Abstract
Tertiary structure of supercoiled DNA is a significant factor in a number of genetic functions and is apparently affected by environmental conditions. We applied atomic force microscopy (AFM) for imaging the supercoiled DNA deposited at different ionic conditions. We have employed a technique for the sample preparation that permits high-resolution AFM imaging of DNA bound to the surface in buffer solutions without drying the sample (AFM in situ). The AFM data show that at low ionic strength, DNA molecules are loosely interwound supercoils with an irregular shape. Plectonemic superhelices are formed in high-concentration, near-physiological salt solutions. At such ionic conditions, superhelical loops are typically separated by regions of close helix–helix contacts. The data obtained show directly and unambiguously that overall geometry of supercoiled DNA depends dramatically on ionic conditions. This fact and the formation of close contacts between DNA helices are important features of supercoiled DNA related to its biological functions.
DNA supercoiling is thought to play key roles in genetic processes in the cell (1). One fundamental feature of DNA supercoiling is that distantly separated DNA regions can be brought in close proximity. Bringing two distant sites into proximity is required for DNA recombination (2–5), for the control of DNA supercoiling by DNA topoisomerases (6–8), and for gene regulation through DNA looping (9–11). A conventional model for supercoiled DNA is a plectonemic helix in which both strands interwind each other. However, DNA is a highly charged polymer, so that electrostatic repulsion of negatively charged DNA helices opposes folding and especially formation of close contacts between DNA regions. Counterions shield the negative charge of DNA and hence decrease the repulsion between DNA segments. So the following questions arise. Does the geometry of supercoiled DNA depend on ionic strength? If it does, then to what extent do salt conditions change the geometry of supercoiled DNA? Finally, what is the geometry of supercoiled DNA at different ionic conditions? These questions are of great importance because the configurations of supercoiled DNA are ultimately related to its functions.
Monte Carlo simulations of supercoiling (12–18) have shown that ion concentration has a strong effect on conformation of supercoiled DNA. In particular, systematic theoretical studies of the effect of ionic strength on the geometry of supercoiled DNA were performed by Vologodskii and Cozzarelli (15, 16). Experimental data on the salt-dependent geometry of supercoiled DNA are quite controversial. Electron microscopy (EM) was successfully applied to studies of DNA supercoiling, and systematic studies of the structure of supercoiled DNA were performed by Boles et al. (19). They found no changes in geometry of molecules when spreading conditions were changed from a low salt buffer to a buffer containing 10 mM MgCl2, which is equivalent to a high salt solution (20). On the contrary, light scattering (21, 22) indicates that the geometry of supercoiled DNA should be sensitive to ionic conditions. Unfortunately, light scattering data do not allow one to retrieve the exact geometry of supercoiled DNA molecules. Most of the data are interpreted in the framework of a toroidal model for DNA supercoiling rather than a plectonemic model supported by EM studies. Very recently, a thorough study of the effect of ionic conditions on conformations of supercoiled DNA has been performed in the laboratory of N. Cozzarelli (V. Rybenkov, A. Vologodskii and N. R. Cozzarelli, unpublished work). In this study, sedimentation data of supercoiled DNA were analyzed in a broad range of ionic conditions, and the authors concluded that the electrostatic interaction of DNA is the primary determinant of conformations of supercoiled DNA.
The major concern with EM as the main direct visualization method is that the sample preparation procedure for DNA may result in artifacts (15, 18). Application of the cryo-EM technique to studies of supercoiled DNA made it possible to observe changes of the overall geometry of the molecules induced by Mg ions (23, 24). However, evaporation of water during the sample preparation cannot be avoided, so the ionic conditions of the vitrified sample are not well specified.
To study conformation of supercoiled DNA in solution at different ionic conditions, we used atomic force microscopy (AFM; ref. 25). AFM was successfully applied to structural studies of numerous biological molecules and their complexes (26–30). We have developed a substrate preparation procedure (aminopropyl mica: AP-mica; refs. 30–34) permitting the deposition of the sample in a broad range of environmental conditions. The recent development of the instrument achieved in Hansma’s group (tapping mode in liquid; ref. 35) permitted them to resolve helical periodicity of DNA imaged in propanol (36). We have combined the advantages of AP-mica described above with the use of tapping-mode AFM in attempts to visualize supercoiled DNA by imaging in buffer solution without drying the sample.
MATERIALS AND METHODS
DNA Samples.
