Abstract
The mitogen-activated protein kinase (MAPK) cascade plays a crucial role in the transduction of extracellular signals into responses governing growth and differentiation. The effects of a specific inhibitor of the MAPK kinase (MEK)/MAPK pathway (PD98059) on nerve growth factor (NGF)-induced growth arrest and inhibition of cell cycle-dependent kinases (CDKs) have been examined. Treatment of NIH 3T3 cells expressing TRKA with PD98059 dramatically reversed the complete inhibition of growth of these cells caused by NGF. PD98059 also blocked the ability of NGF to inhibit the activities of CDK4 and CDK2, while partially preventing NGF induction of p21Cip1/WAF1. To independently evaluate the involvement of the MEK/MAPK pathway in growth arrest, an inducible activated form of the Raf-1 protooncogene (ΔRAF-1:ER) was expressed in these cells. Activation of ΔRAF-1:ER resulted in a prolonged increase in MAPK activity and growth arrest of these cells, with concomitant induction of p21Cip1/WAF1 and inhibition of CDK2 activity. These effects of ΔRAF-1:ER activation were all reversed by treatment of cells with PD98059. These data indicate that in addition to functioning as a positive effector of growth, stimulation of the MEK/MAPK pathway can result in an inhibition of CDK activity and cell cycle arrest.
Keywords: nerve growth factor, MEK inhibitor PD98059, p21Cip1/WAF1, cell cycle-dependent kinase
The mitogen-activated protein kinase (MAPK) family of serine/threonine protein kinases are involved in a wide range of cellular functions. The binding of GTP to the Ras protein initiates a protein kinase cascade, which leads to MAPK activation through the intervening protein kinases Raf-1 and MEK (1). MAPK activity is elevated in response to proliferative factors such as epidermal growth factor (EGF) (2) and platelet-derived growth factor (3), as well as in response to differentiative factors such as nerve growth factor (NGF) (4). MAPK is known to phosphorylate numerous cytosolic substrates, including the EGF receptor (5), pp90Rsk (6), and phospholipase A2 (7). Following activation, MAPK is translocated to the nucleus (8), where it can phosphorylate transcription factors such as Elk-1, leading to increased transcription of c-Fos (9, 10). A large number of oncogenes share the activation of MAPK as a common pathway (1). Furthermore, constitutive activation of the upstream activator of MAPK, MEK1, results in cellular transformation and increased DNA synthesis in some cells (1, 11). These findings argue for a key role of MAPK in the proliferative process.
A primary interest in growth factor research has been to determine the molecular mechanisms that dictate the specificity of growth factor-receptor function. Many initial cellular responses such as activation of PI3-kinase, tyrosine phosphorylation of SHC proteins, GTP loading of Ras, and the downstream activation of MAPK are shared by NGF and EGF (12). In PC12 cells MAPK activity stimulated by NGF is more sustained relative to EGF (20, 44). The prolonged activation in this cell type correlates well with the NGF-induced survival and differentiation. NGF suppresses the activity of cyclin-dependent kinases (CDKs) in several cell types (13, 14) and this inhibition of CDK activity appears to be critical in the ability of NGF to induce differentiation and act as a survival factor (15). In this report, data are presented that directly link activation of the MAPK pathway to the cell cycle machinery. A novel paradigm is described in which the MEK/MAPK pathway negatively regulates CDK activity and mediates cell cycle arrest.
MATERIALS and METHODS
Cell Culture and Cell Lines.
NIH 3T3 cells expressing human trkA (TRK1) have been described (14) and were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% newborn calf serum and 40 units/ml hygromycin. To generate a high-titer retrovirus coding for ΔRaf-1:ER, Phoenix-A amphotrophic packaging cells (Gary P. Nolan, Stanford University, Palo Alto, CA) were transfected with the retroviral construct pBP3ΔRaf-1:ER (E. Bosch, D. Woods, and M. McMahon, unpublished work). This construct, a derivative of pBabe-puro (16) places the expression of ΔRaf-1:ER (17) under control of the murine leukemia virus promoter with puromycin selection driven by the simian virus 40 early region. Supernatants at 48 hr after transfection were used to infect TRK1 cells in the presence of 6 μg hexadimethrine bromide (Sigma). Forty-eight hours after infection, cells were split into phenol red-free DMEM supplemented with 40 units/ml hygromycin and 5% newborn calf serum, which had been absorbed by treatment with 1 g acid-washed, activated charcoal per 50 ml of serum. Puromycin (Sigma) was added at 1 μg/ml and cells were selected for 5 days. Puromycin-resistant colonies were pooled and the pooled population designated TRK:ER cells. Expression was confirmed by Western blotting with anti-estrogen receptor antibody from Santa Cruz Biotechnology (SC-543).
