Abstract
We present a physical and molecular genetic characterization of Drosophila melanogaster TFIIE (dTFIIE), a component of the basal RNA polymerase II transcription apparatus. We have purified dTFIIE to near homogeneity from nuclear extracts of Drosophila embryos and found that it is composed of two subunits with apparent molecular weights of 55 and 38 kDa. Peptide sequence information derived from the two subunits was used to isolate the corresponding cDNA clones, revealing that dTFIIE and human TFIIE share extensive amino acid similarity. Functional conservation was demonstrated by the ability of bacterially expressed dTFIIE to substitute for human TFIIE in an in vitro transcription assay reconstituted from purified components. Cytological mapping analysis shows that both subunits are encoded by single copy genes located on chromosome III.
The transcription of protein coding genes is a complex process that involves both gene specific and general transcription factors. Analysis of gene transcription in vitro has led to the identification of the general transcription factors TFIIA, TFIIB, TFIID [TATA-box binding protein (TBP) and TBP-associated factors], TFIIE, TFIIF, TFIIH, and RNA polymerase II (Pol II). These factors can assemble at the core promoter in a highly ordered fashion to generate an initiation complex that is competent to direct gene transcription in vitro (1). Promoter binding of TFIID, TFIIA, and TFIIB constitutes the first step in this assembly process and is followed by recruitment of TFIIF and Pol II, while TFIIE and TFIIH enter the initiation complex last.
Our knowledge about the role of TFIIE is primarily based on biochemical studies. Bandshift analysis has revealed that TFIIE is required for recruitment of TFIIH into the initiation complex (2), consistent with its ability to interact directly with both Pol II and TFIIH (3, 4). Functionally, the role of TFIIE and TFIIH appear to be related to the helical state of the template DNA in that TFIIE and TFIIH are dispensable for transcription from certain negatively supercoiled templates but necessary for transcription from linear templates (5). Interestingly, TFIIH contains two helicase activities that may be implicated in transcription by RNA Pol II (reviewed in ref. 6). In vitro studies have suggested that TFIIH helicase activities are required for open complex formation, in particular on template DNA lacking negative supercoils (7, 8). However, it has also been proposed that TFIIE and TFIIH may not be required during initiation, per se, but rather play a role in the transition from initiation to elongation (9). The role of TFIIE and TFIIH has also been addressed by analyzing the fate of the general transcription factors after initiation of transcription (10). These studies indicate that TFIIB and TFIIE dissociate from the template shortly after initiation whereas TFIIH may be released slightly later during the elongation phase. This is consistent with a requirement for TFIIE during initiation and promoter clearance. However, the precise role of TFIIE, besides bridging TFIIH to the initiation complex, is poorly understood.
In an effort to characterize the Drosophila RNA Pol II transcription machinery, several laboratories have systematically fractionated extracts from Drosophila embryos and isolated numerous activities necessary for transcription by Pol II in vitro (11–14). cDNA clones encoding the Drosophila transcription factors TFIIA, TFIIB, TFIID, and TFIIF have been isolated and recently employed to reconstitute transcription of Drosophila genes in vitro (refs. 15 and 16 and references therein). In addition, the genes encoding four subunits of Drosophila Pol II have been identified (17–20). Genetic analysis of the role of some of these factors has been undertaken (20–24) and is likely not only to reveal novel insights into the transcriptional process, but also to establish the in vivo relevance of conclusions derived from biochemical analyzes. As part of our efforts to fully define and characterize the Pol II transcription apparatus, we report the purification and initial characterization of Drosophila TFIIE (dTFIIE), the isolation and analysis of cDNA clones encoding both TFIIE subunits, and the reconstitution of transcription in vitro using purified Drosophila factors.
MATERIALS AND METHODS
Drosophila Growth and Nuclear Extract (NE) Preparation.
Wild-type 0 to 12-h Drosophila melanogaster (Canton-S) embryos were collected from cage stocks using standard protocols. Embryonic NE was prepared using the procedure of Soeller et al. (25) with minor modifications. A motorized homogenizer (model LH-21; Yamato, Orangeburg, NY) was used to disrupt embryos.
Protein Purification and Partial Amino Acid Sequence Determination.
Our standard chromatography buffer (HGKEDP) is 25 mM Hepes·KOH (pH 7.6), 15% (vol/vol) glycerol, KCl at the indicated concentration, 0.1 mM EDTA, 1 mM DTT, and 0.1% (vol/vol) phenylmethylsulfonyl fluoride (from a saturated solution in isopropanol).