Plasmid DNAs were the gift of S. Adhya and T. Aki (National Institutes of Health, Bethesda). Plasmids were purified according to the procedure described in ref. 37. The samples were used in AFM experiments without additional purification. Aliquots of the DNA stock solutions in TE buffer (20 mM Tris·HCl, pH 7.6/1 mM EDTA) were dissolved in 50 μl of appropriate buffer before imaging. The final concentrations of DNA pSA509 and mini plasmid were ca. 0.8 nM and 2 nM DNA molecules, respectively.
Preparation of Samples for AFM.
Throughout this work, only mica functionalized with 3-aminopropyltriethoxy silane (APTES; AP-mica) was used a substrate. The procedure for preparation of AP-mica has been described (31–34). The activation of mica in vapors of APTES for 2 h was sufficient to obtain uniformly modified substrates suitable for deposition of DNA and DNA–protein complexes (30, 34). The AP-mica stored under argon remained active for 4–5 weeks (34). For imaging in situ, 1 × 1 cm pieces of AP-mica were mounted on the sample stage on the microscope (NanoScope III, Digital Instruments, Santa Barbara, CA), and 25 μl of DNA solution was injected into the gap between the glass tip holder and the AP-mica. For imaging in air, 10 μl of the sample was placed onto pieces of AP-mica for 2 min, thoroughly rinsed with deionized water (ModuPure Plus, Continental Water System Corp., San Antonio, TX), and argon-dried.
AFM Imaging.
The NanoScope III MultiMode system (Digital Instruments) operated in tapping mode was used for these experiments.
For imaging in liquid, Si3N4, 100-μm-long NanoProbe tips were used. They were mounted on a glass tip holder for tapping mode (Digital Instruments), and the tip was manually approached to the AP-mica surface mounted on the sample stage on the microscope. Approach was stopped when the distance between the tip and the surface was 20–50 μm. The DNA solution (25 μl) was injected into a small gap between the mica surface and the tip holder, and the tip was brought into tapping range under control of the instrument. The scanning parameters were as follows: frequency, 8–9 kHz; scanning rate, 1.97 Hz. The images were taken in the topographic mode.
For imaging in air, NanoProbe TESP (Digital Instruments) tips were used. The typical tapping frequency was 240–280 kHz, and the scanning rate was 1.97 Hz.
RESULTS
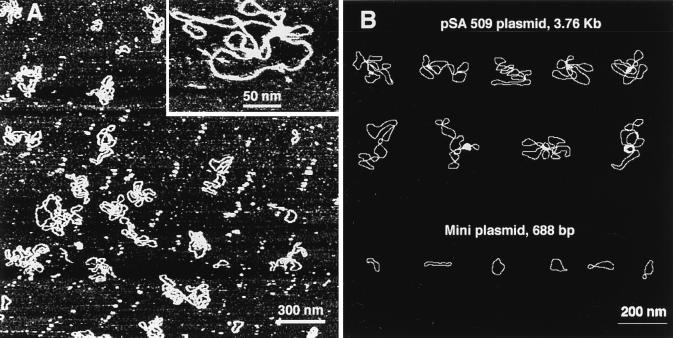
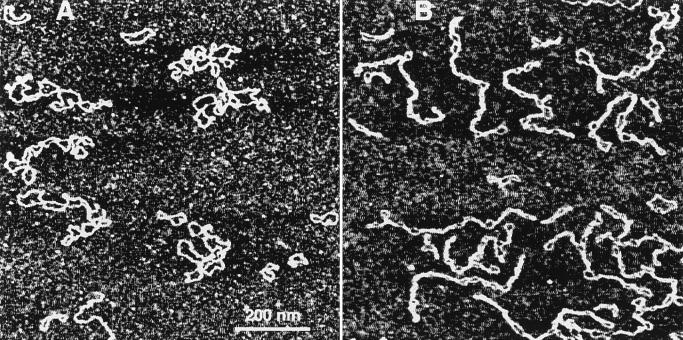
The experimental procedure in these experiments was straightforward. A solution of DNA was injected into a cell, and the molecules bound to the surface were imaged without drying of the sample. A typical large-scale AFM image of the sample in TE buffer is shown in Fig. 1A. A high-resolution image of a DNA molecule obtained by scanning over a small area is shown in Fig. 1A (Inset). Computer traces of typical examples of the molecules of this type are shown in Fig. 1B. The main feature of the sample prepared at these low salt conditions is that the DNA molecules as long as 3.76 kb (pSA509 plasmid) have an irregular geometry, sometimes rosette-like rather than plectonemic superhelical. Mini circles predominately (65%) look like relaxed circles (Fig. 1B).
Figure 1.
