Abstract
Artificially aminoacylated suppressor tRNAs were used to introduce photoreactive amino acids into model mitochondrial precursor proteins to probe the environment along the protein import pathway. Amino acids with benzophenone side chains of various lengths [dl-2-amino-3-(p-benzoylphenyl)propanoic acid (1) and dl-2-amino-5-(p-benzoylphenyl)pentanoic acid (2)] were incorporated at specific sites throughout the cytochrome b2-dihydrofolate reductase fusion proteins, pb2(220)-DHFR and pb2Δ19(220)-DHFR, which were destined for the intermembrane space and the matrix in mitochondria, respectively. In vitro import of pb2(220)-DHFR and pb2Δ19(220)-DHFR bearing 1 or 2 into isolated yeast mitochondria was arrested so that the N terminus reached the intermembrane space or the matrix, respectively, while the DHFR domain remained at the mitochondrial surface. The matrix-targeted pb2Δ19(220)-DHFR was photocrosslinked to Tom40 in the outer membrane, Tim44 in the inner membrane, and Ssc1p in the matrix, suggesting that the protein has an extended conformation in the import channels. On the other hand, incorporation of 2 at various positions in the 50-residue segment of intermembrane-space-targeted pb2(220)-DHFR gave photocrosslinks only to Tom40, suggesting that the segment is not in an extended conformation, but localized near Tom40. The N-terminal portion of pb2(220)-DHFR, but not pb2Δ19(220)-DHFR, was photocrosslinked to an as-yet-unidentified mitochondrial component to generate a 95-kDa crosslinked product.
Keywords: benzophenone amino acid
Protein import into mitochondria is a central process in mitochondrial biogenesis. This process requires coordinated functions of the components in the cytosol, the outer and the inner mitochondrial membranes, and the matrix (for reviews see refs. 1 and 2). Most mitochondrial proteins are synthesized in the cytosol as precursor proteins, and cytosolic chaperones maintain them in transport-competent states (3). In the yeast Saccharomyces cerevisiae, the TOM (the translocase of the outer mitochondrial membrane) complex, consisting of Tom proteins in the mitochondrial outer membrane, mediates recognition, insertion, and translocation of precursor proteins at the outer membrane. Tom20 and Tom22 function as the entry site for virtually all the mitochondrial proteins into the organelles, and Tom40 constitutes a putative protein translocation channel (4). The TIM (the translocase of the inner mitochondrial membrane) complex, consisting of Tim17, Tim23, and Tim44, constitutes a putative import channel in the inner membrane and provides a protein transport pathway to the matrix (5). Tim44 and Ssc1p, a member of heat shock proteins of 70 kDa (hsp70) in the matrix, interact with each other and drive the inward movement of precursor proteins through the inner membrane (6, 7).
The pathways for directing proteins, including cytochromes b2 and c1, to the intermembrane space are in dispute (8, 9). The precursors of cytochromes b2 and c1 possess cleavable bipartite presequences. The first part of the presequence directs the proteins to the mitochondria and the second part mediates sorting to the intermembrane space. In the “conservative-sorting” model, cytochromes b2 and c1 are first translocated across both the outer and the inner membranes to reach the matrix and then are reexported to the intermembrane space. In the “stop-transfer” model, the C-terminal part of the presequence arrests translocation through the inner membrane, so that the mature part crosses only the outer membrane to reach the intermembrane space.
To understand the molecular mechanisms underlying the complex processes of mitochondrial protein import, it is essential to characterize the environment of precursor proteins along the pathway to their destinations. Photocrosslinking has proven a powerful tool for analyzing interactions of precursor proteins during membrane translocation (10). Typically, chemical bifunctional crosslinking reagents are added externally to the system, or instead, modified lysine residues carrying a photocrosslinker are incorporated by means of lysyl tRNA into proteins during cell-free translation. However, it is difficult with these approaches to identify proteins that are in contact with specific regions of a translocating precursor chain.
A method has recently been developed that makes possible the site-specific incorporation of a wide variety of unnatural amino acids into proteins (11, 12). Briefly, an amber suppressor tRNA, which is not a substrate for the endogenous aminoacyl-tRNA synthetases, is chemically modified with an unnatural amino acid. Addition of this acylated suppressor tRNA and a gene containing an amber nonsense codon at the site of interest to an in vitro transcription/translation system yields the desired mutant protein. This method can be used to incorporate photoreactive amino acids at unique positions in proteins. Subsequent photocrosslinking should make it possible to identify components of the import machinery interacting with distinct sites of the polypeptide chain during import. Indeed, this approach has been successfully employed, by using l-3-[p-(trifluoromethyl)-3H-diazirin-3-yl]phenylalanine (Tmd-Phe) as a photoreactive amino acid, for studying the interactions of translocating proteins at the endoplasmic reticulum membrane (13, 14).
In the present study, we have tested amino acids with photoreactive benzophenone side chains of various lengths (1 and 2; see Fig. 1 for structural formulas) for their ability to be site-specifically incorporated into proteins. In comparison to carbene precursors, such as Tmd-Phe, benzophenone derivatives are expected to have advantages in their chemical stability, selective photochemistry and ability to be repeatedly excited by UV illumination without loss of reactivity (15). By using the suppressor-tRNA-mediated method, 1 and 2 were efficiently incorporated at various positions of pb2(220)-DHFR, an intermembrane space-targeting fusion protein consisting of the first 220 residues of the cytochrome b2 precursor (the 80-residue presequence plus 140 residues of the mature protein) and mouse dihydrofolate reductase (DHFR), and pb2 Δ19(220)-DHFR, a matrix-targeting mutant of pb2(220)-DHFR carrying a deletion of 19 amino acid residues (residues 47–65) in the intermembrane space sorting signal (Fig. 1). In vitro import of pb2(220)-DHFR and pb2 Δ19(220)-DHFR into isolated yeast mitochondria was arrested by stabilizing the tertiary structure of the DHFR moiety with methotrexate (Mtx). Subsequently photocrosslinking was performed and several crosslinked products were detected. The appearance of products depended on the position at which 1 or 2 was incorporated in the precursor chains. By analyzing these crosslinked products, it was possible to identify proteins that interacted with various segments of the translocating precursor proteins.