NE was adjusted to 100 mM KCl with HGEDP, centrifuged, and applied onto a phospho-cellulose column previously equilibrated in 125 mM HGKEDP. After washing the column with 125 mM HGKEDP, dTFIIE was eluted with a 325 mM HGKEDP step. The phospho-cellulose 325 mM KCl step fraction was diluted to 75 mM KCl with HGEDP and applied to a DE-52 column previously equilibrated with 75 mM HGKEDP. Bound protein was eluted with a 10 column volume gradient from 75 mM to 500 mM KCl, and dTFIIE eluted as a single peak at 120–150 mM KCl. Fractions containing the highest dTFIIE activity were pooled, diluted to 100 mM KCl with HGEDP, applied to a Q-Superose column, and eluted with an HGKEDP gradient from 100 mM to 500 mM KCl. Active fractions (eluting at 280 mM HGKEDP) were pooled, adjusted to 1.2 M (NH4)2SO4, and chromatographed on Phenyl-Superose. The column was washed with 0.6 M HGAEDP [our standard buffer with (NH4)2SO4 replacing KCl] and a gradient from 0.6 to 0.0 M HGAEDP was applied. dTFIIE eluted at 400–330 mM (NH4)2SO4 as determined by SDS/PAGE and protein silver staining. Peak fractions were electrophoresed on preparative 10% SDS/PAGE and transferred to polyvinylidene difluoride membrane. A slice of the polyvinylidene difluoride membrane was incubated with antiserum previously raised against a dTFIIE containing protein fraction (26) to visualize the two dTFIIE subunits. The remaining membrane was stained with Ponceau S and polypeptides in the size range corresponding to the two TFIIE subunits were excised and digested with trypsin. Tryptic peptides were resolved by reverse-phase HPLC, and N-terminal sequences were determined by Edman degradation.
Transcription Assay Conditions.
As an assay for the purification, native dTFIIE activity was detected by its ability to allow mRNA synthesis in a run-off transcription reaction (11) with the other basal factors and RNA Pol II. The run-off transcription template contained the Drosophila actin 5C promoter, truncated at a PstI site to yield a 460-base run-off product (27). RNA Pol II (28) and DNase inhibitor (29) were prepared and used as described. All other transcription factors were present in a phospho-cellulose fraction prepared from NE [325–425 mM HGKEDP step elution fraction analogous to a 300–400 mM HGKEDP phospho-cellulose step of Kc-cell NE described previously (11)].
To test bacterially produced and reconstituted dTFIIE, transcription reactions were performed with purified components using the Drosophila Adh distal promoter as a template, as described (15). Renatured dTFIIE was used at the amount indicated in the figure legend.
Cloning of dTFIIE cDNAs.
Based on the partial amino acid sequence data, four degenerate primers (two primer pairs) were designed and synthesized for each subunit (see below). “R” represents A or G, “Y” represents T or C, “H” represents A or T or C, “D” represents A or T or G, “N” represents A or G or T or C, and “I” represents inosine. The corresponding peptide sequences are in parenthesis. The oligonucleotides for dTFIIE large subunit are as follows: #1, primer 3835E, TTY ACN GAY TTR GAR GC (FTDLEA); #1, primer 3835F, TTY ACN GAY CTN GAR GC (FTDLEA); #2, primer 3829C, CT YTC NGT CAT CCA DAT (IWMTES); #3, primer 3835C, GAR GCT GAY CAR CTC TTY GAY ATG (EADQLFDM); and #4, primer 3827C, TC DAT GTC IAC AGG YTC NGG YTC (EPEPVDID). The oligonucleotides for dTFIIE small subunit are as follows: #1, primer 3908B, TTY GGT GTN CTN GCN AA (FGVLAK); #2, primer 3918A TC CAT GGC RTC NAC NGT (TVDAMD); #2, primer 3918B, TC CAT GGC RTC NAC YTC (EVDAMD); and #3, primer 3919C, GAR ATH CTA TTY GTN GT (EILFVV); #4, primer 3922, TC RTC NAC TGA RAA RTT (NFSVDD).
In the case of peptide 3918 it was ambiguous whether the sequence was TVD or EVD; therefore two sets of primers were generated (3918A and 3918B).