(A) TE buffer (20 mM Tris·HCl, pH 7.6/1 mM EDTA). (Inset) A fragment of the high-resolution image taken over 380 × 380 nm. The DNA width between the arrows is ≈3 nm. (B) Computer traces of a number of images of pSA509 plasmid (Upper) and mini circles (Lower).
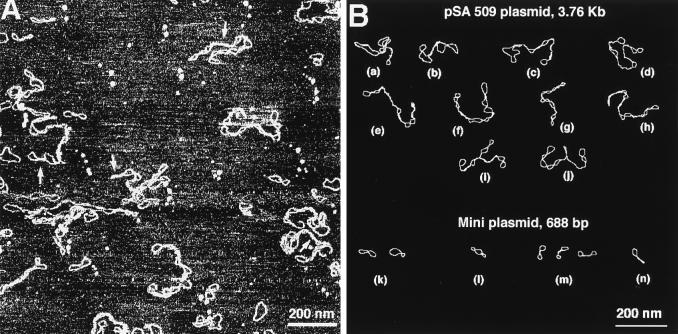
The same sample was imaged in a high salt concentration solution, and a large scan image is shown in Fig. 2A. A general feature of DNA images obtained at these high salt conditions is that DNA molecules have a tightly twisted geometry typical of a conventional plectonemic supercoiled DNA. Computer traces of a number of typical examples of supercoiled mini and pSA509 plasmids imaged at these ionic conditions are shown in Fig. 2B. Mini circles predominantly adopt a figure eight shape (≈80%) with a lesser amount of relaxed molecules (≈20%). Thus the same supercoiled DNA molecules at high ionic strength adopt a geometry that is completely different from the geometry at low salt conditions (see Fig. 1B). We therefore conclude that the geometry of supercoiled DNA depends dramatically on ionic strength. There are several important features of the AFM data obtained at different ionic conditions. The first one is mobility of DNA bound to the surface.
Figure 2.
(A) AFM images of DNA in TE buffer plus 160 mM NaCl. Several regions of a close DNA–DNA contacts are indicated with arrows. (B) Computer traces of a number of high salt images of pSA509 and mini plasmids.
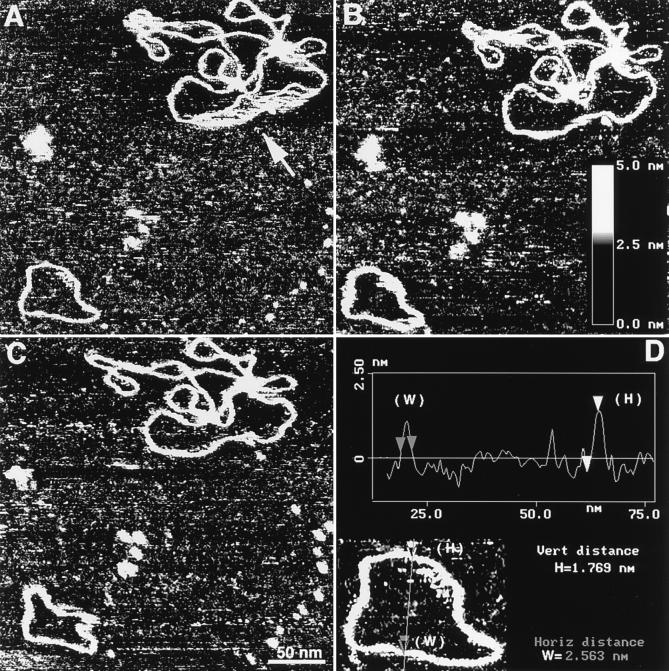
The mobility of DNA at the surface–liquid interface was observed at all ionic conditions, including low salt TE buffer. A set of data illustrating the mobility of DNA bound to AP-mica in TE buffer is shown in Fig. 3. The images are consecutive scans taken with the interval 4.5 min over the same area of 300 × 300 nm with one mini circle and one pSA509 plasmid molecule. An interesting feature of the image in Fig. 3A is a streaky large loop is indicated with an arrow. We believe that this part of the molecule was in a motion during scanning. This loop became bound more stably at a new position because there is no movement of this part of the plasmid during the second scan (Fig. 3B). The mini circle became rather thick and streaky in several parts. We assume that this is the indication of a loose interaction between the molecule and the surface. The image in Fig. 3C shows that the mini circle has changed its shape. Notice that the change of the shape of mini circles took place between the scans; hence this movement of DNA is the result of thermal motion of the molecule itself rather than a tip-induced effect. This means that although electrostatic interaction between the DNA molecules and the AP-mica surface is strong enough to prevent displacement of DNA by the scanning tip, it is not sufficient to withstand the thermal motion of the molecules. Thermal motion may result in desorption of the whole molecule, and such desorption–adsorption cycles were observed for the samples imaged in high salt conditions (data not shown).