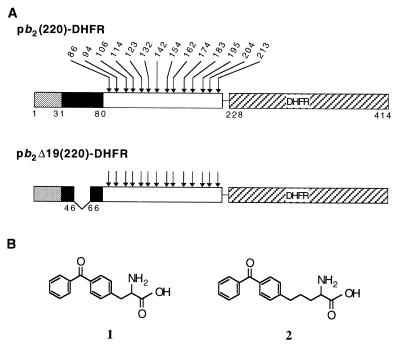
Figure 1.
(A) The fusion protein pb2(220)-DHFR contains the first 220 residues of the cytochrome b2 precursor fused with a 7-residue linker fragment to mouse dihydrofolate reductase (DHFR). The fusion protein pb2 Δ19(220)-DHFR contains the first 220 residues of the cytochrome b2 precursor, lacking residues 47–65 of the presequence, fused with a 7-residue linker fragment to DHFR. The first 31-residue segment of the presequence is cleaved by matrix-localized processing peptidase (MPP) in both proteins, and the second 49-residue segment is cleaved by inner membrane protease I (Imp1p) in pb2(220)-DHFR. The positions of amino acids that were replaced with 1 or 2 are indicated by arrows. The numbering of amino acid residues for pb2(220)-DHFR was used for pb2 Δ19(220)-DHFR so that the crosslinking results (Table 1) can be directly compared. (B) Chemical structures of the photoreactive amino acids dl-2-amino-3-(p-benzoylphenyl)propanoic acid (1) and dl-2-amino-5-(p-benzoylphenyl)pentanoic acid (2).
MATERIALS AND METHODS
Construction of Plasmids.
The CYTB2 gene (donated by G. Reid) was inserted into the EcoRI and HindIII sites of pUC118. The resulting plasmid, pHW1, was then used as template for PCR with primers 5′-CAGGAAACAGCTATGACCATG-3′ and 5′-GCCTCTCGAGAGATCCGCTTGTTTAGTCAAAG-3′ to generate a DNA fragment encoding the first 220 residues of the precursor of cytochrome b2. The amplified 0.7-kb fragment was cut with EcoRI and XhoI and ligated into the EcoRI and XhoI sites of pEH18 (16) (which contains the gene for DHFR) to generate a DNA segment encoding pb2(220)-DHFR (a linker consisting of the seven extra amino acids, DLSRSGI, was inserted). The DNA segment for pb2(220)-DHFR was subsequently subcloned into pGEM-4z (Promega) and mutagenized to generate a derivative of pb2(220)-DHFR, designated pb2D(220), in which the codons for Asp85-Met86 are replaced by those for Glu85-Leu86. The plasmid pb2DΔ19(220), which harbors the gene for pb2Δ19(220)-DHFR, was constructed by replacing a 0.5-kb EcoRI/BamHI fragment of pb2D(220) with a 0.45-kb EcoRI/BamHI fragment of pb2DΔ19(167)), which was constructed as described previously (17). The codons for residues 86, 94, 106, 114, 123, 132, 142, 154, 162, 174, 183, 195, 204, and 213 of pb2(220)-DHFR and pb2Δ19(220)-DHFR were replaced by a TAG codon by using appropriate oligonucleotides according to the procedure of Kunkel et al. (18). The resultant nucleotide replacements were confirmed by sequencing.
Synthesis of Photocrosslinking Compounds.