For both subunits, the first round PCR employed 1 μg of Drosophila cDNA (the DNA/RNA product of first strand synthesis, using random primers and total RNA obtained from 0 to 12-h embryos) and 50 pmol of primers 1 and 2, appropriate for each subunit. Reaction conditions were 20 mM Tris·HCl (pH 8.4), 50 mM KCl, 2 mM MgCl2, 0.2 mM of each dNTPs, and 2 μM of each primer, in a final volume of 25 μl. Touchdown PCR was employed to increase specificity (large subunit, 1°C/cycle ramping from 50°C to 40°C, 30 cycles at 40°C; small subunit, 1°C/cycle ramping from 42°C to 32°C, 30 cycles at 32°C). PCR products were separated into different size ranges and reamplified using nested primers #3 and #4, appropriate for each subunit. Second round touchdown PCR conditions were as follows: large subunit, 1°C/cycle ramping from 60°C to 50°C, 30 cycles at 50°C; and small subunit, 1°C/cycle ramping from 40°C to 30°C, 30 cycles at 30°C. As predicted, a band of ≈215 bp was noted in the large subunit second round reaction, while an 83-bp band was noted in the small subunit second round reaction. Each band was cloned and sequenced.
All PCR- and library-derived cDNA fragments to be used as probes were first excised from vector DNA and subsequently gel purified. DNA fragments (25 ng) were random primer labeled with [α-32P]dATP using the Random Prime It-II labeling kit (Stratagene). Probes were purified using Micro Select G-25 columns (5 Prime → 3 Prime, Inc.).
Two cDNA libraries were used in this study: a 0 to 4-hr embryonic cDNA library was obtained from James Kadonaga (University of California at San Diego), and a Kc cell cDNA library was from David Price (University of Iowa). Both of these libraries were constructed using the ZAP cloning system (Stratagene). Libraries were plated and transferred to membranes following standard methods. Membranes were prehybridized in 0.5% SDS, 2% dextran sulfate, 5× Denhardt’s solution (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA), 5× SSPE [standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA], 50% formamide, and 50 μg/ml boiled salmon sperm DNA for 2 h at 42°C. Membranes were then incubated overnight at 42°C in fresh buffer with denatured DNA probe (3 × 106 cpm per disc). Lastly, membrane discs were washed in high stringency buffer (0.1% SDS/0.1× standard saline citrate), dried, and autoradiographed. Candidate phage were subjected to at least three rounds of repurification and probing. Phagemid excision with candidate clones was carried out using the Lambda ZAP II Cloning Kit (Stratagene).
Northern blot analysis was carried out using standard protocols with total and poly(A)+ mRNA preparations from staged 0 to 12-h wild-type embryos.
Recombinant Protein Production.
An NdeI site was engineered by PCR at the methionine start codon in the large and small subunit cDNA clones and confirmed by sequencing. The large and small subunit cDNAs were inserted into pET19b cut with NdeI–BamHI and NdeI–XhoI, respectively.
Proteins were expressed in BL21 LysS (30) and induced at OD600 of 0.5–0.6 with 0.4 mM isopropyl β-d-thiogalactoside for 1 h. Bacteria were pelleted by centrifugation, resuspended in TEG buffer (1/100 of cell culture volume; 25 mM Tris·HCl pH 7.6/1 mM EDTA/10% glycerol/1 mM DTT/1 mM Na2S2O5/0.1 mM phenylmethylsulfonyl fluoride) and disrupted by sonication or by using a french press. For the small subunit, the insoluble material from the cell lysate was subsequently extracted with TEG containing 0.1 M NaCl, 0.5 M NaCl, and 6 M guanidine·HCl. The majority of the small subunit was only soluble in the presence of guanidine·HCl and was further purified by Ni-agarose chromatography. The column was washed with TEG containing 7 M urea and 40 mM imidazole, and the bulk of TFIIE-S was eluted with TEG containing 7 M urea and 200 mM imidazole.
The pellet from the TEG extraction of TFIIE-L was subjected to three water washes with the bulk of TFIIE-L recovered in the first two water washes. This TFIIE-L was combined with Ni-agarose purified TFIIE-S (in 7 M urea) and renatured by stepwise dialysis against HEMG containing 2 M urea, 0.5 M urea, and finally 0.1 M KCl. Under these conditions, a soluble complex of TFIIE-L and TFIIE-S could be recovered, whereas the two subunits were insoluble when renatured individually.
For antibody production, TFIIE-L from the water extraction was further purified by Ni-agarose chromatography as described for TFIIE-S.
Antibodies.
Rabbits were immunized with 100 μg Ni-agarose purified TFIIE-S or TFIIE-L in Ribi adjuvant (Ribi Immunochem Research) and boosted twice. Serum was used for Western blot analysis at 10,000-fold dilution or coupled to Protein-G Sepharose for immunoprecipitations.
RESULTS
Purification and Properties of dTFIIE.