Figure 3.
DNA motion during scanning (A–C). AFM images in TE buffer over a 300 × 300 nm area. The images in A–C were taken with an interval of 4.5 min. The section of the plasmid DNA that was in motion during scanning of the image in A is indicated with an arrow. Z-range for all the images is indicated by the Inset in Fig. 3B. D illustrates the procedure for measurements width (W) and height (H) of DNA by a cross-section option of the NanoScope III software. The data were obtained for the mini circle shown in Fig. 3A. The measurements were done for the part of the molecule that was stable during scanning.
Another interesting feature of the AFM images of DNA in solution is the thickness and height of the DNA strands. Both parameters can be determined by the use of the cross-section option of the NanoScope software, and one example is shown in Fig. 3D. The width of DNA molecules determined from different cross-sections of images shown in Fig. 3 is 2.8 ± 0.8 nm. (Because the mobility of molecules should result in widening of the images, the measurements of the DNA width were done for non-streaky and hence stably bound parts of plasmid DNA in Fig. 3.) The AFM width of DNA is very close to 2 nm, the crystallographic width of B-DNA, keeping in mind inevitable widening of DNA due to a finite size of the AFM probes (28–33). It is remarkable that images of DNA at such high resolution can be obtained by scanning in buffer solution of fully hydrated samples that were never dried. Such high-resolution data for individual DNA molecules have never been obtained with AFM for any type of water solutions (28–30). Similar resolution was only obtained for individual DNA molecules imaged in propanol (36) and for closely packed DNA molecules assembled on a highly charged cationic bilayer surface (38). The height of the DNA strands was 1.74 ± 0.37 nm, another characteristic of DNA that is very close to the B-form helix diameter.
The third important feature was found for the samples imaged at high salt solutions. In the majority of cases, supercoiled loops are divided by regions where both DNA strands are in a close contact with each other. These features are indicated by small arrows in Fig. 2A and are easily recognized in Fig. 2B (molecules a–j). The regions of a tight contact between DNA segments are formed in mini circles as well. These sites may be found between two loops (Fig. 2B, image m). A region of tight twisting may be attached to a loop leading to formation of molecules with a racket-like geometry (Fig. 2B, image n).
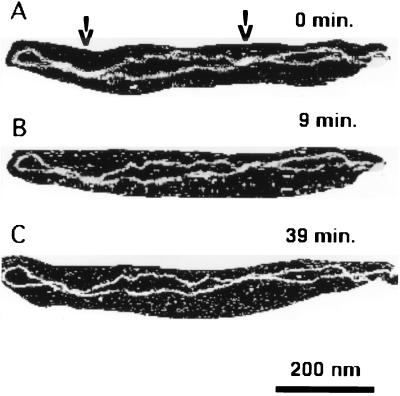
What is the structure of DNA at the regions? Are the DNA strands tightly interwound or do they simply stick to each other without interwinding? An analysis of AFM data showed that the mean number of loops is 8–9. This number is lower than the 16 nodes expected for molecules with a natural supercoiling density (σ = −0.05; ref. 19), indicating that interwinding of DNA strands at the regions of close lateral contacts takes place. The height and the width measurements of these DNA regions (unpublished work) also indicate interwinding of the DNA segments at the regions of close contacts. The direct data are shown in Fig. 4. They illustrate the evolution of the shape of a DNA molecule as a function of time (the scanning was performed in solution with an intermediate amount of salt, 80 mM NaCl). Two elongated, thick regions of close DNA–DNA contacts are indicated with arrows in Fig. 4. The DNA strands in these regions move apart slightly, leading to formation of small loops (cf. Fig. 4 A–C).
Figure 4.
AFM images of an individual DNA molecule, demonstrating segmental movement of the molecule at the solid–liquid interface. The images were taken in TE buffer with 80 mM NaCl. The images in B and C were taken 9 and 39 min, respectively, after the image in A was recorded. The regions of close DNA–DNA contacts are indicated with arrows.