Assay of Cellular Proliferation.
TRK1 or TRK:ER cells were seeded at 1 × 104 cells per 35-mm plate and grown for 4 days in normal growth medium or medium supplemented as described. At 4 days after plating cells were harvested by trypsinization and counted microscopically using a hemocytometer. All experiments were counted in duplicate and data represent at least two independent experiments.
MAPK Kinase Activity.
TRK1 or TRK:ER cells were plated in 60-mm dishes (1.5 × 106 cells per dish) and treated as described for 16 hr, followed by washing with ice-cold phosphate-buffered saline and lysis in 1 ml of MIPA buffer (50 mM Hepes, pH 7.5/1% Nonidet P-40/0.5% deoxycholate/150 mM NaCl/50 mM NaF/1 mM para-nitrophenyl phosphate/1 mM Na3VO4/10 μg/ml aprotinin/10 μg/ml leupeptin/1 mM benzamidine). Lysates were clarified by centrifugation and incubated with nonimmune rabbit serum or 1 μg each of polyclonal antibodies to Erk1 and Erk2 (Santa Cruz Biotechnology; SC-093 and SC-154, respectively). Immune complexes were bound to protein A-Sepharose beads and washed two times with MIPA buffer and one time with kinase wash buffer (25 mM Hepes, pH 7.4/10 mM MgCl2/1 mM EGTA). A kinase mix consisting of Hepes (pH 8.0), 10 mM MgCl2, 1 mM dithiothreitol, 50 μM ATP (10 μCi [γ-32P] ATP per reaction; 1 Ci = 37 GBq) and 0.3 mg/ml myelin basic protein, was added to each sample. After 10 min, reactions were stopped by the addition of SDS sample buffer, boiled, and separated by SDS/PAGE. Gels were fixed and dried, and phosphorylated myelin basic protein was visualized by autoradiography. The extent of phosphorylation was quantitated using a PhosphorImager (Molecular Dynamics). Equivalent precipitation of MAPK was routinely evaluated using anti-MAPK polyclonal antibodies (a generous gift of A. Saltiel, Parke–Davis, Ann Arbor, MI).
CDK Activity.
Cells were grown in 100-mm culture dishes and treated for 16 hr with either vehicle, NGF, or a combination of NGF and the MEK inhibitor PD98059, as indicated. CDK activities were measured following lysis in 1 ml of buffer comprised of 20 mM Hepes (pH 7.4), 150 mM NaCl, 0.5% Nonidet P-40, 10 μg/ml aprotinin, and 10 μg/ml leupeptin. Clarified extracts were incubated with 1.5 μg of anti-CDK2 antibody (Santa Cruz Biotechnology; SC-163) or 1.5 μg of anti-CDK4 antibody (Santa Cruz Biotechnology; SC-260) and immune complexes were recovered with protein A-Sepharose, washed twice with lysis buffer and once with kinase buffer (25 mM Hepes, pH 7.4/10 mM MgCl2/1 mM EGTA). Pellets were resuspended in 25 μl of kinase buffer containing either 4 μg histone H1 (Boehringer Mannheim) or glutathione S-transferase-Rb (aa 379–928) to measure CDK2 and CDK4 activity, respectively. Reactions were terminated at 10 min by the addition of sample buffer and boiling. Kinase reactions were analyzed by SDS/PAGE and dried gels were exposed to film to visualize proteins. Reactions were quantified using a PhosphorImager (Molecular Dynamics). Equal amounts of CDKs were precipitated in each experiment as determined by Western blot analysis (not shown).
Western Blotting.
TRK1 or TRK:ER cells growing in 35-mm dishes were treated as described for 16 hr. Cells were washed twice with 2 ml of phosphate buffered saline (pH 7.4) prior to lysis in 100 μl of sample buffer and immediate boiling. Protein (20 μg) was separated by SDS/PAGE and transferred to nitrocellulose, prior to probing with the appropriate antibodies. Antibodies to p16, p27Kip, cyclin A, cyclin D, cyclin E, cyclin H, and CDK7 were from Santa Cruz Biotechnology and were used at a dilution of 1:250–500. Antibody to p21Cip1/WAF1 was obtained from PharMingen and used at a dilution of 1:1000. Bound antibodies were visualized with [125I]-protein-A followed by autoradiography.