N-Alloc-dl-2-amino-5-(p-benzoylphenyl)pentanoic acid (6) (Alloc = allyloxycarbonyl). A stirred mixture of benzoyl chloride (5 ml, 43 mmol) and aluminum chloride (3.4 g, 25 mmol) was heated to 60°C. After 20 min, (3-bromopropyl)benzene (3.98 g, 20 mmol) was added very carefully and the reaction mixture was heated with stirring for an additional 4 hr. The mixture was cooled to 0°C, quenched with aqueous 2 M HCl, and extracted with CH2Cl2 three times. The combined organic layers were washed with saturated aqueous NaHCO3 and water, dried over anhydrous MgSO4, filtered, and concentrated in vacuo. The crude product was purified by flash column chromatography (4:1 hexane/EtOAc, Rf = 0.75) on silica gel to give pure p-(3-bromopropyl)benzophenone (3) in 55% yield as a brown oil: 1H NMR (C2HCl3) δ 7.80 (2H, d, J = 7.9), 7.76 (2H, d, J = 8.2), 7.60 (1H, t, J = 7.3), 7.49 (2H, t, J = 7.3), 7.32 (2H, d, J = 8.1), 3.43 (2H, t, J = 6.5), 2.89 (2H, t, J = 7.2), 2.23 (2H, m). To a stirred solution of t-butyl N-(benzylidene)glycinate (1.6 g, 7.3 mmol) (19) in 100 ml of dry tetrahydrofuran (THF) was added sodium bis(trimethylsilyl)amide (7.5 ml, 7.5 mmol, 1.0 M solution in THF) at −78°C. After 20 min, a solution of 3 (1.7 g, 5.61 mmol) in 10 ml of THF was added slowly at the same temperature. The reaction mixture was stirred overnight at room temperature, then quenched by addition of saturated aqueous NH4Cl, and extracted with CH2Cl2 three times. Solvent was removed in vacuo and the crude product was purified by flash column chromatography (4:1 hexane/EtOAc, Rf = 0.02) on silica gel to give pure t-butyl dl-2-amino-5-(p-benzoylphenyl)pentanoate (4) in 51% yield as a brown oil: 1H NMR (C2HCl3) δ 7.78 (2H, d, J = 7.0), 7.73 (2H, d, J = 8.2), 7.57 (1H, t, J = 7.4), 7.47 (2H, t, J = 7.3), 7.28 (2H, d, J = 8.2), 3.33 (1H, dd, J = 7.1, 4.5), 2.72 (2H, m), 1.80–1.75 (2H, m), 1.74–1.68 (2H, m), 1.50 (9H, s). To a stirred solution of 4 (630 mg, 1.78 mmol) and triethylamine (0.5 g, 4.9 mmol) in 30 ml of dioxane was added allyl chloroformate (236 mg, 2.0 mmol) at 0°C. After 30 min at room temperature, the reaction was quenched by addition of aqueous 1 M NaHSO4, and the mixture was extracted with CH2Cl2 three times. Solvent was removed in vacuo and the crude product was purified by flash column chromatography (4:1 hexane/EtOAc, Rf = 0.47) on silica gel to give t-butyl N-Alloc-dl-2-amino-5-(p-benzoylphenyl)pentanoate (5) as a yellow oil. The free acid of 5 was generated by adding 5 (730 mg, 1.67 mmol) and anisole (0.5 ml) to 20% trifluoroacetic acid in CH2Cl2 (15 ml). The reaction mixture was stirred overnight at room temperature. The volatile material was removed in vacuo and the residue was directly purified by flash column chromatography (4:1 hexane/EtOAc, Rf = 0.1) on silica gel to give dl-N-Alloc-2-amino-5-(p-benzoylphenyl)pentanoic acid (6) in 70% yield as a very viscous liquid: 1H NMR (C2HCl3) δ 7.78 (2H, d, J = 7.0), 7.73 (2H, d, J = 8.2), 7.56 (1H, t, J = 6.2), 7.47 (2H, t, J = 7.8), 7.27 (2H, d, J = 8.2), 5.90 (1H, m), 5.30 (1H, d, J = 17.2), 5.21 (1H, d, J = 10.4), 4.57 (2H, d, J = 5.4), 4.44 (1H, m), 2.8–2.7 (2H, m), 2.09–1.92 (1H, m), 1.90–1.85 (3H, m); high-resolution MS, calculated 382.1655, found 382.1645 (M+1).
5′-Phospho-2′-deoxycitidylyl(3′-5′)-2′(3′)-O-[dl-2-amino-5-(p-benzoylphenyl)pentanoyl]adenosine (9). N-Alloc-dl-2-amino-5-(p-benzoylphenyl)pentanoate cyanomethyl ester (7) was prepared as described (20) in 90% yield: 1H NMR (C2HCl3) δ 7.78 (2H, d, J = 8.2), 7.74 (2H, d, J = 7.9), 7.58 (1H, t, J = 7.5), 7.47 (2H, t, J = 7.7), 7.26 (2H, d, J = 8.05), 5.89 (1H, m), 5.30 (1H, d, J = 17.7), 5.22 (1H, d, J = 10.4), 4.82 (1H, d, J = 15.7), 4.71 (1H, d, J = 15.6), 4.56 (2H, d, J = 4.4), 4.40 (1H, m), 2.72 (2H, m), 1.94 (1H, m), 1.70 (3H, m). The activated ester was ligated to the dinucleotide pdCpA to afford 5′-phospho-2′-deoxycitidylyl(3′-5′)-2′(3′)-O-[N-Alloc-dl-2-amino-5-(p-benzoylphenyl)pentanoyl]adenosine (8) (20). The Alloc protecting group was removed by dissolving 8 (28 μmol), acetic acid (0.28 ml) and N-methylmorpholine (87 μl) in dimethylformamide (0.9 ml). The solution was degassed with nitrogen for 3 min, tetrakis(triphenylphosphine)palladium (10 mg, 8.65 μmol) was added, and then the reaction mixture was stirred under a nitrogen atmosphere at room temperature for 1.5 hr. The reaction was quenched by addition of 8 ml of 50 mM NH4OAc, pH 4.5/CH3CN (1:1), and the mixture was directly purified by preparative HPLC as described (20): high-resolution MS, calculated 916.2432, found 916.2425 (M+1).
5′-Phospho-2′-deoxycitidylyl(3′-5′)-2′(3′)-O-[dl-2-amino-5-(p-benzoylphenyl)propanoyl]adenosine (12). N-Alloc-dl-2-amino-5-(p-benzoylphenyl)propanoate cyanomethyl ester (10) was prepared by literature procedure (20) from the corresponding amino acid, which has previously been synthesized (15): 1H NMR (C2HCl3) δ 7.80 (2H, d, J = 2.5), 7.78 (2H, d, J = 2.5), 7.60 (1H, t, J = 7.4), 7.50 (2H, t, J = 7.5), 7.29 (2H, d, J = 8.1), 5.9 (1H, m), 5.30 (1H, d, J = 17.1), 5.24 (1H, d, J = 10.5), 4.84 (1H, d, J = 15.7), 4.78 (1H, m), 4.72 (1H, d, J = 15.7), 4.58 (2H, d, J = 5.6), 3.31–3.20 (2H, m). 5′-Phospho-2′-deoxycitidylyl(3′-5′)-2′(3′)-O-[N-Alloc-dl-2-amino-5-(p-benzoylphenyl)propanoyl]adenosine (11) was prepared as described (20) and the Alloc deprotection was performed as described above: high-resolution MS, calculated 888.2119, found 888.2129 (M+1).