Technically, the procedure of AFM imaging in air is much simpler than in solution, but DNA may change its conformation during the steps of the sample preparation followed by the deposition of the sample. We investigated possible artifacts connected with the sample preparation procedure by direct comparison of the results obtained in liquid and in air. AFM images of dried samples obtained by deposition from low salt and a high salt buffer solutions are shown in Fig. 5. DNA molecules of the sample prepared in TE buffer (Fig. 5A) adopt a loose geometry and generally the shape of these molecules is very similar to that for the sample imaged in solution (cf. Fig. 1). The DNA deposited from the high salt concentration solution (Fig. 5B) look like plectonemic supercoiled molecules, and generally these images are very similar to those obtained in solution (cf. Figs. 2 and 5B). We thus conclude that DNA molecules remain at their place and withstand the rinsing–drying steps of the sample preparation procedure (30–34). We performed measurements of the width and the height of the DNA strands for images obtained in air. These numbers are 4.94 ± 0.78 nm and 1.11 ± 0.21 nm for the width and the height, respectively (for the sample deposited from TE buffer). The larger width of DNA imaged in air in comparison with that for imaged in TE buffer (4.94 nm vs. 2.8 nm) is explained by the capillary effect, a phenomenon reported earlier in our work and the work of others (28–32). The height of dried DNA is smaller than for hydrated sample (1.11 nm vs. 1.74 nm). The height of DNA in AFM images in air and in solutions varies between ≈0.5 and ≈1 nm (39), and this deficit in height was attributed to adhesion effects (33), tip-induced compression of the DNA helix, or formation of adlayer on the surface (ref. 39 and references therein). It is important that all these artifacts complicating the interpretation of the AFM data can be eliminated by imaging in situ.
Figure 5.
AFM images of a dried sample taken in air. Deposition from TE buffer (A) and from 160 mM NaCl in TE buffer (B). Both images were taken over a 950 × 950 nm area.
DISCUSSION
The AFM data shown above demonstrate directly that the geometry of supercoiled DNA depends dramatically on ionic strength. However, since we image DNA molecules bound to the substrate surface, the question of the extent to which binding to the substrate changes the geometry of DNA molecules arises. Because the methods described in this paper were never before applied to studies of conformation of supercoiled DNA, we discuss this issue in more detail.
We used as substrate AP-mica, which was obtained by treatment of freshly cleaved mica with 3-aminopropyltriethoxy silane to attach positively charged amino groups to the mica surface (30–34). The AP-mica has the following characteristics, which are important for understanding the process of binding DNA to this surface.
First, the bonds between AP moieties and the mica surface are stable. The idea of preparation of positively charged AP-mica is similar to that widely used for preparation of sorbents for affinity and HPLC chromatography (refs. 30, 31, and 34 and references therein). The covalent Si-O-Si bonds are very stable; their hydrolysis accelerates at extreme acidic pH and by extraction with boiling water for several hours (40). Second, AP-mica interacts with DNA electrostatically since the DNA molecules can be desorbed from AP-mica by incubation in high concentrated salt solution (0.5–2 M NaCl; ref. 31). Finally, AP-mica is a surface with a low density of positively charged amino groups. This is because (i) the procedure of preparation of AP-mica (reaction of mica with APTES) takes place at a very low concentration of chemical in vapor and at ambient conditions (30–34) and (ii) mica is a rather inert material, so only a limited number of hydroxyl groups are available for reaction with APTES (30, 41). These conditions permitted us to obtain a smooth and flat surface that can be used as a substrate for AFM studies of DNA (31). From the AFM data (imaging of short DNA molecules and the samples densely coated with DNA; ref. 30) we estimate the mean distance between AP moieties as 10–30 nm. The direct measurements of the surface density of amino groups on AP-mica by the peroxidase-biotin-streptavidin assay (42) give a rather close number, 25 ± 10 nm (P. Hinterdorfer, personal communication). We used the Gouy–Chapman theory (43) to calculate the surface potential of AP-mica. We obtained the value of 2 mV for the surface potential for the density of AP groups σ = 1/100 nm2 at 0.1 M salt concentration and ≈7 mV for 0.01 M salt solution. Even the last figure is considerably less than 25 mV, the potential for a monovalent ion in solution at room temperature (42). These estimates indicate that AP-mica is a surface with a low charge density, leading to the conclusion that the concentration of ions near the surface is slightly different from their bulk concentration.
Keeping in mind the features of AP-mica described above, we suggest the following mechanism of binding of DNA to this surface. The DNA molecules approach to the surface and interact with AP-mica electrostatically by forming contacts with amino groups at the AP-mica surface. Probably, multiple contacts between the molecule and the surface are formed simultaneously, and after that other segments of DNA stick to the surface, leading to eventual stable binding of the whole molecule. The fact that AP-mica is a surface with a low charge density, has two main consequences. First, the DNA molecules retain their almost unperturbed configuration even in the immediate vicinity of the surface. So one may expect that the configuration of DNA molecules bound to the surface should be close to their geometry in solution. Second, because of the limited number of contacts between the DNA molecules and the surface, loosely bound segments can dissociate and move to another position, leading to a dynamic behavior of DNA molecules at AP-mica surface. We did observe such segmental motion of DNA. Moreover, the effect was higher in concentrated salt solutions. The last two observations indicate directly a weak interaction between DNA molecules and the AP-mica surface and thus prove the assumption we made above, i.e., the conformation of DNA molecules remains almost unchanged as they approach and bind to AP-mica.