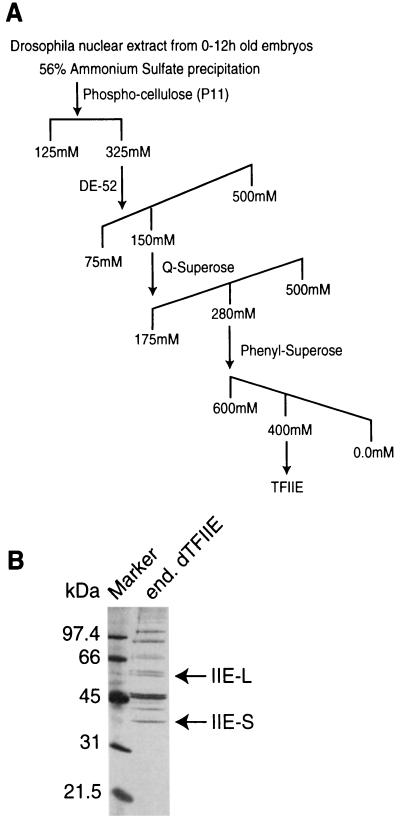
Several laboratories have isolated an activity from Drosophila embryo NEs that can functionally substitute for the human RNA Pol II transcription factor TFIIE, and this activity has therefore been termed dTFIIE (refs. 12 and 14, and A.J.S., X.W., C. George, and W.Z., unpublished data). We have purified dTFIIE by conventional chromatography as outlined in Fig. 1A. After the last column, the purity of the pool of fractions containing dTFIIE activity was analyzed by SDS/PAGE and visualized by silver staining (Fig. 1B). Polypeptides potentially corresponding to the subunits of dTFIIE (IIE-L and IIE-S in Fig. 1B) were identified by Western blot analysis using an antiserum previously raised against a protein fraction containing purified dTFIIE activity (data not shown and ref. 26). As the final purification step, the pool of dTFIIE activity was subjected to preparative SDS/PAGE and electroblotted onto polyvinylidene difluoride membrane. The polypeptide bands corresponding to the two subunits of dTFIIE were excised and the amino-terminal sequence of tryptic peptides was determined. We obtained three peptide sequences from dTFIIE-L and four peptide sequences from dTFIIE-S that showed extensive sequence similarity to the large and small subunits of hTFIIE, respectively (Fig. 2).
Figure 1.
(A) Outline of fractionation scheme for purification of dTFIIE (see Materials and Methods for details). (B) Silver stained SDS/PAGE of dTFIIE fraction used for preparative SDS/PAGE before microsequencing. The arrows indicate the polypeptides corresponding to the large (IIE-L) and small (IIE-S) dTFIIE subunits that were used to generate tryptic fragments for microsequencing.
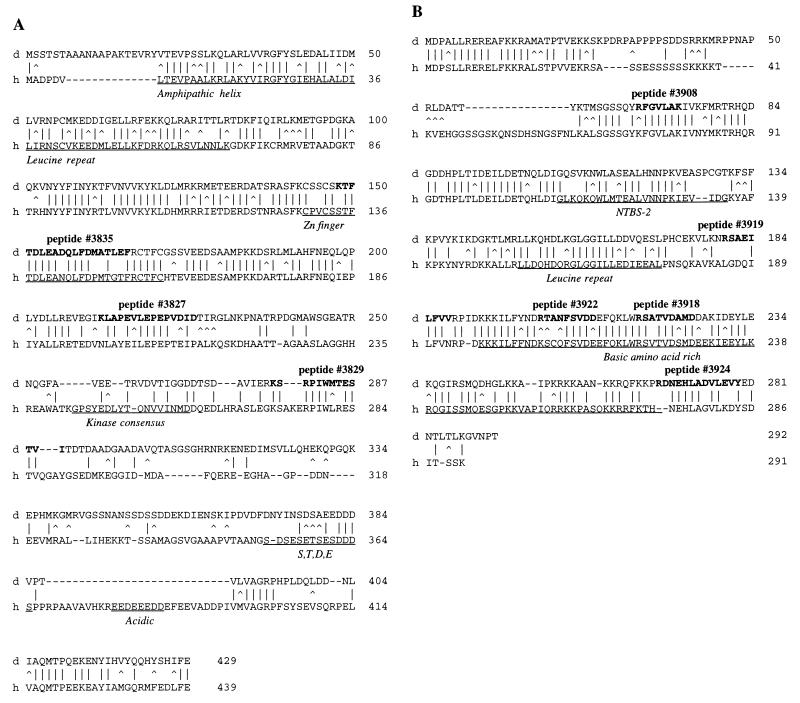
Figure 2.
