Abstract
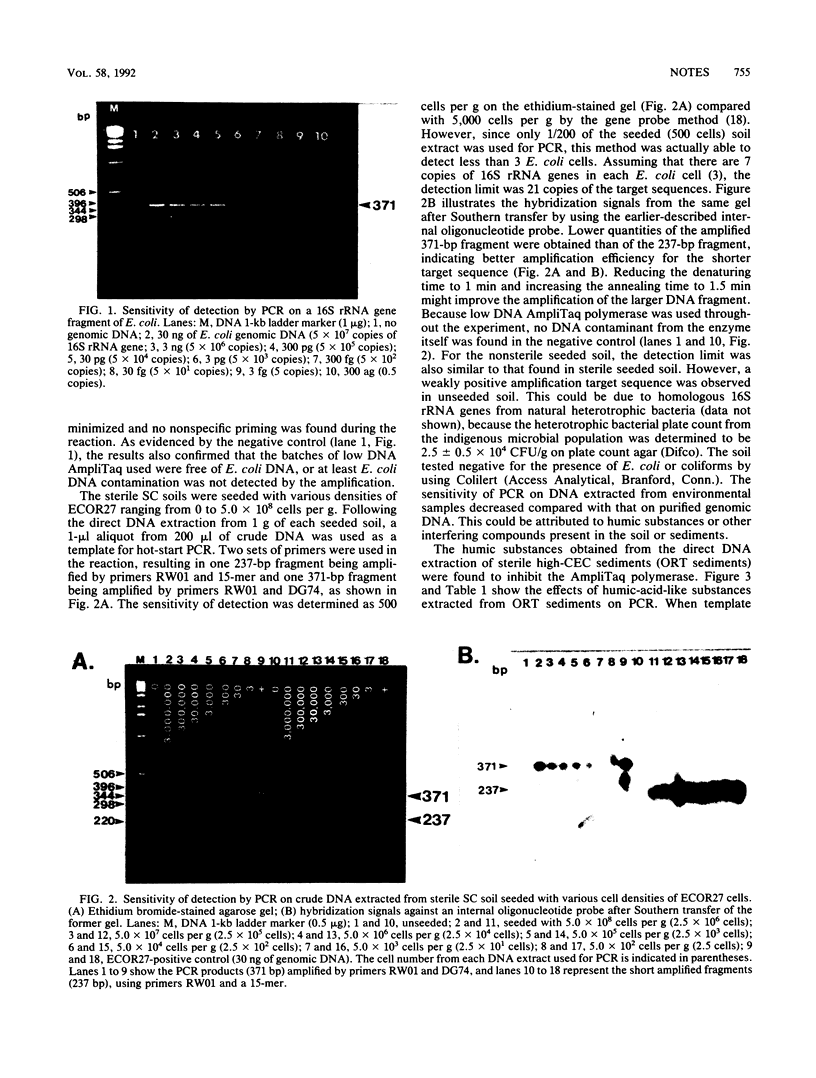
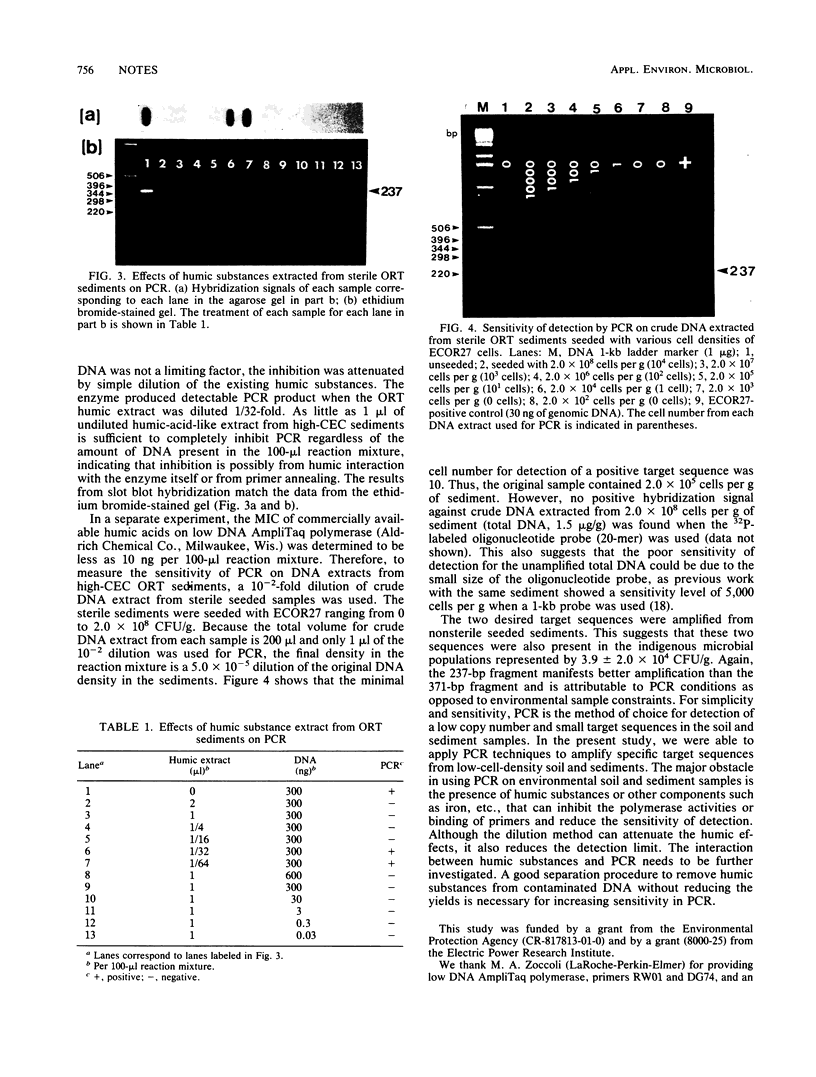
Polymerase chain reaction was used to amplify the low copy number of two 16S ribosomal gene fragments from soil and sediment extracts. Total DNA for polymerase chain reaction was extracted from 1 g of seeded or unseeded samples by a rapid freeze-and-thaw method. Amplified DNA fragments can be detected in DNA fractions isolated from seeded soil containing less than 3 Escherichia coli cells and from seeded sediments containing less than 10 cells. This research demonstrated that coupling polymerase chain reaction to direct DNA extraction improves sensitivity by 1 and 2 orders of magnitude for sediments and soils, respectively. This technique could become a powerful tool for genetic ecology studies.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bej A. K., Mahbubani M. H., Atlas R. M. Detection of viable Legionella pneumophila in water by polymerase chain reaction and gene probe methods. Appl Environ Microbiol. 1991 Feb;57(2):597–600. doi: 10.1128/aem.57.2.597-600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bej A. K., Steffan R. J., DiCesare J., Haff L., Atlas R. M. Detection of coliform bacteria in water by polymerase chain reaction and gene probes. Appl Environ Microbiol. 1990 Feb;56(2):307–314. doi: 10.1128/aem.56.2.307-314.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- D'Aquila R. T., Bechtel L. J., Videler J. A., Eron J. J., Gorczyca P., Kaplan J. C. Maximizing sensitivity and specificity of PCR by pre-amplification heating. Nucleic Acids Res. 1991 Jul 11;19(13):3749–3749. doi: 10.1093/nar/19.13.