Abstract
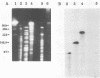
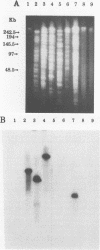
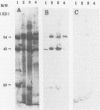
Restriction fragment length polymorphisms, Western blot (immunoblot) analysis, and fluorescence-labelled signature probes were used for the characterization of methanotrophic bacteria as well as for the identification of methanotrophs which contained the soluble methane monooxygenase (MMO) gene and were able to degrade trichloroethylene (TCE). The gene encoding a soluble MMO component B protein from Methylosinus trichosporium OB3b was cloned. It contained a 2.2-kb EcoRI fragment. With this cloned component B gene as probe, methanotroph types I, II, and X and environmental and bioreactor samples were screened for the presence of the gene encoding soluble MMO. Fragments produced by digestion of DNA with rare cutting restriction endonucleases were separated by pulsed-field gel electrophoresis and transferred to Zeta-Probe membrane (Bio-Rad) for Southern blot analysis. Samples were also analyzed for the presence of soluble MMO by Western blot analysis and the ability to degrade TCE. The physiological groups of methanotrophs in each sample were determined by hybridizing cells with fluorescence-labelled signature probes. Among twelve pure or mixed cultures, DNA fragments of seven methanotrophs hybridized with the soluble MMO B gene probe. When grown in media with limited copper, all of these bacteria degraded TCE. All of them are type II methanotrophs. The soluble MMO component B gene of the type X methanotroph, Methylococcus capsulatus Bath, did not hybridize to the M. trichosporium OB3b soluble MMO component B gene probe, although M. capsulatus Bath also produces a soluble MMO.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen L. N., Hanson R. S. Construction of broad-host-range cosmid cloning vectors: identification of genes necessary for growth of Methylobacterium organophilum on methanol. J Bacteriol. 1985 Mar;161(3):955–962. doi: 10.1128/jb.161.3.955-962.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Cohen L., McCarty P. L. Product toxicity and cometabolic competitive inhibition modeling of chloroform and trichloroethylene transformation by methanotrophic resting cells. Appl Environ Microbiol. 1991 Apr;57(4):1031–1037. doi: 10.1128/aem.57.4.1031-1037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzar H., Moore L. W. Complementary methodologies to identify specific agrobacterium strains. Appl Environ Microbiol. 1987 Nov;53(11):2660–2665. doi: 10.1128/aem.53.11.2660-2665.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusseau G. A., Tsien H. C., Hanson R. S., Wackett L. P. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation. 1990;1(1):19–29. doi: 10.1007/BF00117048. [DOI] [PubMed] [Google Scholar]
- Cardy D. L., Laidler V., Salmond G. P., Murrell J. C. Molecular analysis of the methane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol Microbiol. 1991 Feb;5(2):335–342. doi: 10.1111/j.1365-2958.1991.tb02114.x. [DOI] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Colby J., Dalton H. Resolution of the methane mono-oxygenase of Methylococcus capsulatus (Bath) into three components. Purification and properties of component C, a flavoprotein. Biochem J. 1978 May 1;171(2):461–468. doi: 10.1042/bj1710461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox B. G., Froland W. A., Dege J. E., Lipscomb J. D. Methane monooxygenase from Methylosinus trichosporium OB3b. Purification and properties of a three-component system with high specific activity from a type II methanotroph. J Biol Chem. 1989 Jun 15;264(17):10023–10033. [PubMed] [Google Scholar]
- Fox B. G., Lipscomb J. D. Purification of a high specific activity methane monooxygenase hydroxylase component from a type II methanotroph. Biochem Biophys Res Commun. 1988 Jul 15;154(1):165–170. doi: 10.1016/0006-291x(88)90665-1. [DOI] [PubMed] [Google Scholar]
