).
The evolutionary origin of sequences related to the rRNA sequences may be more similar to that of gene control elements and DNA motifs that regulate transcription rather than to that of other types of repetitive sequences. In both cases, there are large numbers of these sequences, and in the case of gene control elements, location is often important for function. In the case of the four 18S rRNA sequences, it is not yet clear that their nonrandom location within genes (see Fig. 2) is significant. Even in the absence of function, however, mechanisms such as gene conversion (21, 22) could maintain the sequence similarities observed among these rRNA-like sequences.
Implications for the Control of Gene Expression.
The functional implications of the present observations remain to be explored. The rRNA-like sequences may function as cis-regulatory elements and affect the expression or stability of mRNAs that contain them by interacting with ribosomes, rRNA, or ribosomal proteins. Experiments to explore these possible interactions may uncover new modes of regulating translation in the cell that differentially distinguish between messages carrying different rRNA-like sequences.
Sequence complementarities with 18S rRNA have been postulated as components in mechanisms by which the small subunit of the ribosome recognizes and interacts with some mRNAs (reviewed in ref. 23). It has also been suggested that all mRNAs contain short GC-rich sequences (7–14 bp) termed clinger fragments that are complementary to 13 regions of the 18S rRNA and that function to attach mRNAs to the 18S rRNA in the small subunit of the ribosome, thereby increasing their chances of being translated (10). However, the sequence similarities we identified were much more extensive than those described previously (10) and were present in both orientations. Moreover, while two of the four 18S rRNA sequence similarities (18S-a and 18S-b, see Fig. 1), identified in hundreds to thousands of genes, contained three clinger fragments, the remaining two (18S-c and 18S-d) did not overlap with clinger fragments. In other studies, it has been suggested that a short sequence complementary to the 3′ region of the 18S rRNA sequence allows the 40S ribosomal subunit to bind to the 5′ UTR of viral RNAs in a cap-independent manner through an internal ribosome entry sequence, or a ribosome landing pad (e.g., see refs. 24–27). This mechanism of translation initiation from an internal ribosome entry sequence is also known to occur in a number of cellular mRNAs (e.g., see refs. 28 and 29).
It is particularly intriguing that sequences complementary to the 18S rRNA (18S-d) were identified immediately upstream of the termination codons of a number of genes (Fig. 3). A comparison of this 18S rRNA sequence to the 16S rRNA sequence of E. coli localizes this sequence to an area that is situated immediately upstream of the sequences required for UGA-mediated translation termination in bacteria (30). The location of these sequences at the termination codons of a number of mRNAs suggests that they may interact with the 18S rRNA in the ribosome and be involved in regulating some aspect of the termination of translation.
In our analysis, a small number of genes were found to contain a sequence that is very similar or identical to the 22-nt α-sarcin domain of the 28S rRNA (nucleotides 4245–4267). This short sequence is highly conserved in all large subunit rRNAs, is involved in binding elongation factors, and is the target of two specific ribosome inactivating proteins, α-sarcin and ricin (reviewed in ref. 31). The α-sarcin sequence is located on the exterior surface of the ribosome and can interact with other RNAs or proteins that affect ribosome function (32, 33). The possibility of cellular RNAs that contain rRNA-like sequences interacting with the exterior surface of ribosomes is particularly interesting in light of a recent report in which three antisense sequences were inserted into an expansion segment of the Tetrahymena 28S rRNA that was known to be located on the exterior surface of the ribosome; these modified ribosomes were able to inhibit gene expression apparently at the level of translation (34).
In addition to interactions within the ribosome, the possibility of interactions of rRNA-like sequences with ribosomal proteins or other molecules outside of the ribosome must be considered. It is possible that some of the rRNA-like sequences discussed here may serve to localize messages to particular locations in the cell. While it is presently unclear how much rRNA or rRNA fragments exist outside ribosomes (35), it is known that some ribosomal proteins have various functions outside of intact ribosomes (reviewed in ref. 36). For example, in E. coli, it has been demonstrated that several ribosomal proteins bind to their own mRNAs and inhibit their translation (reviewed in ref. 37). Human ribosomal protein S14 has been shown to inhibit transcription of its own gene (38), and yeast ribosomal protein L32 has been demonstrated to be involved in regulating the splicing of an intron in the L32 mRNA (39). Both the present findings and the examples in the literature strongly warrant experimental tests of the hypothesis that rRNA-like sequences may interact with rRNA or ribosomal proteins and thereby serve as cis-regulatory elements.
Acknowledgments
We thank Mr. Andrew Benedict for excellent technical assistance and Joseph Gally and George Miklos for critical reading of the manuscript. This research was supported by U.S. Public Health Service Grant HD-09635-23 to G.M.E. G.M.E. is a consultant to Becton Dickinson and Company.
Footnotes
Abbreviation: UTR, untranslated region.