RESULTS
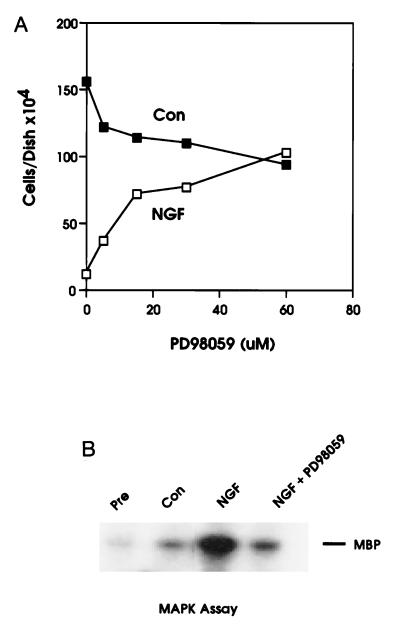
We have previously reported that NGF induces cell cycle arrest at the G1 stage in an NIH 3T3 cell line expressing trkA (TRK1 cells) (14). NGF treatment of these cells also resulted in a sustained activation of MAPK (14). To investigate the role of the MEK/MAPK pathway in NGF-dependent growth arrest, we examined the effects of a specific inhibitor of MEK, PD98059 (18), on this process. The MEK inhibitor PD98059 has shown exquisite specificity toward MEK1 relative to all other serine/threonine and tyrosine kinases assayed to date, including the closely related family members MKK3 and MKK4 (19). Surprisingly, addition of PD98059 to TRK1 cells cultured in the presence of NGF brought about a substantial, dose-dependent reversal of the NGF-induced inhibition of growth (Fig. 1). PD98059 partially inhibited the serum-dependent growth of control cells. NGF-stimulated MAPK activity was inhibited 85–95% by treatment of cells with PD98059 (Fig. 1B).
Figure 1.
Reversal of NGF-mediated growth arrest by the MEK-specific inhibitor PD98059. (A) Treatment with PD98059 produces a dose-dependent reversal of the growth arrest induced by NGF treatment. TRK1 cells were grown for 4 days in normal growth medium (▪) or growth medium supplemented with 20 nM NGF added (□). Vehicle [dimethyl sulfoxide (DMSO), 0.2%] or increasing concentrations of PD98059 were included throughout the 4-day incubation as indicated. Cell numbers represent the average of duplicate determinations. Data are representative of two independent experiments. (B) PD98059 blocks NGF stimulation of MAPK activity. Cells were grown in complete growth medium in the presence or absence of 20 nM NGF (as indicated) for 16 hr following addition of vehicle (DMSO, 0.2%) or 40 μM PD98059.
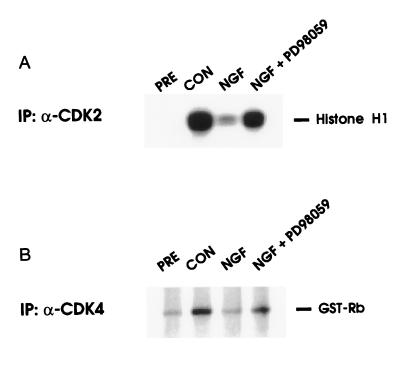
Since NGF has been shown to inhibit CDK2 and CDK4 activity in TRK1 cells (14) and PC12 cells (13), we tested whether the MEK/MAPK pathway might regulate this effect, again using the MEK inhibitor (Fig. 2). NGF alone almost completely inhibits CDK2 and CDK4 activity in TRK1 cells growing in normal serum containing medium (14). Addition of MEK inhibitor to cells reversed the NGF inhibition, with CDK2 and CDK4 activity returning to approximately 50% of control levels, suggesting that the MEK/MAPK pathway plays a significant role in capacity of NGF to suppress the activities of CDK2 and CDK4.
Figure 2.
MEK inhibitor PD09059 prevents inhibition of CDK by NGF. TRK1 cells were grown for 16 hr in normal growth medium in the presence or absence of 20 nM NGF. Simultaneous to the addition of NGF, cells were treated with vehicle (DMSO, 0.2%) or 40 μM PD98059. The activity of the CDKs was assayed by immunoprecipitation kinase assay (12), using histone H1 (A) or glutathione S-transferase-Rb (aa 379–928) (B) as substrates. Similar results were obtained in at least four independent experiments.