However, this is an idealized model that does not consider the inevitable nonuniformity of surface modification. Although the experiments with high concentrated DNA solutions showed that the AP-mica surface is modified rather uniformly (34), clustering of AP-groups may take place. It is possible that such irregularities lead to formation of DNA molecules with a rosette-like shape that were observed in Fig. 1.
We have shown that imaging of dried samples gives results very similar to those obtained by imaging in solution. However, before drying, we rinsed the specimens with water to remove salt components, the step that may result in changes of the DNA geometry. But we have shown that mobility of DNA at AP-mica is a rather slow process. For example, it took almost 10 min to observe the change in the conformation of a mini circle in Fig. 4 (TE buffer). Because the rinsing step typically lasts less than 1 min, the molecules do not have sufficient time to change their conformations. In addition, rinsing with water is accompanied by increase of the Debye length and consequently leads to stabilization of DNA–AP-mica links and thus to further slowing of the desorption process. However, a local movement of DNA segments may occur even during a short period of time. This local mobility may explain why the regions of close contacts between DNA segments for dry samples are shorter than those observed for samples imaged in buffer solutions (cf. Figs. 2 and 5B).
The fact observed in this paper that the shape of supercoiled DNA is very sensitive to ionic strength is consistent with the results of Monte Carlo computer simulations (15, 16, 18). Moreover there is a striking similarity between our data and the theoretical data of (15, 16) in terms of shapes of molecules at low and high salt concentration conditions. However, our data show that winding of DNA molecules into plectonemic superhelix at physiological ionic conditions (high salt solution) is accompanied by the formation of regions of tight contacts between DNA helices. The simulated molecules do not have these features. The regions of close DNA–DNA contacts are easily identified and can hardly be attributed to artifacts of the DNA–substrate interactions. The formation of the region of extensive DNA–DNA contacts (called collapsed DNA) was observed in cryo-EM images of supercoiled DNA vitrified from the solution containing 10 mM Mg2+ (23, 24). Our experiments (unpublished work) indicate that the supercoiled loops in the presence of Mg ions are also often separated by the regions of tight DNA–DNA contacts, although the effect is less than that observed in cryo-EM studies (24). Electrostatic repulsion of the DNA strands can be overwhelmed by their attraction at high ionic conditions and in particular in the presence of divalent and multivalent cations (for a recent review, see ref. 44 and references therein). This effect of cations may be increased by DNA supercoiling. Although neutralization of phosphate charges is the main requirement for formation of close DNA–DNA contacts, bringing DNA segments into close proximity is an entropically unfavorable process. The regions of tight DNA–DNA contacts were found in theoretical works (17, 18) that use the B-spline chain representation of the DNA helix. But direct computations performed by Vologodskii and Cozzarelli (15) showed that the Monte Carlo results are quite insensitive to the type of modeling of the DNA chain. Therefore, other assumptions made in refs. 17 and 18 are responsible for the difference between the results obtained in these works and those in refs. 15 and 16. Supercoiling of DNA may compensate corresponding entropic costs. We think that future AFM experiments with DNA molecules with a specific number of supercoils (individual topoisomers) and the direct comparison of these results with computer simulations would be extremely helpful in understanding this specific phenomenon as well as many other structural and energetic features of supercoiled DNA. The majority of computer models of supercoiled DNA do not deal with the effects of the DNA sequences. However, it is well known that intrinsically curved DNA regions or regions with enhanced flexibility are important elements of regulatory regions, sites for binding of specific proteins and for DNA folding (1). Progress has been recently made in theoretical modeling of the effect of curved elements on structure and intramolecular motion of supercoiled DNA (45–48). Because AFM is capable of monitoring the intramolecular dynamics of DNA, the study of the effects of curved elements on structure (DNA branching in the first place) and dynamics of supercoiled DNA is a challenge for AFM in situ.
