Abstract
Bacteriophage T7 DNA polymerase shares extensive sequence homology with Escherichia coli DNA polymerase I. However, in vivo, E. coli DNA polymerase I is involved primarily in the repair of DNA whereas T7 DNA polymerase is responsible for the replication of the viral genome. In accord with these roles, T7 DNA polymerase is highly processive while E. coli DNA polymerase I has low processivity. The high processivity of T7 DNA polymerase is achieved through tight binding to its processivity factor, E. coli thioredoxin. We have identified a unique 76-residue domain in T7 DNA polymerase responsible for this interaction. Insertion of this domain into the homologous site in E. coli DNA polymerase I results in a dramatic increase in the processivity of the chimeric DNA polymerase, a phenomenon that is dependent upon its binding to thioredoxin.
Keywords: DNA replication, T7 gene 5 protein, Klenow DNA polymerase
High processivity is an important attribute of all replicative DNA polymerases (1). Processivity is defined as the number of nucleotides polymerized by a DNA polymerase during a single association–dissociation cycle with the primer–template. In general, processivity is achieved through the interaction of the DNA polymerase with a class of proteins known as processivity factors.
The replication system of phage T7 provides an attractive model for studying the replication of a chromosome, in part due to the economy of the proteins involved. T7 DNA polymerase, the product of the viral gene 5, by itself has low processivity. It dissociates from a primer–template after the incorporation of <15 nt (2). Upon infection of Escherichia coli, T7 annexes a host protein, thioredoxin, to serve as its processivity factor (3, 4). T7 DNA polymerase and thioredoxin bind in a one-to-one complex with an apparent dissociation constant of 5 nM (5). The binding of thioredoxin to T7 DNA polymerase increases the affinity of the polymerase specifically to a primer–template by 80-fold (6). A consequence of the increased affinity for a primer–template is the ability of T7 DNA polymerase to extend a primer on single-stranded DNA (ssDNA) by thousands of nucleotides without dissociating (2).
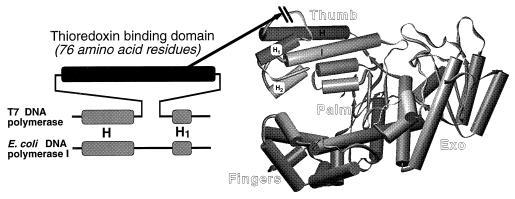
All known DNA polymerases can be classified into four families on the basis of their amino acid sequence similarities (7). T7 DNA polymerase is a member of the “Pol I” family that includes E. coli DNA polymerase I, Thermus aquaticus (Taq) DNA polymerase, mitochondrial DNA polymerase, and the DNA polymerases from phages T5, SP01, and SP02. E. coli DNA polymerase I, the paradigm of this family, is a repair-type DNA polymerase, and as such has low processivity. It dissociates from a primer–template after the incorporation of about 20 nt (8). In contrast to T7 DNA polymerase, it does not associate with any known accessory proteins. The three-dimensional structure of the large fragment of E. coli DNA polymerase I (Klenow DNA polymerase) is known (9, 10). The polymerase active site and DNA binding domain are in the carboxyl-terminal half of the molecule, and the 3′ to 5′ proofreading exonuclease activity is located in a separate domain at the amino-terminal half. Comparison of the sequence of T7 DNA polymerase to E. coli DNA polymerase I reveals a 76-amino acid residue segment in T7 DNA polymerase (residues 258–333) that is not present in E. coli DNA polymerase I. In the structure of E. coli DNA polymerase I this region corresponds to an insert between helices H and H1 (Fig. 1). These two helices are located in a region referred to as the “thumb” of the molecule, a region that is thought to be important for interaction with duplex DNA (10).
Figure 1.
Location of the thioredoxin binding domain (TBD) in T7 and Klenow–TBD DNA polymerases. (Right) The x-ray crystal structure of Klenow DNA polymerase is shown (10), modeled using the program setor (11). (Left) The alignment of the H–H1 region in T7 DNA polymerase and E. coli DNA polymerase I is shown (7). The unique 76-residue region in T7 DNA polymerase, located between helices H and H1, is indicated by the dark rectangle. In the chimeric Klenow–TBD DNA polymerase, this domain has been inserted at the tip of the thumb with the deletion of seven residues from E. coli DNA polymerase, as indicated by the arrow.