In Vitro Synthesis of pb2(220)-DHFR and pb2Δ19(220)-DHFR Containing 1 or 2.
The aminoacylated suppressor tRNA was prepared as described previously (11). The proteins pb2(220)-DHFR and pb2 Δ19(220)-DHFR were synthesized by coupled transcription/translation in reticulocyte lysate (21) in the presence of [35S]methionine and aminoacylated suppressor tRNA at 170 μg/ml.
In Vitro Import of pb2(220)-DHFR and pb2Δ19(220)-DHFR into Isolated Yeast Mitochondria and Photocrosslinking.
Mitochondria were isolated from yeast strain D273-10B as described by Daum et al. (22). For mitochondrial protein import, mitochondria (0.5 mg/ml protein) were incubated with 5 μl of translation products containing the fusion proteins in 100 μl of import buffer (250 mM sucrose/10 mM Mops–KOH, pH 7.2/80 mM KCl/5 mM MgCl2/2.5 mM KPi/5 mM dithiothreitol/5 mM methionine/1% BSA/2 mM NADH) for 20 min at 30°C. When translocation intermediates were being generated, radiolabeled fusion proteins in reticulocyte lysate were preincubated with 1 μM Mtx and 1 mM NADPH in import buffer for 15 min on ice. For ATP depletion, the mitochondria were posttreated with 40 units/ml apyrase and 20 μM oligomycin for 10 min at 30°C after import. After the import reaction, the mitochondria were isolated by centrifugation and washed once with washing buffer (250 mM sucrose/10 mM Mops–KOH, pH 7.2/80 mM KCl). Washing buffer for the translocation intermediates contained 1 μM Mtx. Crosslinking of translocation intermediates was performed as follows. The mitochondrial pellets were resuspended in washing buffer with 1 μM Mtx (to 0.5 mg/ml mitochondrial protein) and were UV irradiated for 5 min on ice at a distance of 1–2 cm from a 365-nm UV lamp (15 W, CSL-15A, Cosmo Bio, Tokyo). The mitochondria were recovered by centrifugation and subjected to SDS/PAGE followed by visualization with an image-analyzing system BAS2000 (Fuji Film).
Immunoprecipitation.
After photocrosslinking, mitochondria were reisolated by centrifugation, dissolved in 1% SDS/1 mM phenylmethylsulfonyl fluoride/20 mM Tris·HCl, pH 8.0/150 mM NaCl, and boiled for 5 min. The solubilized mitochondria were clarified by centrifugation, diluted 20-fold with 0.5% Triton X-100/1 mM phenylmethylsulfonyl fluoride/20 mM Tris·HCl, pH 8.0/150 mM NaCl. The samples were incubated with antibodies against Tom40, Tim44, or Ssc1p with gentle mixing overnight at 4°C and subsequently with staphylococcal protein A-Sepharose for 2 hr at room temperature. The immunoprecipitates were washed twice with 0.5% Triton X-100/20 mM Tris·HCl, pH 8.0/150 mM NaCl and once with 20 mM Tris·HCl, pH 8.0/150 mM NaCl, and were analyzed by SDS/PAGE and visualization with image-analyzing system BAS2000.
RESULTS
The Fusion Proteins Bearing 1 or 2 Were Arrested on the Import Pathways.
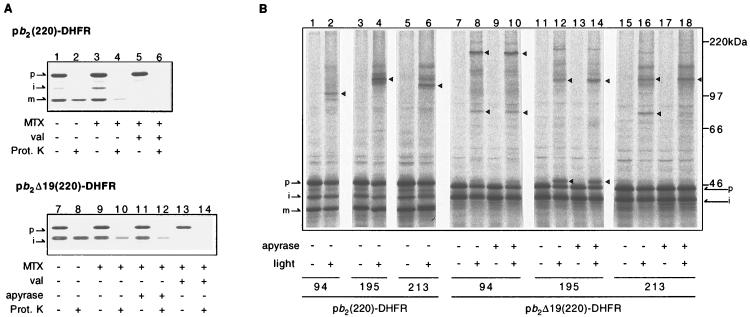
Codons in the genes for pb2(220)-DHFR and pb2 Δ19(220)-DHFR were replaced with the amber nonsense codon, TAG, so that 1 or 2 could be incorporated every ≈10 residues between residues 86 and 213 of pb2(220)-DHFR and pb2Δ19(220)-DHFR (Fig. 1). The modified DNAs were added to the reticulocyte lysate cell-free transcription/translation system containing [35S]methionine and suppressor tRNAs chemically charged with 1 or 2. Radiolabeled pb2(220)-DHFR and pb2Δ19(220)-DHFR bearing 1 or 2 were synthesized with suppression efficiencies of ≤60%. These fusion proteins are readily taken up by isolated yeast mitochondria and reach the intermembrane space and the matrix, respectively (Fig. 2A, lanes 1, 2, 7, and 8). This suggests that the presence of 1 or 2 does not affect import into mitochondria and subsequent intramitochondrial sorting of the fusion proteins.
