Abstract
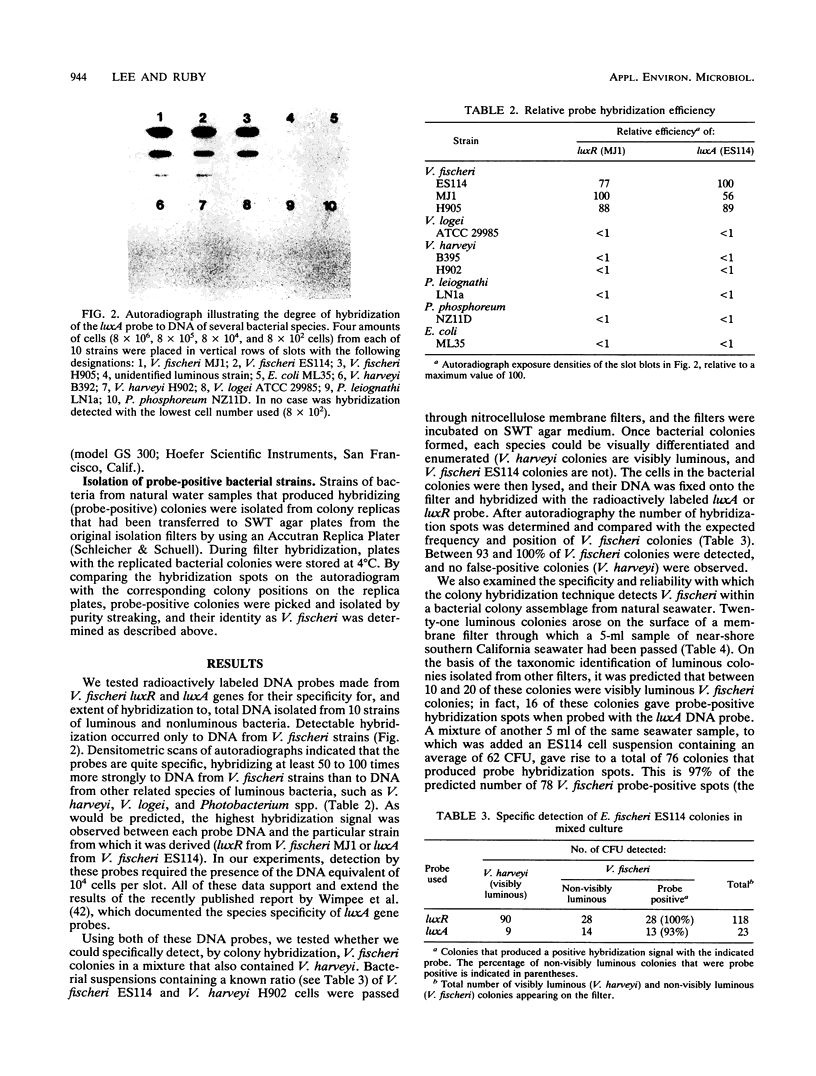
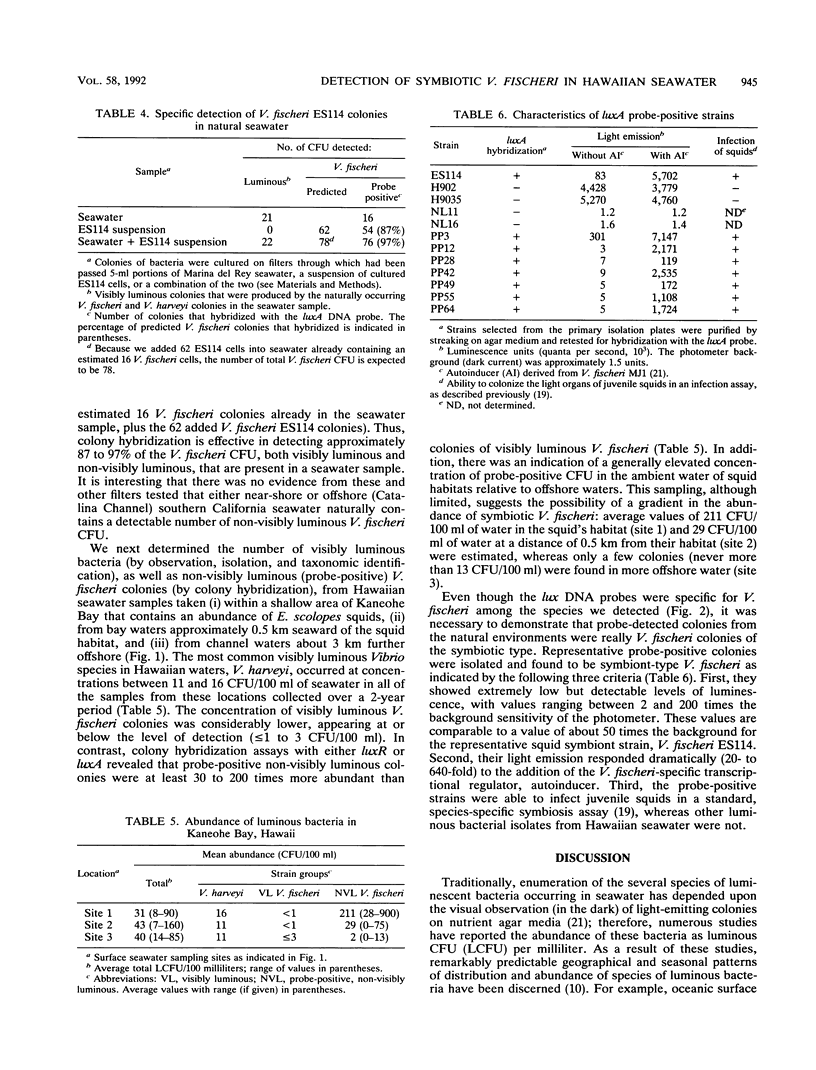
Symbiotic bacteria that inhabit the light-emitting organ of the Hawaiian squid Euprymna scolopes are distinctive from typical Vibrio fischeri organisms in that they are not visibly luminous when grown in laboratory culture. Therefore, the abundance of these bacteria in seawater samples cannot be estimated simply by identifying them among luminous colonies that arise on nutrient agar plates. Instead, we have used luxR and polymerase chain reaction generated luxA gene probes to identify both luminous and non-visibly luminous V. fischeri colonies by DNA-DNA hybridization. The probes were specific, hybridizing at least 50 to 100 times more strongly to immobilized DNAs from V. fischeri strains than to those of pure cultures of other related species. Thus, even non-visibly luminous V. fischeri colonies could be identified among colonies obtained from natural seawater samples by their probe-positive reaction. Bacteria in seawater samples, obtained either within or distant from squid habitats, were collected on membrane filters and incubated until colonies appeared. The filters were then observed for visibly luminous V. fischeri colonies and hybridized with the lux gene probes to determine the number of total V. fischeri colonies (both luminous and non-visibly luminous). We detected no significant differences in the abundance of luminous V. fischeri CFU in any of the water samples observed (≤1 to 3 CFU/100 ml). However, probe-positive colonies of V. fischeri (up to 900 CFU/100 ml) were found only in seawater collected from within the natural habitats of the squids. A number of criteria were used to confirm that these probe-positive strains were indistinguishable from symbiotic V. fischeri. Therefore, the luxA and luxR gene probes were species specific and gave a reliable estimate of the number of culturable V. fischeri colonies in natural water samples.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baumann P., Baumann L., Woolkalis M. J., Bang S. S. Evolutionary relationships in vibrio and Photobacterium: a basis for a natural classification. Annu Rev Microbiol. 1983;37:369–398. doi: 10.1146/annurev.mi.37.100183.002101. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher K. J., Ruby E. G. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990 Jul;172(7):3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaola A., Hopkins L. H., Peeler J. T., Wentz B., McPhearson R. M. Incidence of Vibrio parahaemolyticus in U.S. coastal waters and oysters. Appl Environ Microbiol. 1990 Aug;56(8):2299–2302. doi: 10.1128/aem.56.8.2299-2302.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap P. V. Osmotic control of luminescence and growth in Photobacterium leiognathi from ponyfish light organs. Arch Microbiol. 1985 Feb;141(1):44–50. doi: 10.1007/BF00446738. [DOI] [PubMed] [Google Scholar]
- Kaplan H. B., Greenberg E. P. Overproduction and purification of the luxR gene product: Transcriptional activator of the Vibrio fischeri luminescence system. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6639–6643. doi: 10.1073/pnas.84.19.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M. J., Ruby E. G. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991 Dec 6;254(5037):1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- Nealson K. H., Hastings J. W. Bacterial bioluminescence: its control and ecological significance. Microbiol Rev. 1979 Dec;43(4):496–518. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'brien C. H., Sizemore R. K. Distribution of the Luminous Bacterium Beneckea harveyi in a Semitropical Estuarine Environment. Appl Environ Microbiol. 1979 Nov;38(5):928–933. doi: 10.1128/aem.38.5.928-933.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J. D., Nilsson L., Kjelleberg S. Formation of nonculturable Vibrio vulnificus cells and its relationship to the starvation state. Appl Environ Microbiol. 1991 Sep;57(9):2640–2644. doi: 10.1128/aem.57.9.2640-2644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orndorff S. A., Colwell R. R. Distribution and identification of luminous bacteria from the sargasso sea. Appl Environ Microbiol. 1980 May;39(5):983–987. doi: 10.1128/aem.39.5.983-987.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen R. J., Borman P. A rapid biochemical method for purifying high molecular weight bacterial chromosomal DNA for restriction enzyme analysis. Nucleic Acids Res. 1987 Apr 24;15(8):3631–3631. doi: 10.1093/nar/15.8.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer L. M., Colwell R. R. Detection of luciferase gene sequence in nonluminescent Vibrio cholerae by colony hybridization and polymerase chain reaction. Appl Environ Microbiol. 