Several lines of evidence suggest that this unique domain in T7 DNA polymerase plays a role in the interaction with thioredoxin and is responsible for conferring processivity on the nucleotide polymerization reaction. For example, this domain of T7 DNA polymerase is more susceptible to proteolytic attack than other domains of the protein, a phenomenon that is inhibited by the presence of thioredoxin (2, 12). Mutations within the DNA sequence encoding this domain affect the ability of T7 DNA polymerase to bind to thioredoxin (refs. 13 and 14; X.-M. Yang and C.C.R., unpublished data). In one instance, a suppressor mutation, which allows T7 phage to use a genetically altered thioredoxin, resides on the fringe of this domain of T7 DNA polymerase (Glu-319 → Lys). Biochemical evidence shows that this particular suppressor mutation in the T7 gene 5 restores the ability of the T7 DNA polymerase to interact with this particular mutant thioredoxin, suggesting that the sites of these two mutations may represent a contact point between the two proteins (14).
One approach to understanding the structural basis for functional differences between homologous enzymes is to characterize the properties of active site hybrids. We previously used this approach to define a single residue in Pol I-type polymerases critical for their ability to distinguish between deoxy- and dideoxynucleotides (15). Using this strategy, we have examined the properties of a chimeric DNA polymerase in which this 76-amino acid residue domain from T7 DNA polymerase has been placed into the homologous site in E. coli DNA polymerase I. We show that this domain in T7 DNA polymerase is responsible for interacting with thioredoxin to confer processivity on the polymerase reaction.
METHODS
Plasmid Constructions.
The gene for the large fragment of E. coli DNA polymerase I (Klenow DNA polymerase) was derived from the plasmid pCJ55 (16) and inserted into the plasmid pT7-5 under the control of a T7 RNA polymerase promoter to generate pKLN-0 (provided by Stephen Notarnicola, Harvard Medical School). Into this plasmid the region encoding the TBD from T7 gene 5 (amino acid residues 258–333) was inserted, replacing the region encoding amino acids 571–577 in Klenow DNA polymerase. The chimeric polymerase was constructed in three PCR steps using a procedure modified from Sarkar and Sommer (17). The DNA sequence of the entire insert and the junctions of the final plasmid were verified by DNA sequencing.
Purification of the Klenow–TBD DNA Polymerase.
The Klenow–TBD polymerase was overproduced in BL21(DE3)/pLysS (Novagen). A cell lysate prepared using standard procedures was mixed with Phenyl Sepharose (Pharmacia) equilibrated with 30 mM Tris·HCl (pH 7.5), 1 mM DTT, 2 mM EDTA, and 1 M ammonium sulfate. The resin was washed with the same buffer in the absence of ammonium sulfate, and the protein was eluted with 20% acetonitrile in 30 mM Tris·HCl (pH 7.5), 1 mM DTT, and 2 mM EDTA. The polymerase was purified further by fast protein liquid chromatography (FPLC) Mono Q column chromatography (Pharmacia) using a buffer containing 30 mM Tris·HCl (pH 7.5), 1 mM DTT, 2 mM EDTA, and 10% glycerol after elution with a linear NaCl gradient at ≈160 mM. Purified Klenow–TBD DNA polymerase was stored at −20°C in 20 mM KPO4 (pH 7.5), 0.1 mM DTT, 1 mM EDTA, and 50% glycerol.
Exonuclease Assay.
Exonuclease activity was determined by a modification of previously described procedures (5, 13). For the determination of 3′ to 5′ double-stranded DNA (dsDNA) exonuclease activity, the template consisted of M13 mGP1-2 primed with a 22-mer oligonucleotide that was extended an average of 240 nt in the presence of [α-32P]dATP. The reactions contained 0.6 pmol (in nucleotide equivalents) of labeled template. Klenow DNA polymerase and Klenow–TBD DNA polymerase were at 3 nM, and T7 DNA polymerase at 6 nM. When present, the concentration of thioredoxin was 8 μM. For the determination of ssDNA exonuclease activity, a uniformly labeled 900-bp DNA fragment was synthesized by performing a PCR in the presence of [methyl-3H]dTTP, as described (13). The reactions contained 0.15 nmol (in nucleotide equivalents) of the template, either 30 mM Klenow DNA polymerase, 30 nM Klenow–TBD DNA polymerase, or 3 nM T7 DNA polymerase. When present, thioredoxin was at a concentration of 2 μM. One unit of exonuclease activity catalyzes the release of one pmol of total nucleotides into an acid soluble form in one min. 100% ssDNA exonuclease activity corresponds to 55,000 units/μg, and 100% dsDNA exonuclease activity corresponds to 55 units/μg.
DNA Polymerase Assay.
DNA polymerase activity was measured in an assay based on previously described procedures (2, 13). The primer–template was single-stranded M13 mGP1-2 DNA annealed with a 20-mer oligonucleotide. The reactions contained 20 nM primed m13 mGP1-2, 2 nM DNA polymerase, and, when present, 2 μM thioredoxin. The reactions were carried out at 37°C for either 5 min (T7 DNA polymerase) or 15 min (Klenow or Klenow–TBD DNA polymerase). One unit of DNA polymerase activity catalyzes the incorporation of one pmol of total nucleotide into a form retained by DE81 filter paper in 1 min.