Figure 2.
Translocation intermediates of pb2(220)-DHFR and pb2Δ19(220)-DHFR containing 2 and their crosslinked products. (A) In vitro import of pb2(220)-DHFR (lanes 1–6) or pb2Δ19(220)-DHFR (lanes 7–14) containing 2 at position 195 into isolated yeast mitochondria. The fusion proteins were preincubated in the presence (lanes 3–6 and 9–14) or absence (lanes 1, 2, 7, and 8) of 1 μM Mtx and 1 mM NADPH, and were subsequently incubated with mitochondria for 20 min at 30°C in the absence (lanes 1–4 and 7–12) or presence (lanes 5, 6, 13, and 14) of 10 μg/ml valinomycin. In lanes 11 and 12, ATP was depleted by further incubation of mitochondria with 40 units/ml apyrase and 20 μM oligomycin for 10 min at 30°C. Mitochondria were reisolated by centrifugation, washed once with washing buffer, and suspended in the same buffer. The samples were halved, and incubated for 30 min on ice with (even-numbered lanes) or without (odd-numbered lanes) 100 μg/ml proteinase K. (B) Fusion proteins containing 2 at indicated positions were arrested as translocation intermediates by inhibiting the unfolding of the DHFR moiety by Mtx. In lanes 9, 10, 13, 14, 17, and 18, ATP was depleted after the import but prior to UV irradiation. The samples were UV irradiated for 5 min on ice (even-numbered lanes). Triangles indicate major crosslinked products that were detected only in the presence of Mtx and only in the absence of valinomycin. MTX, methotrexate; val, valinomycin (dissipating the membrane potential across the inner membrane); Prot. K, proteinase K; light, UV irradiation; p, precursor-size form; i, intermediate-size form; m, mature-size form.
The fusion proteins pb2(220)-DHFR and pb2Δ19(220)-DHFR can be arrested as “translocation intermediates” spanning mitochondrial membrane(s) when unfolding of the DHFR moiety is inhibited by binding of Mtx, a high-affinity ligand for DHFR (17). Upon incubation with isolated mitochondria in the presence of Mtx, pb2(220)-DHFR is processed in a potential-dependent manner to the intermediate-size form by MPP and, subsequently, to the mature-size form by Imp1p, suggesting that pb2(220)-DHFR reaches the intermembrane space (Fig. 2A, lanes 3 and 5). However, pb2(220)-DHFR is still susceptible to digestion by an externally added protease, indicating that the DHFR–Mtx complex remains outside the mitochondria (Fig. 2A, lane 4). According to the conservative-sorting model, the arrested pb2(220)-DHFR should form a loop that spans three membranes, whereas according to the stop-transfer model it should span only the outer membrane.
Upon incubation with isolated mitochondria in the presence of Mtx, pb2Δ19(220)-DHFR is processed in a potential-dependent manner by MPP to the intermediate-size form, suggesting that pb2Δ19(220)-DHFR reaches the matrix (Fig. 2A, lanes 9, 11, and 13). Like pb2(220)-DHFR, pb2Δ19(220)-DHFR is accessible to externally added protease, although the presence of ATP renders the fusion protein partly resistant to digestion (lanes 10 and 12), probably due to closer contact of the DHFR moiety to the outer membrane than in the absence of ATP (23). The pb2Δ19(220)-DHFR translocation intermediate thus spans both the outer and the inner membranes. The translocation intermediates of pb2(220)-DHFR and pb2Δ19(220)-DHFR were able to complete translocation when Mtx was removed by isolating the mitochondria and resuspending them in buffer lacking Mtx (ref. 24; data not shown).
The Translocation Intermediates Were Photocrosslinked to the Components Mediating Mitochondrial Protein Import.
The translocation intermediates of pb2(220)-DHFR and pb2Δ19(220)-DHFR containing 1 or 2 were subjected to UV irradiation. The resulting crosslinked products were identified on SDS/PAGE by their reduced mobility relative to noncrosslinked polypeptides (Fig. 2B). Crosslinking efficiency was ≤10%. Generation of the crosslinked products required UV irradiation and the presence of a membrane potential across the inner membrane, and it was dependent on the position of the photoreactive amino acid along the polypeptide chain. By comparing the crosslinking patterns with and without metal chelators, which inhibit MPP but not Imp1p (25), we found that most of the crosslinked products arose from the intermediate-size or the mature-size forms but not from the precursor forms of the fusion proteins (Table 1). The presence of ATP during UV irradiation also affected crosslinking with pb2Δ19(220)-DHFR (Fig. 2B, lanes 7–18). The crosslinking efficiency and suppression efficiencies of 1 and 2 varied with the positions at which they were incorporated in the proteins. For simplicity, only the results with 2 are shown.
Table 1.