References
- 1.Mauro V P, Krushel L A, Cunningham B A, Edelman G M. J Cell Biol. 1992;119:191–202. doi: 10.1083/jcb.119.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crossin K L, Prieto A L, Mauro V P. In: Tenascin and Counteradhesive Molecules of the ECM. Crossin K L, editor. Amsterdam: Harwood; 1996. pp. 23–46. [Google Scholar]
- 3.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Rairkar A, Rubino H M, Lockard R E. Biochemistry. 1988;27:582–592. doi: 10.1021/bi00402a013. [DOI] [PubMed] [Google Scholar]
- 6.Holmberg L, Melander Y, Nygard O. Nucleic Acids Res. 1994;22:1374–1382. doi: 10.1093/nar/22.8.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain S K, Crampton J, Gonzalez I L, Schmickel R D, Drysdale J W. Biochem Biophys Res Commun. 1985;131:863–867. doi: 10.1016/0006-291x(85)91319-1. [DOI] [PubMed] [Google Scholar]
- 8.Dooley S, Farber U, Welter C, Theisinger B, Blin N. Gene. 1992;110:263–264. doi: 10.1016/0378-1119(92)90659-d. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida T, Schneider E L, Mori N. Gene. 1994;151:253–255. doi: 10.1016/0378-1119(94)90666-1. [DOI] [PubMed] [Google Scholar]
- 10.Matveeva O V, Shabalina S A. Nucleic Acids Res. 1993;21:1007–1011. doi: 10.1093/nar/21.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maxwell E S. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 12.Bachellerie J-P, Michot B, Nicoloso M, Balakin A, Ni J, Fournier M J. Trends Biochem Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- 13.Kiss-Laszio Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 14.Chan Y-L, Gutell R, Noller H F, Wool I G. J Biol Chem. 1984;259:224–230. [PubMed] [Google Scholar]
- 15.Hancock J M, Tautz D, Dover G A. Mol Biol Evol. 1988;5:393–414. doi: 10.1093/oxfordjournals.molbev.a040501. [DOI] [PubMed] [Google Scholar]
- 16.Gerbi S A. In: Ribosomal RNA. Zimmerman R A, Dahlberg A E, editors. New York: CRC; 1996. pp. 71–87. [Google Scholar]
- 17.Tenson T, DeBlasio A, Mankin A. Proc Natl Acad Sci USA. 1996;93:5641–5646. doi: 10.1073/pnas.93.11.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiner A M, Deininger P L, Efstratiadis A. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 19.Dandeker T, Sibbald P R. Nucleic Acids Res. 1990;18:4719. doi: 10.1093/nar/18.16.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eul J, Graessmann M, Graessmann A. EMBO J. 1995;14:3226–3235. doi: 10.1002/j.1460-2075.1995.tb07325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edelman G M, Gally J A. Brookhaven Symp Biol. 1968;21:328–344. [PubMed] [Google Scholar]
- 22.Kass D H, Batzer M A, Deininger P L. Mol Cell Biol. 1995;15:19–25. doi: 10.1128/mcb.15.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonenberg N. Gene Expression. 1993;3:317–323. [PMC free article] [PubMed] [Google Scholar]
- 24.Le S-Y, Chen J-H, Sonenberg N M, Maizel J V. Virology. 1992;191:858–866. doi: 10.1016/0042-6822(92)90261-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilipenko E V, Gmyl A P, Maslova S V, Svitkin Y V, Sinyakov A N, Agol V I. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- 26.Pestova T V, Hellen C U T, Wimmer E. J Virol. 1991;65:6194–6204. doi: 10.1128/jvi.65.11.6194-6204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholson R, Pelletier J, Le S-Y, Sonenberg N. J Virol. 1991;65:5886–5894. doi: 10.1128/jvi.65.11.5886-5894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vagner S, Gensac M C, Maret A, Almaric F, Prats H, Prats A C. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teerink H, Voorma H O, Thomas A A. Biochim Biophys Acta. 1995;1264:403–408. doi: 10.1016/0167-4781(95)00185-9. [DOI] [PubMed] [Google Scholar]
- 30.Prescott C D, Kleuvers B, Goringer H U. Biochimie. 1991;73:1121–1129. doi: 10.1016/0300-9084(91)90155-t. [DOI] [PubMed] [Google Scholar]
- 31.Szewczak A A, Moore P B. J Mol Biol. 1995;247:81–98. doi: 10.1006/jmbi.1994.0124. [DOI] [PubMed] [Google Scholar]
- 32.Brigotti M, Lorenzetti R, Denaro M, Carnicelli D, Montanaro L, Sperti S. Biochem Mol Biol Int. 1993;31:897–903. [PubMed] [Google Scholar]
- 33.Saxena S K, Ackerman E J. J Biol Chem. 1990;265:3263–3269. [PubMed] [Google Scholar]
- 34.Sweeney R, Fan Q, Yao M-C. Proc Natl Acad Sci USA. 1996;93:8518–8523. doi: 10.1073/pnas.93.16.8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fontoura B M, Sorokina E A, Ebenezer D, Carroll R B. Mol Cell Biol. 1992;12:5145–5151. doi: 10.1128/mcb.12.11.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wool I G. Trends Biochem Sci. 1996;21:164–165. [PubMed] [Google Scholar]
- 37.Draper D E. Trends Biochem Sci. 1989;14:335–338. doi: 10.1016/0968-0004(89)90167-9. [DOI] [PubMed] [Google Scholar]
- 38.Tasheva E S, Roufa D J. Genes Dev. 1995;9:304–316. doi: 10.1101/gad.9.3.304. [DOI] [PubMed] [Google Scholar]
- 39.Vilardell J, Warner J R. Genes Dev. 1994;8:211–220. doi: 10.1101/gad.8.2.211. [DOI] [PubMed] [Google Scholar]