Processivity Assay.
Processivity was measured by a modification of the previously described procedure (2). The primer consisted of a 22-mer oligonucleotide that was 32P-labeled at its 5′ end, and annealed to the single-stranded circular M13 mGP1-2 DNA template, then purified by gel filtration through a Sepharose CL6B column. The reaction mixture contained 3 nM of 32P-labeled primer–template molecules, and enzyme at either 0.06 or 0.012 nM, corresponding to a molar ratio of primer–template to DNA polymerase ratio of 50:1 or 250:1, respectively. Thioredoxin, when present, was at 20 μM. DNA products were separated by denaturing PAGE using 8% polyacrylamide gels containing 8 M urea.
Surface Plasmon Resonance.
Surface plasmon resonance (18) was carried out using a BIAcore instrument (Pharmacia). Antithioredoxin mAbs were covalently bound to a chip via their amine groups using procedures described by the manufacturer, with a 7-min pulse of the mAb, diluted to 50 μg/ml in 5 mM maleate (pH 7.0). The monoclonal antithioredoxin antibodies were generously provided by John McCoy (Genetics Institute, Cambridge, MA). The flow buffer contained 10 mM Hepes (pH 7.5), 150 mM NaCl, 3.4 mM EDTA, 0.01% Tween 20, 0.1 mM DTT, and 0.5% glycerol. The flow rate used in all the experiments was 5 μl/min. Thioredoxin was bound to the mAb by a 1-min pulse of 40 μg/ml thioredoxin.
RESULTS
Design of a Chimeric E. coli DNA Polymerase I Containing the TBD of T7 DNA Polymerase.
Our strategy in testing the hypothesis that the unique region in T7 DNA polymerase is involved in binding thioredoxin was to insert this region into the large fragment of E. coli DNA polymerase I (Klenow DNA polymerase), and then characterize the properties of the hybrid enzyme. In constructing the Klenow–TBD DNA polymerase we considered both the extensive sequence homology in the carboxyl-terminal regions of the two enzymes (7) and the crystal structure of Klenow DNA polymerase (10). Alignment of the homologous regions in the two proteins shows that the region unique to T7 DNA polymerase consists of the 76-amino acid residues from 258 to 333. The corresponding region in E. coli DNA polymerase I is represented by the 7 residues from 571 to 577. This region is flanked by the helices H and H1 located at the tip of the thumb portion of the molecule (Fig. 1).
We constructed a recombinant molecule in which the DNA encoding amino acid residues 258–333 of T7 DNA polymerase were substituted for the DNA encoding amino acid residues 571–577 of E. coli DNA polymerase I. The remaining 598 residues encoded by the gene were from E. coli DNA polymerase I. After overexpression of this chimeric gene under the control of a T7 RNA polymerase promoter and lysis of the induced cells, most of the overproduced protein was soluble. We purified this protein using standard hydrophobic and anion-exchange chromatography, and the presence of the hybrid protein during purification was monitored by both SDS/PAGE analysis and DNA polymerase activity. As judged by SDS/PAGE, the purified Klenow–TBD DNA polymerase, migrating as a single species of the expected Mr of 76,000, was >80% pure (data not shown).
Effect of Thioredoxin on the Polymerase and Exonuclease Activities of Klenow–TBD DNA Polymerase.
The DNA polymerase activity of Klenow–TBD DNA polymerase was compared with that of Klenow and T7 DNA polymerases in the presence and absence of thioredoxin (Table 1). Klenow–TBD DNA polymerase was stimulated ≈8-fold by the presence of thioredoxin whereas the activity of Klenow DNA polymerase was unaffected. T7 DNA polymerase, as expected, was stimulated >250-fold. Insertion of the TBD into Klenow DNA polymerase resulted in the hybrid polymerase having reduced specific activity in the absence of thioredoxin (5-fold lower then Klenow). Yet its specific activity remains much higher than that of T7 DNA polymerase in the absence of thioredoxin. In the presence of thioredoxin, the activity of Klenow–TBD DNA polymerase is higher than that of Klenow DNA polymerase and is only 3-fold lower than that of T7 DNA polymerase–thioredoxin complex.
Table 1.
Effect of thioredoxin on DNA polymerase activities
| Polymerase | Specific activity, units/mg
|
|
|---|---|---|
| − thioredoxin | + thioredoxin | |
| Klenow DNA polymerase | 2100 | 2,200 |
| T7 DNA polymerase | <4 | 10,000 |
| Klenow–TBD1 DNA polymerase | 400 | 3,400 |
Polymerase activities were measured on primed single-stranded M13 mGP1-2 DNA using a 10-fold molar excess of template over enzyme as described.