(A) Comparison of amino acid sequences for the large subunit of TFIIE from Drosophila (d) and human (h). Vertical bars indicate alignment of identical residues, while carrots indicates similar residues. Amino acids said to be “similar” are as follows: A,S,T; D,E; N,Q; R,K; I,L,M,V; and F,Y,W. Boldfaced amino acids in dTFIIE specify the peptide sequences obtained by microsequencing. Underlined amino acids specify sequence motifs in hTFIIE: amphipathic helix; leucine repeat domain; zinc finger domain; kinase consensus domain; S,T,D,E domain; and acidic domain. (B) Comparison of amino acid sequences for the small subunit of TFIIE from Drosophila (d) and human (h). Vertical bars indicate alignment of identical residues, while carrots indicate similar residues (see A for details). Boldfaced amino acids in dTFIIE specify the peptide sequences obtained by microsequencing. Underlined amino acids specify sequence motifs in hTFIIE: nucleotide binding site-2 (NTBS-2); leucine repeat domain; and basic amino acid-rich domain.
Molecular Cloning and cDNA Analysis.
To isolate cDNAs encoding the two dTFIIE subunits, we employed a nested PCR approach using degenerate oligonucleotide primers that were designed based upon the amino acid sequence information obtained from each subunit. As a template we used first-strand cDNA that was prepared from total RNA isolated from 0 to 12-h Drosophila embryos. We isolated partial cDNA fragments corresponding to each subunit, and these fragments were used to screen Drosophila cDNA libraries. Isolates nominally large enough to encode each of the two subunits of dTFIIE were sequenced.
The cDNA, DPA11, corresponding to the large subunit, has a total length of 1587 bp and contains an ORF encoding a protein of 429 amino acids (Fig. 2A) with a predicted molecular weight of 48 kDa. The amino acid sequence exhibits 44% identity and 59% similarity with the large subunit of hTFIIE. Most of the conserved residues are within the amino-proximal portion (residues 21–232 of dTFIIE-L) while the carboxyl-terminal half shows little homology. A similar pattern is observed when comparing the sequences of the human and Xenopus TFIIE large subunits (31–33). The amino-proximal region of hTFIIE56 has been characterized functionally and shown to be required for transcriptional activity, association with hTFIIH, as well as stimulation of phosphorylation of the carboxyl-terminal domain of Pol II (34). The zinc finger domain in hTFIIE56, which is required for transcriptional activity in vitro (35), is also present in dTFIIE-L and maps within the conserved region (Fig. 2A). These comparisons reveal that functionally important regions identified within hTFIIE56 are highly conserved in the Drosophila homologue. By contrast, the leucine repeat present in hTFIIE56 is less well conserved in the Drosophila homologue (Fig. 2A). Other structural motifs such as the proposed kinase consensus (residues 245–270) and the S,T,D,E-rich region (residues 352–365) in the human protein are not evident in the Drosophila protein.
The cDNA, DPB3, corresponding to the small subunit, has a total length of 1055 bp and includes an ORF encoding a protein of 292 amino acids (Fig. 2B) giving a calculated molecular weight of 33 kDa. Drosophila TFIIE-S and human TFIIE34 exhibit extensive homology (56% identity and 71% similarity) throughout most of the protein and only residues 27–60 and the carboxyl-terminal 9 amino acids of dTFIIE-S show little homology with the human sequence. A similar pattern is observed when comparing Xenopus and human TFIIE sequences (36, 37). The conserved region includes structural motifs such as the leucine repeat and the putative nucleotide binding site 2 (NTBS2), which can be aligned with the Drosophila subunit with 50% identity (Fig. 2B) as well as the basic region near the carboxy terminus (human residues 197–274 and Drosophila residues 193–267) (33).
Partial cDNA clones were used to identify dTFIIE transcripts by Northern blot analysis of polyadenylated RNA purified from 0 to 12-h embryos. One species of RNA was identified for each subunit: an approximate 2000-base transcript for the large subunit and a 1300-base transcript for the small subunit (data not shown). Both mRNAs are longer than the isolated cDNAs, suggesting that the cDNAs reported here may not be full length although they appear to contain the entire ORFs.
Chromosomal mapping of the genes encoding both subunits was carried out using partial cDNA isolates for each subunit in hybridizations to polytene chromosomes of the larval salivary gland. The large subunit gene maps to the 68D/E region on chromosome III, while the small subunit gene maps to chromosome III at the 64B region.
Expression and Analysis of Recombinant dTFIIE.