CONCLUSIONS
The AFM experiments demonstrate directly that the tertiary structure of supercoiled DNA changes easily and is dramatically affected by variations in ionic strength. Our data show that winding of DNA molecules into plectonemic superhelices is accompanied by formation of regions of close contacts between DNA helices. Formation of such close contacts is required in DNA recombination and long-range transcription regulation processes. Furthermore, the observed ability of supercoiled DNA to easily change shape may, by increasing the number of possible DNA–DNA contacts, accelerate searching, orientation, and recognition of interacting regions. Therefore, the features of supercoiled DNA discussed in this work may be of help in explaining the role of DNA supercoiling in genetic processes, as widely discussed in literature (1–24). Finally, it is important to note that our results were obtained using AFM, a method new to structural biology. Our sample preparation procedure (30–34), permitting high-resolution imaging of biological objects in situ at broad ionic conditions, including physiological ones, opens new areas of applications of AFM in situ to biological studies, e.g., direct high-resolution imaging of biochemical processes.
Acknowledgments
We thank S. Adhya and T. Aki for providing us with the samples of DNA. We are also grateful to N. R. Cozzarelli, S. Adhya, S. Garges, S. M. Lindsay, W. K. Olson, and A. V. Vologodskii for valuable comments and discussion of the results. We especially appreciated the critical comments of the reviewers. This work was supported by grants from the National Institutes of Health (1R43 GM 54991-01) and Digital Instruments, Inc.
Footnotes
Abbreviations: EM, electron microscopy; AFM, atomic force microscopy; APTES, 3-aminopropyltriethoxy silane; AP-mica, aminopropyl mica.
References
- 1.Sinden R R. DNA Structure and Function. San Diego: Academic; 1994. pp. 95–128. [Google Scholar]
- 2.Craigie R, Mizuuchi K. Cell. 1986;45:793–800. doi: 10.1016/0092-8674(86)90554-4. [DOI] [PubMed] [Google Scholar]
- 3.Benjamin H W, Cozzarelli N R. Proc Robert A Welch Found Conf Chem Res. 1986;29:107–126. [Google Scholar]
- 4.Echols H. J Biol Chem. 1986;265:14687–14700. [Google Scholar]
- 5.Sumners D W, Ernst C, Spengler S J, Cozzarelli N R. Q Rev Biophys. 1995;28:253–313. doi: 10.1017/s0033583500003498. [DOI] [PubMed] [Google Scholar]
- 6.Wang J C. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 7.Gellert M, Nash H. Nature (London) 1987;325:401–404. doi: 10.1038/325401a0. [DOI] [PubMed] [Google Scholar]
- 8.Zechiedrich E L, Osheroff N. EMBO J. 1990;9:4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adhya S. Annu Rev Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- 10.Hochschild A. DNA Topology and Its Biological Effects. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 107–116. [Google Scholar]
- 11.Schleif R. Annu Rev Biochem. 1992;61:199–233. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 12.Klenin K V, Vologodskii A V, Anshelevich V V, Dykhne A M, Frank-Kamenetskii M D. J Mol Biol. 1991;217:413–419. doi: 10.1016/0022-2836(91)90745-r. [DOI] [PubMed] [Google Scholar]
- 13.Vologodskii A V, Levene S D, Klenin K V, Frank-Kamenetskii M D, Cozzarelli N R. J Mol Biol. 1992;227:1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- 14.Tesi M C, Janse van Rensburg E J, Orlandi E D, Sumners D W, Whittington S G. Phys Rev E. 1994;49:868–872. doi: 10.1103/physreve.49.868. [DOI] [PubMed] [Google Scholar]
- 15.Vologodskii A V, Cozzarelli N R. Annu Rev Biophys Biomol Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- 16.Vologodskii A V, Cozzarelli N R. Biophys J. 1996;70:2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenley M O, Olson W K, Tobias I, Manning G S. Biophys Chem. 1994;50:255–271. doi: 10.1016/0301-4622(93)e0094-l. [DOI] [PubMed] [Google Scholar]
- 18.Schlick T, Li B, Olson W K. Biophys J. 1994;67:2146–2166. doi: 10.1016/S0006-3495(94)80732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boles T C, White J H, Cozzarelli N R. J Mol Biol. 1990;213:931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- 20.Shaw S Y, Wang J C. Science. 1993;260:533–536. doi: 10.1126/science.8475384. [DOI] [PubMed] [Google Scholar]