Both T7 DNA polymerase and E. coli DNA polymerase I have a 3′ to 5′ exonuclease activity. The exonuclease activity of T7 DNA polymerase is several hundred times more active than that of E. coli DNA polymerase I (2). Thioredoxin increases the exonuclease activity of T7 DNA polymerase on dsDNA by up to several hundred fold, but it has little effect on the activity on ssDNA (2, 19). In light of these differences, it was of interest to determine the level of exonuclease activity of Klenow–TBD DNA polymerase and also to examine the effect of thioredoxin on this activity.
The ssDNA exonuclease activity of T7 DNA polymerase was 40-fold higher than that observed for Klenow DNA polymerase. The ssDNA exonuclease activity of Klenow–TBD DNA polymerase was 40% higher than that of the Klenow DNA polymerase (Table 2). Thioredoxin did not affect the level of ssDNA exonuclease activity in these three enzymes. The dsDNA exonuclease activity of T7 DNA polymerase was 20-fold higher than that of Klenow DNA polymerase, in the absence of thioredoxin. The Klenow–TBD DNA polymerase, in the absence of thioredoxin, had the same level of this activity as the Klenow DNA polymerase. Although thioredoxin had no effect on the dsDNA exonuclease activity of Klenow DNA polymerase, it stimulated this activity in T7 DNA polymerase 15-fold, and in Klenow–TBD DNA polymerase by 50% (Table 2). The fact that the stimulation of the dsDNA exonuclease activity of T7 DNA polymerase by thioredoxin observed here is less than that previously reported (2, 19) is likely due to differences in the substrates used in the different studies.
Table 2.
Effect of thioredoxin (Trx) on ssDNA and dsDNA exonuclease activities
| Polymerase | Specific activity, Units/mg
|
|||
|---|---|---|---|---|
| ssDNA
|
dsDNA
|
|||
| −Trx | +Trx | −Trx | +Trx | |
| Klenow DNA polymerase | [100] | 100 | [100] | 100 |
| T7 DNA polymerase | 3900 | 3800 | 2100 | 32,000 |
| Klenow–TBD DNA polymerase | 140 | 140 | 100 | 150 |
Exonuclease activities were measured on uniformly labeled ssDNA or dsDNA in the presence or absence of thioredoxin as described. Units are defined as the percentage of activity to that observed for the Klenow DNA polymerase in the absence of thioredoxin (in brackets).
The fact that thioredoxin does not stimulate the dsDNA exonuclease activity of the Klenow–TBD DNA polymerase to nearly the same extent as it does T7 DNA polymerase (1.5- versus 15-fold) indicates that there are important differences between the two enzymes in the mechanism by which they carry out exonucleolytic degradation. On the other hand, the fact that both the ssDNA and dsDNA exonuclease activities of the Klenow–TBD are very similar to the values observed for the Klenow DNA polymerase provides supporting evidence that the TBD insert has not significantly altered the folding of the protein.
Measurement of the Observed Equilibrium Dissociation Constant Between the Klenow–TBD DNA Polymerase and Thioredoxin.
The stimulation of DNA polymerase activity by thioredoxin implies the formation of a polymerase/thioredoxin complex and permits the determination of an observed equilibrium dissociation constant for the two proteins (5, 13). The amount of DNA synthesis that is above that observed in the absence of thioredoxin is assumed to be proportional to the amount of polymerase/thioredoxin complex formed. The experimental conditions are such that the amount of bound thioredoxin is negligible when compared with the total amount of thioredoxin present, and a Scatchard plot is used to determine the observed equilibrium dissociation constant. We used this method to determine the relative affinities of the Klenow–TBD and T7 DNA polymerases for thioredoxin.
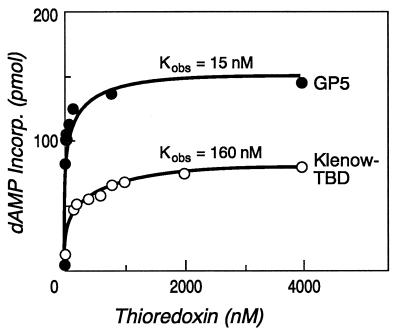
The presence of increasing concentrations of thioredoxin stimulates the DNA polymerase activity of both T7 and Klenow–TBD DNA polymerases (Fig. 2). The observed equilibrium dissociation constants derived from the data was 15 nM for the T7 DNA polymerase/thioredoxin interaction, in close agreement with the values previously reported of 5 nM at 20°C and 20 nM at 40°C (5). The observed equilibrium dissociation constant for the Klenow–TBD DNA polymerase/thioredoxin interaction was 160 nM.
Figure 2.