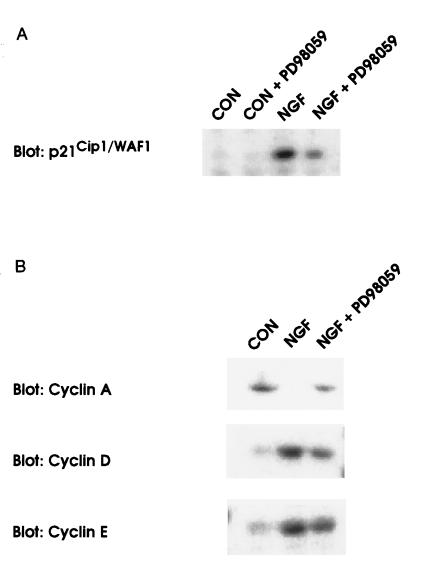
NGF has been shown to increase levels of the CDK inhibitor protein p21Cip1/WAF1 in PC12 cells (13) and TRK1 cells (14). To investigate the mechanism through which the MEK/MAPK pathway might inhibit CDK activity, the effect of PD98059 on the NGF-dependent increase in p21Cip1/WAF1 protein levels was determined. As shown in Fig. 3, treatment of TRK1 cells with the MEK inhibitor caused an approximate 50% reduction in the levels of p21Cip1/WAF1 protein induced by NGF. Levels of the CDK inhibitors p16INK4 (21) and p27KIP1 (22) were not effected by NGF or PD98059 treatment (not shown). The effects of PD98059 on levels of some of the other cell cycle components known to be involved in the regulation of CDK4 or CDK2 activity are also shown (Fig. 3). PD98059 treatment of cells prevented the elevation of cyclin D1 and cyclin E levels caused by NGF and dramatically reversed the down-regulation of cyclin A protein levels found in response to NGF treatment. Levels of CDK2, CDK4, CDK7, and cyclin H were not appreciably affected by NGF or PD98059 treatment (not shown).
Figure 3.
Effects of the MEK inhibitor, PD98059, on NGF-mediated p21Cip1/WAF1 induction and changes in cyclin expression. Levels of p21Cip1/WAF1 (A) or cyclin proteins involved in the activation of CDK2 and CDK4 (B) were analyzed by Western blot analysis of whole cell lysates (20) following treatment with NGF (20 nM) or with a combination of NGF and the MEK inhibitor, PD98059 (40 μM) for 16 hr. Similar results were found in at least three independent experiments.
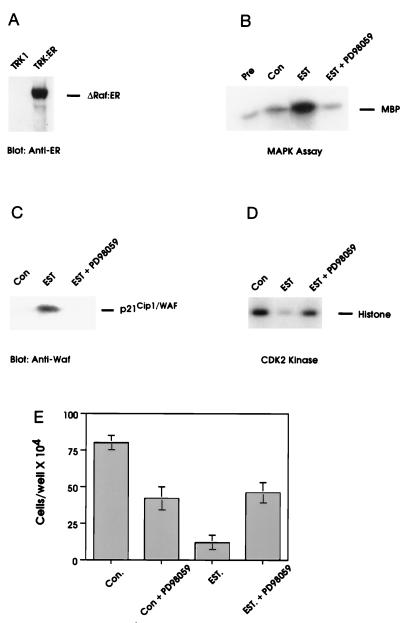
The persistent elevation of MAPK activity caused by NGF and the pronounced effects of the MEK inhibitor on NGF action suggested that the MEK/MAPK pathway was necessary for the cell cycle arrest mediated by NGF. We therefore sought to investigate whether the MEK/MAPK pathway was sufficient to induce cell cycle arrest. The kinase domain of the Raf-1 protooncogene (an upstream activator of MEK) fused to the estrogen binding domain of the estrogen receptor (ΔRaf-1:ER) is an estrogen-inducible form of Raf-1, through which MAPK activity can be regulated (17). We expressed ΔRaf-1:ER in TRK1 cells (TRK:ER cells, Fig. 4A) and examined the effects of estrogen treatment on MAPK activity and cellular proliferation. Treatment of TRK:ER cells with estradiol resulted in a prolonged activation of MAPK activity (Fig. 4B). In conjunction, estrogen treatment elevated levels of p21Cip1/WAF1 protein (Fig. 4C), and inhibited CDK2 activity and cell proliferation (Fig. 4 D and E). Estrogen had no effect on the growth of parental TRK1 cells (not shown). The estrogen-dependent MAPK activity, p21Cip1/WAF1 induction, CDK2 inhibition, and growth arrest could all be reversed by the addition of the MEK inhibitor, PD98059 (Fig. 4 B–E).