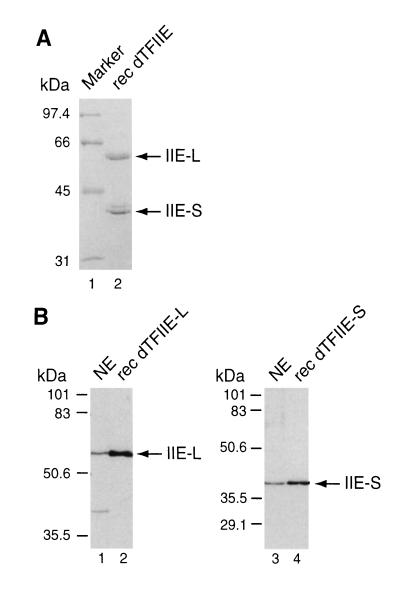
We next sought to determine if the ORFs identified in DPA11 and DPB3 encode full-length proteins that are functionally equivalent to native dTFIIE. To obtain recombinant protein for antibody production and in vitro assays, the two cDNAs were fused to N-terminal poly-histidine tags, expressed in bacteria and purified under denaturing conditions. The small and large subunits were combined and renatured by stepwise dialysis, yielding a complex that was soluble under native conditions (Fig. 3A). The formation of a complex was verified by a coimmunoprecipitation experiment in which the small subunit could be coprecipitated with the large subunit (using the antiserum raised against dTFIIE-L) and vice versa (data not shown).
Figure 3.
(A) Histidine-tagged dTFIIE-L and dTFIIE-S were expressed individually in bacteria, extracted, and purified as described. The two subunits were combined and renatured by stepwise dialysis. Soluble dTFIIE complex was subjected to SDS/PAGE and stained with Coomassie blue (lane 2). The mobility of the large (IIE-L) and small (IIE-S) subunits are indicated. The identity of the band directly above IIE-S is unknown and most likely a bacterial contaminant. (B) Western blot analyses of bacterially expressed untagged dTFIIE-L (lane 2), dTFIIE-S (lane 4), and Drosophila embryo NE (lanes 1 and 3). Blots were probed with antibodies raised against recombinant dTFIIE-L (lanes 1 and 2) and dTFIIE-S (lanes 3 and 4).
To determine if the two cDNAs encode full-length proteins, the sizes of the recombinant proteins were compared with those of dTFIIE from embryo NEs by Western blot analysis. For this analysis, we expressed untagged versions of dTFIIE-L and dTFIIE-S in bacteria. As it appears from Fig. 3B, recombinant dTFIIE-L and dTFIIE-S comigrate with the endogenous subunits, suggesting that the two cDNAs encode full-length proteins.
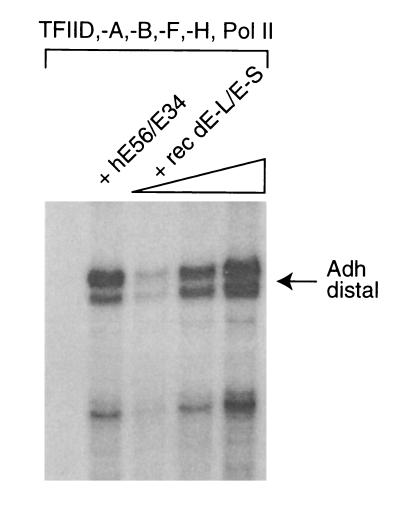
To test if the bacterially expressed dTFIIE was functional, we performed in vitro transcription experiments using a system reconstituted with highly purified dTFIIH, dPol II, recombinant dTFIIA, dTFIIB, dTFIIF, and partially purified dTFIID (15). Using this set of factors, transcription from the Drosophila Adh distal promoter is completely dependent on the addition of exogenous hTFIIE (Fig. 4, lanes 1 and 2). Moreover, recombinant dTFIIE can fully substitute for hTFIIE in mediating transcription, suggesting that the cDNAs presented here encode the Drosophila homologues of the two hTFIIE subunits.
Figure 4.
Reconstitution of transcription using recombinant dTFIIE. The transcription factors TFIID, -A, -B, -F, -H, and RNA pol II were included in all reactions. TFIIE was omitted in lane 1. Recombinant hTFIIE was included in lane 2. Increasing amounts of dTFIIE was included in lanes 3–5 (2 ng, 5 ng, and 60 ng of renatured dTFIIE shown in Fig. 3A). The Drosophila Adh distal promoter was used as a transcription template (supercoiled plasmid), and RNA synthesis was analyzed by primer extension analysis.