Summary and quantification of crosslinking with pb2(220)-DHFR and pb2Δ19(220)-DHFR
| Position* | pb2(220)-DHFR crosslinking, %
|
pb2Δ19(220)-DHFR crosslinking, %
|
|||||
|---|---|---|---|---|---|---|---|
| To Tom40 | To Tim44 | To Ssc1p | To X | To Tom40 | To Tim44 | To Ssc1p | |
| 86 | − | − | − | − | − | − | − |
| 94 | − | − | − | i ≈10† | − | − | − |
| 114 | − | − | − | i ≈ 3† | − | − | − |
| 123 | − | − | − | − | − | − | − |
| 132 | − | − | − | − | ND | ND | ND |
| 154 | ND | ND | ND | ND | − | − | − |
| 162 | i ≈ 10† | − | − | − | − | − | i ≈ 3† |
| 183 | i ≈ 10‡ | − | − | − | ND | ND | ND |
| 186 | i ≈ 10‡ | ||||||
| m ≈ 10§ | |||||||
| 195 | i ≈ 5‡ | − | − | − | − | i ≈ 5‡ | − |
| m ≈ 5§ | |||||||
| 204 | ? 5–10 | − | − | − | − | − | − |
| 213 | i ≈ 4‡ | − | − | − | i ≈ 2† | − | − |
| m ≈ 3§ | |||||||
The radioactivity in the bands was quantified with image-analyzing system BAS2000 (Fuji Film). The radioactivity before crosslinking in the intermediate-size or the mature-size forms of the fusion proteins from which crosslinked products arose was taken as 100%. −, No crosslinking was detected; i, the intermediate-size form; m, the mature-size form. ND, crosslinking was not detected because of low efficiencies of suppression or the accumulation of translocating intermediates. The data for positions 106, 142, and 174 are all ND and are therefore omitted from the table.
Position, residue number of introduced 2.
The crosslinked products arose from the intermediate-size forms of the fusion proteins, since the presence of metal chelators (5 mM EDTA + 1 mM o-phenanthroline) caused a shift of the band to higher molecular mass by more than ≈3 kDa or changed the band into a smear.
The crosslinked products arose from the intermediate-size forms of the fusion proteins, since the presence of metal chelators caused a shift of the band to higher molecular mass by ≈3 kDa.
The crosslinked products arose from the mature-size form of the fusion protein since, it appeared just below the one arising from the intermediate-size form.
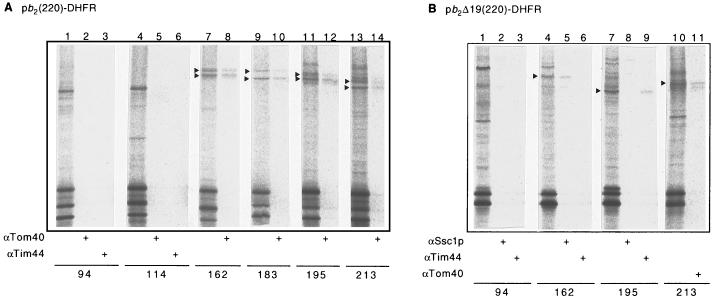
To identify the components in mitochondria that were crosslinked to the translocation intermediates after UV irradiation, the mitochondria were solubilized with detergent and subjected to immunoprecipitation with antibodies directed against the proteins that are known to mediate protein translocation across the mitochondrial membranes (Fig. 3). In the case of pb2(220)-DHFR, we observed a crosslinked product of 95 kDa when 2 was incorporated at residues 94 and 114 (Fig. 3A, lanes 1 and 4), and crosslinked products of 90–100 kDa when 2 was incorporated at residues 162, 183, 195, and 213 (Fig. 3A, lanes 7, 9, 11, and 13). The crosslinked products of 90–100 kDa were efficiently immunoprecipitated by antibodies against Tom40 (Fig. 3A, lanes 8, 10, 12, and 14). In the case of pb2Δ19(220)-DHFR, we found crosslinked products of 130, 89, and 97 kDa when 2 was incorporated at residues 162, 195, and 213, respectively (Fig. 3B, lanes 4, 7, and 10). The crosslinked product of 130 kDa (2 at residue 162) was immunoprecipitated with antibodies against Ssc1p; the product of 89 kDa (2 at residue 195), with antibodies against Tim44; and the product of 97 kDa (2 at residue 213), with antibodies against Tom40 (Fig. 3B, lanes 5, 9, and 11). These results are summarized in Table 1. Fig. 4 shows possible working models for the membrane topology of the pb2(220)-DHFR and pb2Δ19(220)-DHFR intermediates accounting for the results of the photocrosslinking experiments.
Figure 3.
Crosslinked products with pb2(220)-DHFR (A) or pb2Δ19(220)-DHFR (B) containing 2 at the indicated positions were analyzed by immunoprecipitation with antibodies against Tom40, Tim44, and Ssc1p. The major crosslinked products immunoprecipitated with one of these antibodies are indicated with arrowheads. αTom40, antibodies against Tom40; αTim44, antibodies against Tim44; αSsc1p, antibodies against Ssc1p.
Figure 4.
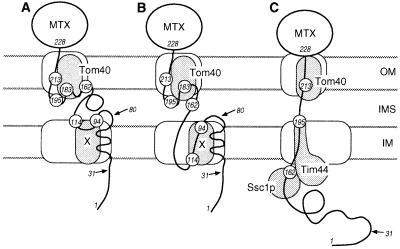
Working models for the interactions of the translocation intermediates with the mitochondrial import machinery in the outer and the inner membrane. (A) The translocation intermediate of pb2(220)-DHFR in the “stop-transfer” model. (B) The translocation intermediate of pb2(220)-DHFR in the “conservative-sorting” model. (C) The translocation intermediate of pb2Δ19(220)-DHFR. The numbers indicate positions of incorporated 2, and the arrows indicate the processing sites. MTX, methotrexate; OM, the outer membrane; IMS, the intermembrane space; IM, the inner membrane.