Figure 4.
Activation of the MEK/MAPK pathway by ΔRaf-1:ER results in p21Cip1/WAF1 induction, inhibition of CDK2 activity, and growth arrest. (A) TRK:ER cells were analyzed for the expression of ΔRaf-1:ER protein by blotting with anti-estrogen receptor antibody. (B–D) TRK:ER cells were pretreated for 5 min with either DMSO (0.1%) or PD98059 (30 μM) prior to addition of vehicle (0.1% ethanol) or 1 μM 17-β-estradiol for 16 hr. Cells were then analyzed for MAPK activity (B), whole cell levels of p21Cip1/WAF1 (C), and CDK2 activity (D) as described. (E) TRK:ER cells were seeded at 1 × 104 cells per dish and pretreated for 5 min with either vehicle (0.1% DMSO) or 30 μM PD98059, prior to the addition of 0.1% ethanol or 1 μM 17-β-estradiol for 4 days. Cells were counted in duplicate using a hemocytometer. For all data shown, similar results were obtained in at least three independent experiments.
DISCUSSION
Although the MEK/MAPK pathway has been shown to function in the stimulation of cellular proliferation (1, 11, 23, 24), we provide evidence that a sustained increase in MAPK activity can lead to inhibition of CDK activity and growth arrest. These effects may be mediated through induction of the CDK inhibitor p21Cip1/WAF1, although a significant amount of p21Cip1/WAF1 remains in PD98059 treated cells, which have overcome NGF-dependent cell cycle block. Inhibition by p21Cip1/WAF1 has been suggested to occur by a stoichiometric mechanism in which low levels may activate CDK activity and high levels inhibit the same (25). If so, the 50% reduction in p21Cip1/WAF1 protein levels caused by PD98059 treatment could be sufficient to allow recovery of CDK activity and cell cycle progression. Whereas we cannot rule out that the MEK/MAPK pathway-mediated reduction of cyclin A levels is the primary event in NGF-dependent growth arrest, it seems likely that this is a secondary event, as recent data suggests that the G1/S transition is necessary for the synthesis of cyclin A (26). In any case, NGF-dependent up-regulation of p21Cip1/WAF1 protein appears to be only partially mediated through the MEK/MAPK pathway. In contrast, p21Cip1/WAF1 induction in response to ΔRaf-1:ER activation is completely sensitive to inhibition of MEK. This is the first example of long-term induction of p21Cip1/WAF1 protein through stimulation of the MEK/MAPK pathway.
A role for the MEK/MAPK pathway in growth arrest or cellular differentiation is not unprecedented. MAPK is critical for differentiation of R7 photoreceptor cells from immature precursor cells in Drosophila (27) and for maturation of vulval precursor cells in Caenorhabditis elegans (28). In addition, it has been shown that MAPK is necessary for differentiation of thymocytes, but not thymocyte proliferation (29). There is also a striking parallel between the NGF-dependent pathway for cell cycle arrest described here and the yeast pheromone pathway (30). As for NGF, pheromone treatment results in cell cycle arrest at G1, mediated by yeast homologues of MEK and MAPK (Ste7 and Fus3/Kss1, respectively, in Saccharomyces cerevisiae). Yeast CDK activity is inhibited through the action of a transcriptionally regulated CDK inhibitor protein (Far1) (31). Furthermore, in studies examining cooperativity between oncogenes, it was found that introduction of the v-Ras oncogene into REF52 rat cells (32) or Schwann cells (33) resulted in growth arrest and that a second “nuclear” oncogene, such as Myc was necessary for cellular transformation (33, 34). Similarly, activation of ΔRaf-1:ER in C2-3T3 cells caused “morphological transformation,” but actually inhibited growth factor-stimulated mitogenesis (35). Numerous other studies have shown that expression of oncogenic Ras or Raf can transform or confer a growth advantage to cells without concomitant elevation of MAPK activity (36–38).