DISCUSSION
We have presented evidence for the identification of two Drosophila genes that encode both subunits of the transcription factor TFIIE. The isolated cDNAs reveal extensive amino acid sequence homology with the corresponding human TFIIE subunits, particularly within regions known to harbor important functional domains. In addition, we have reconstituted transcription in vitro using bacterially expressed dTFIIE in combination with other recombinant or purified endogenous Drosophila factors. Future analysis of dTFIIE, in vitro and in vivo, may help to uncover important aspects of the role of TFIIE in the transcriptional process.
The function of TFIIE is generally considered to be linked to that of TFIIH for several reasons. First, TFIIH contains carboxyl-terminal domain kinase activity that may be stimulated by TFIIE (38). Second, the role of TFIIE and TFIIH is related to the helical state of the template DNA, as both TFIIE and TFIIH are required for transcription from linear, but not supercoiled templates (5). However, in our transcription system we observe a requirement for TFIIE even when using supercoiled templates. This phenomenon may rely on the nature of the transcription system and, in particular, the use of TFIID rather than TATA-box binding protein (TBP). A requirement for TFIIE in the presence of TFIID has also been observed by Tyree et al. (39) and could suggests that TBP-associated factors in the TFIID complex may impose a requirement for TFIIE activity. Indeed, TFIIE has been reported to interact with TFIID, but the role of TBP-associated factors in this interaction is unknown (4). Future analyses of putative communication between TBP-associated factors and TFIIE may reveal novel properties of TFIIE in the transcriptional process.
Analyses of the role of TFIIE and TFIIH during transcription in vitro has suggested that TFIIE and TFIIH mediate the transition from an initiating to an elongating state of the Pol II enzyme (9). Importantly, this transition from a promoter bound to an elongating form of Pol II constitutes a critical step in the transcriptional regulation of several genes including the Drosophila Hsp70 gene (40). In vivo, the Hsp70 promoter contains an initiated but stalled polymerase that can be released upon heat shock. The release of Pol II correlates with carboxyl-terminal domain phosphorylation (41) that may be accomplished by carboxyl-terminal domain kinase activities present in TFIIH (6). Thus, TFIIE and TFIIH and in particular regulation of their activities may be implicated in controlling release of Pol II upon induction of a heat shock response. However, it is unknown whether TFIIE or TFIIH can serve as targets for heat shock regulation. Nevertheless, the possibility that dTFIIE may be implicated in transcriptional regulation can now be tested, and, more importantly, the potential to isolate specific alleles of dTFIIE that affect Hsp70 gene transcription might help elucidate the role of TFIIE in promoter clearance and transcriptional regulation.
In the light of a functional relationship between TFIIE and TFIIH, it has also been proposed that TFIIE may play a role in the coupling of transcription and DNA repair (4). In mammals, Drosophila, and yeast, several TFIIH subunits have been implicated in nucleotide excision repair (6). It would be of great interest to determine if TFIIE plays a role in conjunction with TFIIH either to reveal a DNA lesion and recruit the DNA repair machinery or to assist Pol II in resuming transcription after DNA repair. The potential of identifying specific alleles of dTFIIE may provide insights into this putative aspect of TFIIE function.
Acknowledgments
We would like to thank John Lis for cytological mapping studies and Karen Goodrich for DNA sequencing. We would also like to thank Mary Maxon, Cathy Thut, Tom O’Brien, and Jon Oliner for critical reading of the manuscript and various members of the Tjian lab for helpful discussions. This work was supported by a grant to W.Z. from the American Cancer Society (NP-800).