DISCUSSION
In the present study, amino acids with benzophenone side chains of various lengths (1 and 2) were incorporated site-specifically into the fusion proteins pb2(220)-DHFR and pb2Δ19(220)-DHFR. The translocation intermediates of pb2(220)-DHFR and pb2Δ19(220)-DHFR were efficiently photocrosslinked to Tom40, Tim44, and/or Ssc1p, whose involvement in protein import into mitochondria has been established (Table 1). These results demonstrate the validity of this approach for studying the environment of translocating precursor proteins. Importantly, analyses of crosslinked products allowed us to identify which segments of the translocating precursor chain are in contact with each component of the translocation machinery. The intermediate-size form of pb2Δ19(220)-DHFR with 2 incorporated at position 213 was crosslinked to Tom40 in the outer membrane, protein with 2 at position 195 was crosslinked to Tim44 in the inner membrane, and protein with 2 at position 162 was crosslinked to Ssc1p in the matrix. Since the DHFR moiety starts at position 228, the 66-residue segment between positions 162 and 228 must span both the outer and the inner membranes. The thickness of mitochondrial contact sites assessed by electron microscopy is 18–20 nm (26). A 66-amino acid peptide corresponds to ≈9.9 nm in length in the α-helical conformation and ≈22 nm in the extended β-strand conformation (27). Therefore, the polypeptide chain of pb2Δ19(220)-DHFR must have an extended conformation in the import channels (Fig. 4). This supports previous observations that the DHFR fusion proteins which are complexed with Mtx and arrested at translocation contact sites require ≈50 residues to span the outer and the inner membranes and have access to MPP in the matrix, and an additional ≈30-40 residues to be held stably in the import channels (probably by Ssc1p binding in the matrix) (28, 29). The distance between residues 195 and 228, which would be 12 nm in the extended β-strand conformation, is not sufficient to span the two membranes. Previous studies showed that the bulk of Tim44 is located on the matrix side and only the C-terminal tail is exposed on the intermembrane side (30, 31). Therefore, protein with 2 at position 195 appears to be in contact with the C-terminal tail of Tim44 near the intermembrane space side, not the matrix side, of the inner membrane (Fig. 4).
Surprisingly, the pattern of crosslinking for pb2(220)-DHFR differs significantly from that for pb2Δ19(220)-DHFR (Table 1). In the case of the pb2(220)-DHFR intermediate, proteins containing 2 at positions between 162 and 213 were all crosslinked to Tom40 in the outer membrane, while in the case of pb2Δ19(220)-DHFR, the protein with 2 at positions 195 and 162 was crosslinked to Tim44 in the inner membrane and to Ssc1p in the matrix, respectively. It is possible that pb2(220)-DHFR randomly slides back and forth in the TOM channel in the outer membrane so that the segment between residues 162 and 213 is transiently accessible to Tom40. However, this explanation seems unlikely, because protease treatment of the pb2(220)-DHFR intermediate (mainly in the mature-size form) results in a major 17-kDa fragment inside the mitochondria (data not shown). This size is in good agreement with a fragment of 147 amino acids (16.5 kDa), expected for the cytochrome b2 part (residues 81–220 plus the 7-residue linker) of the fusion protein. A more likely explanation is that the pb2(220)-DHFR intermediate does not have an extended conformation, but its 50-residue segment (residues 162–213) is close to Tom40 (Fig. 4). The extended conformation of pb2Δ19(220)-DHFR, but not pb2(220)-DHFR, is likely stabilized by binding of Ssc1p to the N-terminal portion of pb2Δ19(220)-DHFR in the matrix. These results suggest that the N-terminal regions of pb2Δ19(220)-DHFR and pb2(220)-DHFR, even though they have translocated across the outer membrane and reached their destinations, still affect the interactions of the C-terminal regions of the fusion proteins with Tom proteins.
The partner of the major crosslinked product of 95 kDa observed for the intermediate-size form of pb2(220)-DHFR with 2 incorporated at positions 94 and 114 (Fig. 3A, lanes 1 and 4) has not been identified yet. The crosslinked product with protein X was not extracted by treatment with sodium carbonate, pH 11.5, suggesting that protein X is an integral membrane protein (data not shown). Models of possible membrane topology for the pb2(220)-DHFR intermediate (Fig. 4) suggest that this crosslinked partner (X) is likely an inner membrane protein. Since the crosslinked product with protein X is observed only with pb2(220)-DHFR, not with pb2Δ19(220)-DHFR, protein X may be involved in the sorting of pb2(220)-DHFR to the intermembrane space. In the stop-transfer model, protein X may be a protein that recognizes the sorting signal to arrest the translocation. In the conservative-sorting model, protein X may constitute a putative export machinery. We are currently trying to characterize this novel crosslink partner, protein X.
Acknowledgments
We thank T. Yano for technical assistance and G. Reid, B. S. Glick, and G. Schatz for the cytochrome b2 gene. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture (T.E.), by a grant for the Biodesign Research Program for the Institute of Physical and Chemical Research (T.E.), and by a grant from the National Institutes of Health (Grant No. GM49220) (P.G.S.). T.K. is a Research Fellow of the Japan Society for the Promotion of Science. P.G.S. is a Howard Hughes Medical Institute Investigator.
Footnotes
Abbreviations: Tmd-Phe, l-3-[p-(trifluoromethyl)-3H-diazirin-3-yl]phenylalanine; 1, dl-2-amino-3-(p-benzoylphenyl)propanoic acid; 2, dl-2-amino-5-(p-benzoylphenyl)pentanoic acid; DHFR, dihydrofolate reductase; pb2(220)-DHFR, a fusion protein consisting of the first 220 residues of the cytochrome b2 precursor (the 80-residue presequence plus 140 residues of the mature protein) and mouse DHFR; pb2Δ19(220)-DHFR, a mutant of pb2(220)-DHFR carrying a deletion of 19 amino acid residues (residues 47–65) in the intermembrane-space sorting signal; Mtx, methotrexate; Alloc-, allyloxycarbonyl-; MPP, matrix-localized processing peptidase; Imp1p, inner membrane protease I.