In contrast, treatment of cells with most mitogenic stimuli results in a transient, perhaps biphasic, increase in MAPK activity (1). In some instances, this type of increase appears to be required for mitogenesis (23, 39). Constitutive activation of the MAPK pathway has been shown to occur in some cell types transformed by the Ras or Raf oncogenes (36, 40, 41). Our data indicating that the MEK inhibitor PD98059 partially inhibits the growth of TRK1 cells in serum containing medium also supports a role for the MEK/MAPK pathway in proliferation.
These data can be reconciled in part if the ultimate cellular response (i.e., cell cycle arrest versus cellular proliferation) to activation of the MEK/MAPK pathway depends on the strength and duration of the MAPK signal. Transient or cyclical activation may contribute to cell cycle progression, whereas sustained high levels of activity may result in cell cycle arrest. Lack of cell cycle arrest in some tumor cell lines exhibiting “constitutive” activation of MAPK could be due to relatively low levels of chronic MAPK activity or to some compensatory mechanism that circumvents the MAPK-induced cell cycle block such as deregulation of expression of c-myc (as discussed above).
There are several plausible contexts in which MEK/MAPK-mediated cell cycle arrest might be of physiologically relevant. (i) It could act to sense inappropriate signaling from the Ras pathway serving as a dominant inhibitory signal to preclude cell division. (ii) The MEK/MAPK pathway may contribute to G1 arrest in response to ionizing radiation since ionizing radiation has recently been shown to cause activation of Raf-1 activity (42). (iii) In the PC12 cell model, NGF-induced neuronal differentiation is accompanied by a prolonged increase in MAPK activity (4) and by inhibition of CDK activities (13) much as we have found for TRK1 cells treated with NGF. Activation of the MEK/MAPK pathway may be required for the cell cycle arrest accompanying terminal differentiation of PC12 cells. The general applicability of this paradigm to mammalian cell differentiation remains to be determined. (iv) The MEK/MAPK pathway may also regulate apoptosis in some cell types since expression of constitutively activated MEK protects PC12 cells from apoptosis following NGF withdrawal (43). This protection may involve inhibition of CDK activity because CDK inhibitors can also protect against apoptosis due to NGF withdrawal (15). Future experiments will be directed toward testing these hypotheses.
Acknowledgments
We thank M. McMahon for plasmid pBPΔRAF-1:ER, G. Nolan for Phoenix packaging cells, E. Harlow for glutathione S-transferase-Rb, and A. Saltiel and A. Fattaey for helpful discussions. K.M.P. is supported by a National Research Service Award fellowship from the National Institutes of Health.
Footnotes
Abbreviations: MAPK, mitogen-activated protein kinase; EGF, epidermal growth factor; NGF, nerve growth factor; CDK, cell cycle-dependent kinase.
References
- 1.Waskiewicz A J, Cooper J A. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 2.Ahn N G, Weiel J E, Chan C P, Krebs E G. J Biol Chem. 1990;265:11487–11494. [PubMed] [Google Scholar]
- 3.Rossomando A J, Payne D M, Weber M J, Sturgill T W. Proc Natl Acad Sci USA. 1989;86:6940–6943. doi: 10.1073/pnas.86.18.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyasaka T, Chao M V, Sherline P, Saltiel A R. J Biol Chem. 1990;265:4730–4735. [PubMed] [Google Scholar]
- 5.Takishima K, Griswold-Prenner I, Ingebritsen T, Rosner M R. Proc Natl Acad Sci USA. 1991;88:2520–2524. doi: 10.1073/pnas.88.6.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sturgill T W, Ray L B, Erikson E, Maller J L. Nature (London) 1988;334:715–718. doi: 10.1038/334715a0. [DOI] [PubMed] [Google Scholar]
- 7.Lin L L, Wartmann M, Lin A Y, Knopf J L, Seth A, Davis R J. Cell. 1993;72:269–278. doi: 10.1016/0092-8674(93)90666-e. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez F A, Seth A, Raden D L, Bowman D S, Fay F S, Davis R J. J Cell Biol. 1993;122:1089–1101. doi: 10.1083/jcb.122.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb M H, Shaw P E. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Treisman R. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 11.Marshall C J. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 12.Schlessinger J. Curr Opin Genet Dev. 1994;4:25–30. doi: 10.1016/0959-437x(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 13.Yan G Z, Ziff E B. J Neurosci. 1995;15:6200–6212. doi: 10.1523/JNEUROSCI.15-09-06200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decker S J. J Biol Chem. 1995;270:30841–30844. doi: 10.1074/jbc.270.52.30841. [DOI] [PubMed] [Google Scholar]