Stimulation of DNA synthesis by thioredoxin. The amount of DNA synthesis in the presence of increasing concentrations of thioredoxin was determined in reactions catalyzed by either T7 DNA polymerase (GP5) or Klenow–TBD DNA polymerase (Klenow–TBD). The amount of [32P]dAMP incorporated into primed single-stranded M13 mGP1-2 DNA was determined as described. The molar ratio of DNA polymerase to primer–template was 1:10 for all reactions.
Physical Analysis of the Klenow–TBD DNA Polymerase/Thioredoxin Interaction.
In corroboration of the indirect method described in the previous section to determine an observed equilibrium dissociation constant, we have measured directly the interaction between Klenow–TBD DNA polymerase and thioredoxin by surface plasmon resonance. In this method, thioredoxin is bound to a solid support on a sensor chip, and subsequently, its interaction with Klenow–TBD DNA polymerase is observed by a change in the surface plasmon resonance on the chip’s surface when thioredoxin physically binds to the polymerase.
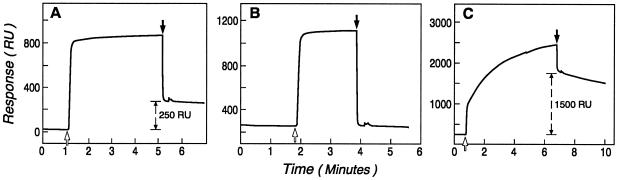
Thioredoxin was bound to the chip’s surface via its interaction with antithioredoxin mAbs that were covalently attached to the chip. The bound thioredoxin presents a uniform population of molecules for binding. In the experiment shown in Fig. 3A, thioredoxin binds to the mAb-coated chip. This binding is manifested by an increase of 250 resonance units (RU). The binding between thioredoxin and the mAb is extremely stable, and there is negligible decay of the thioredoxin–mAb complex during the course of an experiment. Klenow DNA polymerase does not bind to the thioredoxin-covered chip (Fig. 3B). However, as evidenced by the 1500 RU increase, Klenow–TBD DNA polymerase does (Fig. 3C). This increase is not observed when the chip contains immobilized mAbs without thioredoxin (data not shown). Therefore the interaction of the Klenow–TBD DNA polymerase is specific for thioredoxin.
Figure 3.
Surface plasmon resonance analysis of Klenow–TBD polymerase/thioredoxin interaction. (A) The binding of thioredoxin to a sensor chip that has antithioredoxin mAbs covalently attached to its surface. The open arrow indicates the start of the injection of thioredoxin, and the filled arrow indicates the start of the buffer wash. The number of RU that remain after the initial sharp decay at the start of the buffer wash are indicated by the discontinuous arrows. (B) The binding of Klenow DNA polymerase to a chip containing thioredoxin bound to the mAbs. (C) The binding of Klenow–TBD DNA polymerase to a chip containing thioredoxin bound to the mAbs.
In surface plasmon resonance, the increase in RU is proportional to the change in the refractive index, which in turn is directly proportional to the change in mass on the chip’s surface (20). In the experiment shown in Fig. 3, there is six times the amount of mass of DNA polymerase bound to the chip (1500 RU units) compared with thioredoxin (250 RU units). Because the ratio of the mass of Klenow–TBD DNA polymerase to thioredoxin is 6.3:1, the molar ratio of the polymerase bound to thioredoxin is 0.95. Neither increasing the injection time (to be certain that equilibrium has been reached) nor increasing the concentration of the Klenow–TBD DNA polymerase injected increased the RU units of bound Klenow–TBD DNA polymerase above 1600. This result strongly suggests that the complex formed is in a one-to-one ratio (data not shown). This value is consistent with the one-to-one stoichiometry of the T7 DNA polymerase/thioredoxin interaction previously reported (3).
Thioredoxin Increases Dramatically the Processivity of Klenow–TBD DNA Polymerase.
The binding of thioredoxin to T7 DNA polymerase increases the polymerase activity by increasing its affinity for a primer–template (6) and thus dramatically increasing the processivity of DNA synthesis (2). It was of interest to determine if this is likewise the explanation for the increased polymerase activity of the Klenow–TBD DNA polymerase in the presence of thioredoxin. We compared the processivity of Klenow, T7, and Klenow–TBD DNA polymerases using a dilution experiment. In this experiment DNA synthesis was carried out on a primer–template with a 5′-labeled primer using a large molar excess of primer–template to polymerase. Thus the DNA synthesis observed from the extension of a given primer was the result of a single binding event between the polymerase and the primer template (Fig. 4).