References
- 1.Lill R, Nargang F E, Neupert W. Curr Opin Cell Biol. 1996;8:505–512. doi: 10.1016/s0955-0674(96)80028-7. [DOI] [PubMed] [Google Scholar]
- 2.Schatz G, Dobberstein B. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 3.Mihara K, Omura T. Trends Cell Biol. 1996;6:104–108. doi: 10.1016/0962-8924(96)81000-2. [DOI] [PubMed] [Google Scholar]
- 4.Lill R, Neupert W. Trends Cell Biol. 1996;6:56–61. doi: 10.1016/0962-8924(96)81015-4. [DOI] [PubMed] [Google Scholar]
- 5.Pfanner N, Craig E A, Meijer M. Trends Biochem Sci. 1994;19:368–372. doi: 10.1016/0968-0004(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 6.Schneider H-C, Berthold J, Bauer M F, Dietmeier K, Guiard B, Brunner M, Neupert W. Nature (London) 1994;371:768–774. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- 7.Horst, M., Azem, A., Schatz, G. & Glick, B. S. (1997) Biochim. Biophys. Acta, in press. [DOI] [PubMed]
- 8.Glick B S, Beasley E M, Schatz G. Trends Biochem Sci. 1992;17:453–459. doi: 10.1016/0968-0004(92)90487-t. [DOI] [PubMed] [Google Scholar]
- 9.Stuart R A, Neupert W. Trends Biochem Sci. 1996;21:261–267. [PubMed] [Google Scholar]
- 10.Martoglio B, Dobberstein B. Trends Cell Biol. 1996;6:142–147. doi: 10.1016/0962-8924(96)10001-5. [DOI] [PubMed] [Google Scholar]
- 11.Ellman J A, Mendel D, Schultz P G. Methods Enzymol. 1991;202:301–336. doi: 10.1016/0076-6879(91)02017-4. [DOI] [PubMed] [Google Scholar]
- 12.Bain J D, Glabe C G, Dix T A, Chamberlain A R. J Am Chem Soc. 1989;111:8013–8014. [Google Scholar]
- 13.High S, Martoglio B, Görlich F, Andersen S S L, Ashford A J, Ginger A, Hartmann E, Prehn S, Rapoport T A, Dobberstein B, Brunner J. J Biol Chem. 1993;268:26745–26751. [PubMed] [Google Scholar]
- 14.Martoglio B, Hofmann M W, Brunner J, Dobberstein B. Cell. 1995;81:207–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- 15.Kauer J C, Erickson-Viitanen S, Wolfe H R, Jr, DeGrado W F. J Biol Chem. 1986;261:10695–10700. [PubMed] [Google Scholar]
- 16.Hurt E C, Pesold-Hurt B, Schatz G. FEBS Lett. 1984;178:306–310. doi: 10.1016/0014-5793(84)80622-5. [DOI] [PubMed] [Google Scholar]
- 17.Koll H, Guiard B, Rassow J, Ostermann J, Horwich A L, Neupert W, Hartl F-U. Cell. 1992;68:1163–1175. doi: 10.1016/0092-8674(92)90086-r. [DOI] [PubMed] [Google Scholar]
- 18.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 19.Stork G, Leong A Y W, Touzin A M. J Org Chem. 1976;41:3491–3493. doi: 10.1021/jo00883a044. [DOI] [PubMed] [Google Scholar]
- 20.Robertson S A, Ellman J A, Schultz P G. J Am Chem Soc. 1991;113:2722–2729. [Google Scholar]
- 21.Jackson R J, Hunt T. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- 22.Daum G, Böhni P C, Schatz G. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 23.Schwarz E, Seytter T, Guiard B, Neupert W. EMBO J. 1993;12:2295–2302. doi: 10.1002/j.1460-2075.1993.tb05883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruhler A, Ono H, Guiard B, Neupert W, Stuart R A. EMBO J. 1995;14:1349–1359. doi: 10.1002/j.1460-2075.1995.tb07121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Böhni P C, Daum G, Schatz G. J Biol Chem. 1983;258:4937–4943. [PubMed] [Google Scholar]
- 26.Rassow J, Guiard B, Wienhues U, Herzog V, Hartl F-U, Neupert W. J Cell Biol. 1989;109:1421–1428. doi: 10.1083/jcb.109.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Creighton T E. Proteins: Structures and Molecular Properties. 2nd Ed. New York: Freeman; 1993. [Google Scholar]
- 28.Rassow J, Hartl F-U, Guiard B, Pfanner N, Neupert W. FEBS Lett. 1990;275:190–194. doi: 10.1016/0014-5793(90)81469-5. [DOI] [PubMed] [Google Scholar]
- 29.Ungermann C, Neupert W, Cyr D M. Science. 1994;266:1250–1253. doi: 10.1126/science.7973708. [DOI] [PubMed] [Google Scholar]
- 30.Maarse A C, Blom J, Grivell L A, Meijer M. EMBO J. 1992;11:3619–3628. doi: 10.1002/j.1460-2075.1992.tb05446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blom J, Kübrich M, Rassow J, Voos W, Dekker P J T, Maarse A C, Meijer M, Pfanner N. Mol Cell Biol. 1993;13:7364–7371. doi: 10.1128/mcb.13.12.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]