1991 May;57(5):1286–1293. doi: 10.1128/aem.57.5.1286-1293.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittenberg S. C., Shilo M. Early host damage in the infection cycle of Bdellovibrio bacteriovorus. J Bacteriol. 1970 Apr;102(1):149–160. doi: 10.1128/jb.102.1.149-160.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak D. B., Colwell R. R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987 Sep;51(3):365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby E. G., Greenberg E. P., Hastings J. W. Planktonic marine luminous bacteria: species distribution in the water column. Appl Environ Microbiol. 1980 Feb;39(2):302–306. doi: 10.1128/aem.39.2.302-306.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby E. G., Nealson K. H. Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica; a model of symbiosis based on bacterial studies. Biol Bull. 1976 Dec;151(3):574–586. doi: 10.2307/1540507. [DOI] [PubMed] [Google Scholar]
- Sayler G. S., Layton A. C. Environmental application of nucleic acid hybridization. Annu Rev Microbiol. 1990;44:625–648. doi: 10.1146/annurev.mi.44.100190.003205. [DOI] [PubMed] [Google Scholar]
- Shilo M., Yetinson T. Physiological characteristics underlying the distribution patterns of luminous bacteria in the mediterranean sea and the gulf of elat. Appl Environ Microbiol. 1979 Oct;38(4):577–584. doi: 10.1128/aem.38.4.577-584.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzman A., Kapoor S., Graham A. F., Meighen E. A. A new Vibrio fischeri lux gene precedes a bidirectional termination site for the lux operon. J Bacteriol. 1990 Dec;172(12):6797–6802. doi: 10.1128/jb.172.12.6797-6802.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimpee C. F., Nadeau T. L., Nealson K. H. Development of species-specific hybridization probes for marine luminous bacteria by using in vitro DNA amplification. Appl Environ Microbiol. 1991 May;57(5):1319–1324. doi: 10.1128/aem.57.5.1319-1324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetinson T., Shilo M. Seasonal and geographic distribution of luminous bacteria in the eastern mediterranean sea and the gulf of elat. Appl Environ Microbiol. 1979 Jun;37(6):1230–1238. doi: 10.1128/aem.37.6.1230-1238.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]