- 21.Campbell A M. Biochem J. 1978;171:281–283. doi: 10.1042/bj1710281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brady G W, Satkowski M, Foos D, Benham C J. J Mol Biol. 1987;195:185–191. doi: 10.1016/0022-2836(87)90335-4. [DOI] [PubMed] [Google Scholar]
- 23.Adrian M, Bordier B H, Wahli W, Stasiak A Z, Stasiak A. EMBO J. 1990;9:4551–4554. doi: 10.1002/j.1460-2075.1990.tb07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bednar J, Furrer P, Stasiak A, Dubochet J, Egelman E H, Bates A D. J Mol Biol. 1994;235:825–847. doi: 10.1006/jmbi.1994.1042. [DOI] [PubMed] [Google Scholar]
- 25.Binnig G, Quate C F, Gerber C H. Phys Rev Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 26.Arscott P, Bloomfield V A. Methods Enzymol. 1992;211:491–502. doi: 10.1016/0076-6879(92)11027-g. [DOI] [PubMed] [Google Scholar]
- 27.Engel A. Annu Rev Biophys Chem. 1991;20:79–108. doi: 10.1146/annurev.bb.20.060191.000455. [DOI] [PubMed] [Google Scholar]
- 28.Bustamante C, Erie D A, Keller D. Curr Opin Struct Biol. 1994;3:750–760. [Google Scholar]
- 29.Hansma H, G, Hoh J. Annu Rev Biophys Biochem Struct. 1994;23:115–139. doi: 10.1146/annurev.bb.23.060194.000555. [DOI] [PubMed] [Google Scholar]
- 30.Lyubchenko Y L, Jacobs B L, Lindsay S M, Stasiak A. Scanning Microsc. 1995;9:705–727. [PubMed] [Google Scholar]
- 31.Lyubchenko Y L, Gall A A, Shlyakhtenko L S, Harrington R E, Jacobs B L, Oden P I, Lindsay S M. J Biomol Struct Dyn. 1992;10:589–606. doi: 10.1080/07391102.1992.10508670. [DOI] [PubMed] [Google Scholar]
- 32.Lyubchenko Y L, Shlyakhtenko L S, Harrington R E, Oden P I, Lindsay S M. Proc Natl Acad Sci USA. 1993;90:2137–2140. doi: 10.1073/pnas.90.6.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyubchenko Y L, Oden P I, Lampner D, Lindsay S M. Nucleic Acids Res. 1993;21:1117–1123. doi: 10.1093/nar/21.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyubchenko, Y. L., Blankenship, R. E., Gall, A. A., Lindsay, S. M., Thiemann, O., Simpson, L. & Shlyakhtenko, L. S. (1996) Scanning Microsc., in press. [PubMed]
- 35.Hansma H G, Sinsheimer R L, Groppe J, Bruice T C, Elings V, Gurley G, Bezanilla M, Mastrangelo I A, Hough P V C, Hansma P K. Scanning. 1993;15:296–299. doi: 10.1002/sca.4950150509. [DOI] [PubMed] [Google Scholar]
- 36.Hansma H G, Laney D E, Bezanilla M, Sinsheimer R L, Hansma P K. Biophys J. 1995;68:672–677. doi: 10.1016/S0006-3495(95)80343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choy H E, Adhya S. Proc Natl Acad Sci USA. 1993;90:472–476. doi: 10.1073/pnas.90.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mou J, Czajkowsky D M, Zhang Y, Shao Z. FEBS Lett. 1995;371:279–282. doi: 10.1016/0014-5793(95)00906-p. [DOI] [PubMed] [Google Scholar]
- 39.Vesenka, J., Marsh, T., Henderson, E. & Vellandi, C. (1996) Scanning Microsc., in press.
- 40.Ishida H, Koenig J L. J Polym Sci Polym Phys Ed. 1980;18:1931–1943. [Google Scholar]
- 41.Parker J L, Cho D L, Claesson P M. J Phys Chem. 1989;93:6121–6125. [Google Scholar]
- 42.Bayer E A, Wilchek M. Methods Enzymol. 1990;184:138–173. doi: 10.1016/0076-6879(90)84268-l. [DOI] [PubMed] [Google Scholar]
- 43.McLaucghlin S. Curr Top Membr Transp. 1977;9:71–144. [Google Scholar]
- 44.Bloomfield V A. Curr Opin Struct Biol. 1996;6:334–341. doi: 10.1016/s0959-440x(96)80052-2. [DOI] [PubMed] [Google Scholar]
- 45.Chirico G, Langowski J. Biophys J. 1996;71:955–971. doi: 10.1016/S0006-3495(96)79299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sprous D, Tan R K-Z, Harvey S C. Biopolymers. 1966;39:243–258. doi: 10.1002/(SICI)1097-0282(199608)39:2%3C243::AID-BIP11%3E3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 47.Tan R K-Z, Sprous D, Harvey S C. Biopolymers. 1996;39:259–278. doi: 10.1002/(sici)1097-0282(199608)39:2<259::aid-bip12>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 48.Sprous D, Harvey S C. Biophys J. 1996;70:1893–1908. doi: 10.1016/S0006-3495(96)79754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]