Footnotes
References
- 1.Zawel L, Reinberg D. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 2.Flores O, Lu H, Reinberg D. J Biol Chem. 1992;267:2786–2793. [PubMed] [Google Scholar]
- 3.Inostroza J, Flores O, Reinberg D. J Biol Chem. 1991;266:9304–9308. [PubMed] [Google Scholar]
- 4.Maxon M E, Goodrich J A, Tjian R. Genes Dev. 1994;8:515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- 5.Parvin J D, Sharp P A. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 6.Seroz T, Hwang J R, Moncollin V, Egly J M. Curr Opin Genet Dev. 1995;5:217–221. doi: 10.1016/0959-437x(95)80011-5. [DOI] [PubMed] [Google Scholar]
- 7.Holstege F C P, van der Vliet P C, Timmers H, Th M. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 8.Dvir A, Garrett K P, Chalut C, Egly J M, Conaway J W, Conaway R C. J Biol Chem. 1996;271:7245–7248. doi: 10.1074/jbc.271.13.7245. [DOI] [PubMed] [Google Scholar]
- 9.Goodrich J A, Tjian R. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 10.Zawel L, Kumar K P, Reinberg D. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 11.Price D H, Sluder A E, Greenleaf A L. J Biol Chem. 1987;262:3244–3255. [PubMed] [Google Scholar]
- 12.Wampler S L, Tyree C M, Kadonaga J T. J Biol Chem. 1990;265:21223–21231. [PubMed] [Google Scholar]
- 13.Dynlacht B D, Hoey T, Tjian R. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 14.Austin R J, Biggin M D. Proc Natl Acad Sci USA. 1996;93:5788–5792. doi: 10.1073/pnas.93.12.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen S K, Tjian R. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 16.Sauer F, Hansen S K, Tjian R. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 17.Falkenburg D, Dworniczak B, Faust D M, Bautz E F K. J Mol Biol. 1987;195:929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- 18.Jokerst R S, Weeks J R, Zehring W A, Greenleaf A L. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 19.Harrison D A, Mortin M A, Corces V G. Mol Cell Biol. 1992;12:928–935. doi: 10.1128/mcb.12.3.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton B J, Mortin M A, Greenleaf A L. Genetics. 1993;134:517–529. doi: 10.1093/genetics/134.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortin M A, Zuerner R, Berger S, Hamilton B J. Genetics. 1992;131:895–903. doi: 10.1093/genetics/131.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Weeks J, Mortin M A, Greenleaf A L. Mol Cell Biol. 1993;13:4214–4222. doi: 10.1128/mcb.13.7.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim W J, Burke L P, Mortin M A. J Mol Biol. 1994;244:12–22. doi: 10.1006/jmbi.1994.1700. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Chafin D, Price D H, Greenleaf A L. J Biol Chem. 1996;271:5993–5999. [PubMed] [Google Scholar]
- 25.Soeller W C, Marshall N F, Kornberg T. Genes Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- 26.Wang X. Ph.D. thesis. Detroit: Wayne State Univ. School of Medicine; 1996. [Google Scholar]
- 27.Kephart D D, Marshall N F, Price D H. Mol Cell Biol. 1992;12:2067–2077. doi: 10.1128/mcb.12.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zehring W A, Lee J M, Weeks J R, Jokerst R S, Greenleaf A L. Proc Natl Acad Sci USA. 1988;85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sluder A E, Price D H, Greenleaf A L. Biochimie. 1987;69:1199–1205. doi: 10.1016/0300-9084(87)90147-7. [DOI] [PubMed] [Google Scholar]
- 30.Studier F W, Rosenberg A, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 31.Ohkuma Y, Sumimoto H, Hoffman A, Shimasaki S, Horikoshi M, Roeder R G. Nature (London) 1991;354:398–401. doi: 10.1038/354398a0. [DOI] [PubMed] [Google Scholar]
- 32.Ohkuma Y, Hashimoto S, Roeder R G, Horikoshi M. Nucleic Acids Res. 1992;20:5838. doi: 10.1093/nar/20.21.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson M G, Inostroza J, Maxon M E, Flores O, Admon A, Reinberg D, Tjian R. Nature (London) 1991;354:369–373. doi: 10.1038/354369a0. [DOI] [PubMed] [Google Scholar]
- 34.Ohkuma Y, Hashimoto S, Wang C K, Horikoshi M, Roeder R G. Mol Cell Biol. 1995;15:4856–4866. doi: 10.1128/mcb.15.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maxon M E, Tjian R. Proc Natl Acad Sci USA. 1994;91:9529–9533. doi: 10.1073/pnas.91.20.9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sumimoto H, Okhuma Y, Sinn E, Kato H, Shimasaki S, Horikoshi M, Roeder R G. Nature (London) 1991;354:401–404. doi: 10.1038/354401a0. [DOI] [PubMed] [Google Scholar]
- 37.Okhuma Y, Hashimoto S, Roeder R G, Horikoshi M. Nucleic Acids Res. 1992;20:4363. doi: 10.1093/nar/20.16.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu H, Zawel L, Fisher L, Egly J M, Reinberg D. Nature (London) 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 39.Tyree C M, George C P, Lira-DeVito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J T. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 40.Fernandez M, O’Brien T, Lis J T. In: The Biology of Heat Shock Proteins and Molecular Chaperones. Morimoto R I, Tissieres A, Georgopoulos C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 375–393. [Google Scholar]
- 41.O’Brien T, Hardin S, Greenleaf A, Lis J T. Nature (London) 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]