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmler G. J., Buffone G. J., Schimbor C. M., May R. A. Detection of cytomegalovirus in urine from newborns by using polymerase chain reaction DNA amplification. J Infect Dis. 1988 Dec;158(6):1177–1184. doi: 10.1093/infdis/158.6.1177. [DOI] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Miller R. H., Feinstone S. M., Unoura M., Kobayashi K., Hattori N., Purcell R. H. Detection of serum hepatitis B virus DNA in patients with chronic hepatitis using the polymerase chain reaction assay. Proc Natl Acad Sci U S A. 1989 Jan;86(1):312–316. doi: 10.1073/pnas.86.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawyer F. C., Stoffel S., Saiki R. K., Myambo K., Drummond R., Gelfand D. H. Isolation, characterization, and expression in Escherichia coli of the DNA polymerase gene from Thermus aquaticus. J Biol Chem. 1989 Apr 15;264(11):6427–6437. [PubMed] [Google Scholar]
- Mahbubani M. H., Bej A. K., Miller R. D., Atlas R. M., DiCesare J. L., Haff L. A. Detection of bacterial mRNA using polymerase chain reaction. Biotechniques. 1991 Jan;10(1):48–49. [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Effects of Hg, CH(3)-Hg, and Temperature on the Expression of Mercury Resistance Genes in Environmental Bacteria. Appl Environ Microbiol. 1990 Nov;56(11):3266–3272. doi: 10.1128/aem.56.11.3266-3272.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991 Apr;57(4):1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernars K., Delfgou E., Soentoro P. S., Notermans S. Successful approach for detection of low numbers of enterotoxigenic Escherichia coli in minced meat by using the polymerase chain reaction. Appl Environ Microbiol. 1991 Jul;57(7):1914–1919. doi: 10.1128/aem.57.7.1914-1919.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]