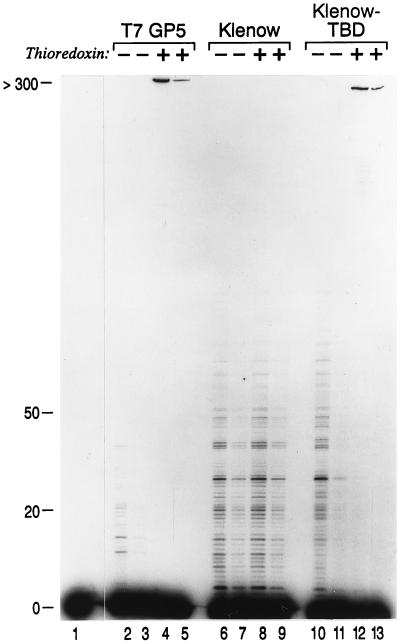
Figure 4.
The effect of thioredoxin on the processivity of Klenow–TBD polymerase. DNA polymerase reactions were carried out using a 5′ 32P-labeled primer annealed to M13 mGP1-2 DNA in the presence and absence of thioredoxin. Lane 1 contains the starting primer–template in the absence of any DNA polymerase. In the even-numbered lanes (bottom) the primer–template was present at a 50-fold molar excess over the DNA polymerase, and in the odd-numbered lanes the primer–template present at a 250-fold molar excess over the DNA polymerase. In lanes 2–5 the reactions were carried out using T7 DNA polymerase (T7 GP5). In lanes 6–9 the reactions were carried out using Klenow DNA polymerase (Klenow), and in lanes 10–13 the reactions were carried out using Klenow–TBD DNA polymerase (Klenow–TBD). The products were separated on a denaturing 8% polyacrylamide gel. After electrophoresis, the gel was dried and autoradiographed. The mobility of primers that have been extended by the indicated numbers of nucleotides is shown on the left.
The processivity of Klenow DNA polymerase was about 20 nt per binding event, a value that was not affected by the presence of thioredoxin (Fig. 4, lanes 6–9). The processivity of T7 DNA polymerase was only several nucleotides per binding event in the absence of thioredoxin but increased to more than 300 nt per binding event in the presence of thioredoxin (Fig. 4, lanes 2–5). These processivity values are consistent with previously published results for these two enzymes (2, 8).
In the absence of thioredoxin, Klenow–TBD DNA polymerase had a processivity similar to that of Klenow DNA polymerase (Fig. 4, lanes 10 and 11). The presence of the thioredoxin increased the processivity of Klenow–TBD DNA polymerase to greater than 300 nt incorporated per binding event (Fig. 4, lanes 12 and 13). Thus, as with T7 DNA polymerase, the binding of thioredoxin to Klenow–TBD DNA polymerase results in a dramatic increase in its processivity, accounting for the observed increase in polymerase activity (Table 1).
DISCUSSION
Although T7 DNA polymerase and E. coli DNA polymerase I share extensive sequence homology they serve different functions in DNA metabolism and, as such, differ significantly in a number of their properties. The property we have focused on in this paper is the ability of T7 DNA polymerase to bind to E. coli thioredoxin and use it as a processivity factor. The T7 DNA polymerase/thioredoxin complex has a processivity of hundreds of nucleotides, considerably higher than that of E. coli DNA polymerase I. The acquisition of processivity by T7 DNA polymerase is not surprising in view of its role in replicating the viral genome.
In the alignment of the homologous regions in the polymerase domains of T7 DNA polymerase and E. coli DNA polymerase I there is a stretch of 76-amino acid residues in T7 DNA polymerase that is not present in E. coli DNA polymerase I. We have shown here that when this region is transferred into the homologous position in the Klenow DNA polymerase, the chimeric DNA polymerase (Klenow–TBD) now carries out highly processive synthesis that is dependent upon its binding to thioredoxin. The fact that the exonuclease activities of the Klenow–TBD DNA polymerase are similar to that of Klenow DNA polymerase and that the hybrid enzyme retains polymerase activity implies that the TBD has folded as an independent domain that has not perturbed significantly the structure of the enzyme.
Although Klenow–TBD DNA polymerase and T7 DNA polymerase both have high processivity in the presence of thioredoxin, the two have a number of important differences in their enzymatic properties. First, in the absence of thioredoxin the polymerase activity of Klenow–TBD DNA polymerase is ≈100-fold higher than that of T7 DNA polymerase (Table 1). The reason for such low activity of T7 DNA polymerase in the absence of thioredoxin is not understood.
A second difference between Klenow–TBD DNA polymerase and T7 DNA polymerase is that the former binds thioredoxin with an apparent 10-fold lower affinity. There are several possible explanations for this observation. One is that the TBD used here constitutes most, but not all, of the interactions between T7 DNA polymerase and thioredoxin. This would imply that other regions in the T7 DNA polymerase molecule, either on the thumb or elsewhere on the molecule, are important for maximal binding to thioredoxin. The appearance of suppressor mutations in other regions of the T7 DNA polymerase that allow T7 phage to use a genetically altered thioredoxin may support this possibility (13). A second possibility is that this TBD is sufficient for maximal binding affinity to T7 DNA polymerase, but, in the context that it has been placed within the Klenow DNA polymerase, there may be structural constraints that have resulted in a slightly aberrant folding. Finally, it is possible that thioredoxin is in fact binding with the same affinity to the Klenow–TBD DNA polymerase as to T7 DNA polymerase, and the differences in the observed binding affinities are a consequence of the indirect method used to measure the binding constants. Regardless, it is clear from the data presented that the TBD used here constitutes most of the essential interactions required for thioredoxin to stimulate the processivity of T7 DNA polymerase.