- 15.Park D S, Farninelli S E, Greene L A. J Biol Chem. 1996;271:8161–8169. doi: 10.1074/jbc.271.14.8161. [DOI] [PubMed] [Google Scholar]
- 16.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuels M L, Weber M J, Bishop J M, McMahon M. Mol Cell Biol. 1993;13:6241–6251. doi: 10.1128/mcb.13.10.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudley D T, Pang L, Decker S J, Bridges A J, Saltiel A R. Proc Natl Acad Sci USA. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alessi D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 20.Traverse S, Gomez N, Paterson H, Marshall C, Cohen P. Biochem J. 1992;288:351–355. doi: 10.1042/bj2880351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamb A, Gruis N A, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian S V, Stockert E, Day R S, Johnson B E, Skolnick M H. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 22.Polyak K, Kato J Y, Solomon M J, Sherr C J, Massague J, Roberts J M, Koff A. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 23.Pages G, Lenormand P, L’Allemain G, Chambard J C, Meloche S, Pouyssegur J. Proc Natl Acad Sci USA. 1993;90:8319–8323. doi: 10.1073/pnas.90.18.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowley S, Paterson H, Kemp P, Marshall C J. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Hannon G J, Beach D. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- 26.Huet X, Rech J, Plet A, Vie’ A, Blanchard J M. Mol Cell Biol. 1996;16:3789–3798. doi: 10.1128/mcb.16.7.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perrimon N. Cell. 1993;74:219–222. doi: 10.1016/0092-8674(93)90412-j. [DOI] [PubMed] [Google Scholar]
- 28.Dickson B, Hafen E. Curr Opin Genet Dev. 1994;4:64–70. doi: 10.1016/0959-437x(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 29.Alberola-Ila J, Forbush K A, Seger R, Krebs E G, Perlmutter R M. Nature (London) 1995;373:620–623. doi: 10.1038/373620a0. [DOI] [PubMed] [Google Scholar]
- 30.Herskowitz I. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 31.Oehelen L J W M, McKinney J D, Cross F R. Mol Cell Biol. 1996;16:2830–2837. doi: 10.1128/mcb.16.6.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohl N E, Ruley H E. Oncogene. 1987;2:41–48. [PubMed] [Google Scholar]
- 33.Ridley A J, Paterson H F, Noble M, Land H. EMBO J. 1988;7:1635–1645. doi: 10.1002/j.1460-2075.1988.tb02990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Land H, Parada L F, Weinberg R A. Nature (London) 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 35.Pritchard C A, Samuels M L, Bosch E, McMahon M. Mol Cell Biol. 1995;15:6430–6432. doi: 10.1128/mcb.15.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallego C, Gupta S K, Heasley L E, Qian N X, Johnson G L. Proc Natl Acad Sci USA. 1992;89:7355–7359. doi: 10.1073/pnas.89.16.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buscher D, Dello-Sbarbra P, Hipskind R A, Rapp U R, Stanley E R, Baccarini M. Oncogene. 1993;8:3323–3332. [PubMed] [Google Scholar]
- 38.Krautwald S, Buscher D, Dent P, Ruthenberg K, Baccarini M. Oncogene. 1995;10:1187–1192. [PubMed] [Google Scholar]
- 39.Kelleher M D, Abe M K, Chao T S, Jain M, Green J M, Solway J, Rosner M R. Am J Physiol. 1995;268:894–901. doi: 10.1152/ajplung.1995.268.6.L894. [DOI] [PubMed] [Google Scholar]
- 40.Kyriakis J M, App H, Zhang X F, Banerjee P, Brautigan D L, Rapp U L, Avruch J. Nature (London) 1992;358:417–421. doi: 10.1038/358417a0. [DOI] [PubMed] [Google Scholar]
- 41.Dent P, Haser W, Haystead T A J, Vincent L, Roberts T M, Sturgill T W. Science. 1992;257:1404–1407. doi: 10.1126/science.1326789. [DOI] [PubMed] [Google Scholar]
- 42.Kasid U, Suy S, Dent P, Ray S, Whiteside T L, Sturgill T W. Nature (London) 1996;382:813–816. doi: 10.1038/382813a0. [DOI] [PubMed] [Google Scholar]
- 43.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 44.Heasley L E, Johnson G L. Mol Biol Cell. 1992;3:545–553. doi: 10.1091/mbc.3.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]