- Hanson R. S., Tsien H. C., Tsuji K., Brusseau G. A., Wackett L. P. Biodegradation of low-molecular-weight halogenated hydrocarbons by methanotrophic bacteria. FEMS Microbiol Rev. 1990 Dec;7(3-4):273–278. doi: 10.1111/j.1574-6968.1990.tb04924.x. [DOI] [PubMed] [Google Scholar]
- Higgins I. J., Hammond R. C., Sariaslani F. S., Best D., Davies M. M., Tryhorn S. E., Taylor F. Biotransformation of hydrocarbons and related compounds by whole organism suspensions of methane-grown methylosinus trichosporium OB 3b. Biochem Biophys Res Commun. 1979 Jul 27;89(2):671–677. doi: 10.1016/0006-291x(79)90682-x. [DOI] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin S. M., Tam P. E., Bastien C. A., Hanson R. S. Genetic and physical analyses of Methylobacterium organophilum XX genes encoding methanol oxidation. J Bacteriol. 1988 Jan;170(1):141–148. doi: 10.1128/jb.170.1.141-148.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielenz J. R., Jackson L. E., O'Gara F., Shanmugam K. T. Fingerprinting bacterial chromosomal DNA with restriction endonuclease EcoRI: comparison of Rhizobium spp. and identification of mutants. Can J Microbiol. 1979 Jul;25(7):803–807. doi: 10.1139/m79-118. [DOI] [PubMed] [Google Scholar]
- Miller R. E., Guengerich F. P. Metabolism of trichloroethylene in isolated hepatocytes, microsomes, and reconstituted enzyme systems containing cytochrome P-450. Cancer Res. 1983 Mar;43(3):1145–1152. [PubMed] [Google Scholar]
- Oldenhuis R., Vink R. L., Janssen D. B., Witholt B. Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl Environ Microbiol. 1989 Nov;55(11):2819–2826. doi: 10.1128/aem.55.11.2819-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed W. M., Dugan P. R. Distribution of Methylomonas methanica and Methylosinus trichosporium in Cleveland Harbor as Determined by an Indirect Fluorescent Antibody-Membrane Filter Technique. Appl Environ Microbiol. 1978 Feb;35(2):422–430. doi: 10.1128/aem.35.2.422-430.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. L., Zidwick M. J., Abebe H. M. Bradyrhizobium japonicum Serocluster 123 and Diversity among Member Isolates. Appl Environ Microbiol. 1986 Jun;51(6):1212–1215. doi: 10.1128/aem.51.6.1212-1215.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Sobral B. W., Sadowsky M. J., Atherly A. G. Genome analysis of Bradyrhizobium japonicum serocluster 123 field isolates by using field inversion gel electrophoresis. Appl Environ Microbiol. 1990 Jun;56(6):1949–1953. doi: 10.1128/aem.56.6.1949-1953.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stainthorpe A. C., Murrell J. C., Salmond G. P., Dalton H., Lees V. Molecular analysis of methane monooxygenase from Methylococcus capsulatus (Bath). Arch Microbiol. 1989;152(2):154–159. doi: 10.1007/BF00456094. [DOI] [PubMed] [Google Scholar]
- Stirling D. I., Dalton H. Properties of the methane mono-oxygenase from extracts of Methylosinus trichosporium OB3b and evidence for its similarity to the enzyme from Methylococcus capsulatus (Bath). Eur J Biochem. 1979 May 2;96(1):205–212. doi: 10.1111/j.1432-1033.1979.tb13030.x. [DOI] [PubMed] [Google Scholar]
- Tsien H. C., Bratina B. J., Tsuji K., Hanson R. S. Use of oligodeoxynucleotide signature probes for identification of physiological groups of methylotrophic bacteria. Appl Environ Microbiol. 1990 Sep;56(9):2858–2865. doi: 10.1128/aem.56.9.2858-2865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Brusseau G. A., Hanson R. S., Waclett L. P. Biodegradation of trichloroethylene by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1989 Dec;55(12):3155–3161. doi: 10.1128/aem.55.12.3155-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T. M., McCarty P. L. Biotransformation of tetrachloroethylene to trichloroethylene, dichloroethylene, vinyl chloride, and carbon dioxide under methanogenic conditions. Appl Environ Microbiol. 1985 May;49(5):1080–1083. doi: 10.1128/aem.49.5.1080-1083.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland M. P., Dalton H. Purification and characterization of component A of the methane monooxygenase from Methylococcus capsulatus (Bath). J Biol Chem. 1984 Jan 10;259(1):53–59. [PubMed] [Google Scholar]