A third difference between Klenow–TBD DNA polymerase and T7 DNA polymerase is their levels of 3′ to 5′ ssDNA and dsDNA exonuclease activities, and the effect of thioredoxin on the latter activity. In the presence of thioredoxin the affinity of T7 DNA polymerase to a primer template is increased greatly (6), resulting in stimulation of the double-stranded exonuclease activity; ssDNA exonuclease activity is not affected. The double-stranded exonuclease activity of T7 DNA polymerase was stimulated 15-fold by the presence of thioredoxin (Table 2). In the Klenow–TBD DNA polymerase, the 3′ to 5′ dsDNA exonuclease activity is identical to that observed in Klenow DNA polymerase, and is only stimulated 1.5-fold by the presence of thioredoxin. The fact that the dsDNA exonuclease activity of Klenow–TBD DNA polymerase is not stimulated by thioredoxin to the same extent as that of T7 DNA polymerase is surprising and may reflect differences in the mechanisms by which these enzymes behave in their exonuclease mode.
What is the mechanism by which thioredoxin confers high processivity on T7 DNA polymerase and the chimeric Klenow–TBD DNA polymerase? On the basis of the comparison between the crystal structures of Klenow DNA polymerase in the presence and absence of DNA it is thought that the thumb region plays an important role in binding Klenow DNA polymerase to duplex DNA (10). The 76-amino acid residue TBD is located between helices H and H1 in the thumb region, and is juxtaposed to the site in Klenow DNA polymerase that is observed to be nested into the minor groove of the duplex DNA in a cocrystal structure (10). This location then places the thioredoxin bound to the TBD in a position well situated to facilitate the clamping of the complex to the dsDNA, thereby increasing processivity. In this model, the DNA polymerase/thioredoxin complex encircles the duplex DNA, with thioredoxin providing one face of the putative clamp. Consistent with this model, we have shown that both T7 DNA polymerase and Klenow DNA polymerase protect ≈14 bp of duplex DNA from exonuclease degradation in a footprint analysis, and T7 DNA polymerase in the presence of thioredoxin protects an additional 7 bp of duplex DNA from exonuclease degradation (S.T. and C.C.R., unpublished data). It should be noted that thioredoxin by itself has no affinity for either ssDNA or dsDNA (5).
Is the mechanism by which thioredoxin stimulates the processivity of T7 DNA polymerase common to other DNA polymerases in the Pol I family? Of the members of this family whose sequence is known, five polymerases, besides T7 DNA polymerase, are responsible for the replication of a chromosome and are thus likely to have high processivity. These DNA polymerases are from bacteriophages SP01, SP02 and T5, mycobacteriophage L5, and mitochondria. T5 DNA polymerase is known to be processive in the absence of any processivity factors (21). When its sequence is aligned with other Pol I-type DNA polymerases, it has an extra 75 amino acid residues at its carboxy terminus (7). This is potentially of interest because in the crystal structure of Klenow DNA polymerase, the carboxy terminus is in close proximity to the thumb domain (10). Thus, it is possible that this domain at the end of the T5 DNA polymerase is serving a role similar to that described above for the TBD/thioredoxin complex in promoting the clamping of the duplex DNA to the thumb domain of the polymerase. The mitochondrial DNA polymerase γ from Drosophila has been purified (22) and has been demonstrated to have high processivity (23). Interestingly, this polymerase is also a large protein (125 kDa) that has a 35-kDa protein associated with it. Alignment of the 125-KDa subunit sequence with that of E. coli DNA polymerase I indicates a high degree of amino acid sequence conservation (24). Bacteriophage SP01 DNA polymerase is the only member of the Pol I family, besides T7 DNA polymerase, that has a significant domain between the regions homologous to the H and H1 helices of Klenow DNA polymerase (7). This domain in SP01 DNA polymerase is 46-amino acid residues in length has only limited sequence homology with the 76-amino acid residue domain found in T7 DNA polymerase. SPO1 DNA polymerase has an additional domain of about 70-amino acid residues at its carboxy terminus, similar to that observed in the T5 and mitochondrial DNA polymerases. While the purification of SP01 DNA polymerase has been reported (25), it is not clear from this work whether the enzyme was processive and whether it may have had associated with it a small processivity factor such as thioredoxin.
It seems likely that once the structures of these diverse, replicative Pol I-type DNA polymerases have been determined, they will be shown to achieve high processivity using mechanisms that will share many of the features used by the thioredoxin/T7 DNA polymerase complex. Thus, the T7 DNA polymerase/thioredoxin complex is a useful model system for understanding the processivity of DNA replication in this polymerase family. It is possible that insertion of the TBD into other relatively nonprocessive DNA polymerases in the DNA Pol I family (such as Taq DNA polymerase) may result in an increase of their processivity that will be dependent upon their binding to thioredoxin.
Acknowledgments
We are grateful to Dr. John McCoy at Genetics Institute for generously providing the antithioredoxin mAbs. We thank Jeff S. Himawan for helpful discussions throughout this work and assistance with the BIAcore experiments. We thank Luis Blanco, Margarita Salas, and Stephen Notarnicola for valuable discussions that led to this work. C.C.R. and S.T. are consultants to Amersham Life Science Inc., which has licenses from Harvard University to commercialize DNA polymerases for use in DNA sequencing. This investigation was supported by Grant AI-06045 from the U.S. Public Health Service and Grant DE-GF02-88ER60688 from the U.S. Department of Energy.
Footnotes
Abbreviations: ssDNA, single-stranded DNA; TBD, thioredoxin binding domain; dsDNA, double-stranded DNA; RU, resonance units.
References
- 1.Kornberg A, Baker T A. DNA Replication. 2nd Ed. New York: Freeman; 1992. [Google Scholar]
- 2.Tabor S, Huber H E, Richardson C C. J Biol Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 3.Modrich P, Richardson C C. J Biol Chem. 1975;250:5515–5522. [PubMed] [Google Scholar]
- 4.Mark D, Richardson C C. Proc Natl Acad Sci USA. 1976;73:780–784. doi: 10.1073/pnas.73.3.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huber H E, Russel M, Model P, Richardson C C. J Biol Chem. 1986;261:15006–15012. [PubMed] [Google Scholar]
- 6.Huber H E, Tabor S, Richardson C C. J Biol Chem. 1987;262:16224–16232. [PubMed] [Google Scholar]
- 7.Braithwaite D K, Ito J. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bambara R A, Uyemura D, Choi T. J Biol Chem. 1978;253:413–423. [PubMed] [Google Scholar]
- 9.Ollis D L, Brick P, Hamlin R, Xuong N G, Steitz T A. Nature (London) 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 10.Beese L, Derbyshire V, Steitz T A. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 11.Evans S V. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 12.Yang X-M, Richardson C C. J Biol Chem. 1996;271:24207–24212. doi: 10.1074/jbc.271.39.24207. [DOI] [PubMed] [Google Scholar]
- 13.Himawan J S, Richardson C C. Proc Natl Acad Sci USA. 1992;89:9774–9778. doi: 10.1073/pnas.89.20.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Himawan J S, Richardson C C. J Biol Chem. 1996;271:19999–20008. doi: 10.1074/jbc.271.33.19999. [DOI] [PubMed] [Google Scholar]
- 15.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joyce C M, Grindley N D F. Proc Natl Acad Sci USA. 1983;80:1830–1834. doi: 10.1073/pnas.80.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkar G, Sommer S S. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 18.Malmqvist M. Nature (London) 1993;361:186–187. doi: 10.1038/361186a0. [DOI] [PubMed] [Google Scholar]
- 19.Adler S, Modrich P. J Biol Chem. 1979;254:11605–11614. [PubMed] [Google Scholar]
- 20.Jonsson U, Fagerstam L, Ivarsson B, Jonhnsson B, Karlsson R, Lundh K, Lofas S, Persson B, Roos H, Ronnberg I, Sjolander S, Stenberg E, Stahlberg R, Urbaniczky C, Ostlin H, Malmqvist M. BioTechniques. 1991;11:620–625. [PubMed] [Google Scholar]
- 21.Das S K, Fujimura R K. J Biol Chem. 1979;254:1227–1232. [PubMed] [Google Scholar]
- 22.Olson M W, Wang Y, Elder R H, Kaguni L S. J Biol Chem. 1995;270:28932–28937. doi: 10.1074/jbc.270.48.28932. [DOI] [PubMed] [Google Scholar]
- 23.Williams A J, Wernette C M, Kaguni L S. J Biol Chem. 1993;268:24855–24862. [PubMed] [Google Scholar]
- 24.Lewis D L, Farr C L, Wang Y, Lagina A T, III, Kaguni L S. J Biol Chem. 1996;271:23389–23394. doi: 10.1074/jbc.271.38.23389. [DOI] [PubMed] [Google Scholar]
- 25.Yehle C O, Ganesan A T. J Biol Chem. 1973;248:7456–7463. [PubMed] [Google Scholar]