Abstract
A significant amount of reactive oxygen species (ROS) is generated during mitochondrial oxidative phosphorylation. Several studies have suggested that mtDNA may accumulate more oxidative DNA damage relative to nuclear DNA. This study used quantitative PCR to examine the formation and repair of hydrogen peroxide-induced DNA damage in a 16.2-kb mitochondrial fragment and a 17.7-kb fragment flanking the β-globin gene. Simian virus 40-transformed fibroblasts treated with 200 μM hydrogen peroxide for 15 or 60 min exhibited 3-fold more damage to the mitochondrial genome compared with the nuclear fragment. Following a 60-min treatment, damage to the nuclear fragment was completely repaired within 1.5 hr, whereas no DNA repair in the mitochondrion was observed. Mitochondrial function, as assayed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide reduction, also showed a sharp decline. These cells displayed arrested-cell growth, large increases in p21 protein levels, and morphological changes consistent with apoptosis. In contrast, when hydrogen peroxide treatments were limited to 15 min, mtDNA damage was repaired with similar kinetics as the nuclear fragment, mitochondrial function was restored, and cells resumed division within 12 hr. These results indicate that mtDNA is a critical cellular target for ROS. A model is presented in which chronic ROS exposure, found in several degenerative diseases associated with aging, leads to decreased mitochondrial function, increased mitochondrial-generated ROS, and persistent mitochondrial DNA damage. Thus persistent mitochondrial DNA damage may serve as a useful biomarker for ROS-associated diseases.
Reactive oxygen species (ROS) are generated from cellular sources such as neutrophils and macrophages (1) and during oxidative phosphorylation in which oxygen is reduced to water through a four-step addition of electrons (2). This reduction generates superoxide anion (O2⨪), which is converted to hydrogen peroxide (H2O2) by superoxide dismutase. H2O2, while relatively inactive, can be reduced to the highly reactive hydroxyl radical (OH·) by a metal ion through the Fenton reaction (2). Nonenzymatic production of the hydroxyl radical is generated during exposure to ionizing radiation (3, 4). Several studies have characterized ROS-induced DNA damage, which includes single- and double-strand breaks, abasic sites, and base damages (5, 6). Other studies have indicated that when compared with nuclear DNA, mtDNA contains an elevated basal level of base damage such as 8-oxoguanine (7). In addition, cellular exposure to certain compounds, such as alloxan, has been shown to induce significant mtDNA damage (8). Oxidative mtDNA damage has been linked to the onset of specific human diseases such as neuronal degeneration and cardiovascular disease (9); it has also been linked to aging (10–12). H2O2, a representative ROS, has recently been shown to induce permanent growth arrest (13, 14) and apoptosis (15–17) in a number of cell types. ROS also induces the expression of p53, which in turn activates p21 (WAF1/CIP1) leading to cell cycle arrest (18, 19).
The repair of ROS-induced base damage in mtDNA and nuclear DNA is believed to occur through a base-excision repair pathway (8, 20). In general, base-excision repair involves the removal of damaged bases by the action of damage-specific DNA glycosylases. For example, the Escherichia coli Fpg protein (MutM gene product) contains both DNA glycosylase and abasic site (AP) lyase activity. The substrate specificity for this protein includes bases containing imidazol ring-open and oxidized purines (5). The resulting AP is further processed by AP-endonucleases, DNA polymerase, and ligase. Currently there is no evidence that mitochondria are capable of performing nucleotide-excision repair.
Identification of oxidative lesions in total cellular DNA and DNA isolated from mitochondria has been performed by HPLC-electrochemical detection (7). These studies have suggested that 8-oxoguanine occurs more frequently in mtDNA compared with nuclear DNA (7); both genomes have been shown to accumulate this adduct with increasing age (11). However, one disadvantage regarding this technique is the requirement for large quantities of nuclear DNA and/or mtDNA, which can be limiting when specific tissues or primary cell cultures are used.
The purpose of this study was to examine the formation and repair of ROS-induced DNA damage in the mitochondrial and nuclear genomes without the need to purify large quantities of DNA or isolate the organelle before DNA purification. To this end a novel quantitative PCR assay was used to measure H2O2-induced DNA damage and repair in a 17.7-kb fragment upstream of the nuclear β-globin gene and a 16.2-kb fragment from the mitochondrial genome. The present study has revealed that mtDNA is more sensitive than nuclear DNA to H2O2-induced damage, and protracted treatment leads to persistent mtDNA damage and loss of mitochondrial function. These cells also demonstrate arrested growth, increased p21 expression, and morphological changes consistent with apoptosis.
MATERIALS AND METHODS
Cells and Cell Culture.
The simian virus 40 (SV40)-transformed human fibroblast cell line, GM00637E (NIGMS Human Genetic Mutant Cell Repository, Coriell Institute, Camden, NJ), used throughout this study was maintained at 37°C and 5% CO2/95% air in Earle’s MEM (GIBCO/BRL) supplemented with 15% fetal bovine serum (HyClone), 2× MEM essential amino acids (Sigma) and nonessential amino acids (GIBCO/BRL), 2× vitamins (GIBCO/BRL), and 2.0 mM l-glutamine (GIBCO/BRL). The cells were routinely split 1:6 every 4–5 days.
Treatment of Cells with H2O2.
SV40-transformed fibroblasts, 1 × 106 unless otherwise indicated, were plated in duplicate in 60-mm dishes 14–16 hr before treatment. H2O2 (30%; Fisher) was diluted into PBS and the concentration was determined by absorbance at 240 nm as described (21). Monolayer cultures were exposed to the indicated concentrations of H2O2 for either 15 or 60 min at 37°C in serum-free medium. For time-course analysis of induced DNA damage, the cells were exposed to 200 μM H2O2 for 5, 15, 30, or 60 min. Control monolayers were mock-treated with serum-free medium alone. Monolayers were washed once with PBS, harvested immediately by brief trypsinization (0.25%), or allowed to recover in the original plating medium (i.e., conditioned medium) for the indicated times.
Characterization of H2O2 Depletion from the Culture Medium.
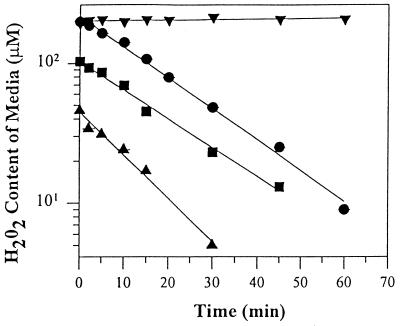
SV40-transformed fibroblasts were plated as described above; however, exposure to H2O2 was performed in both phenol red-free and serum-free medium. H2O2 depletion was monitored by a colorimetric assay that involved the oxidation of o-diansidine dihydrochloride (o-DD) in the presence of horseradish peroxidase (22). Briefly, 100 μl aliquots of the treatment medium were removed at various times and incubated for 1 hr at 37°C with 2.5 units per ml of horseradish peroxidase and 250 μM o-DD. The absorbance was measured at 470 nm against a blank-containing treatment medium without addition of H2O2. The amount of H2O2 remaining in the medium was determined from a standard curve generated with known concentrations of H2O2. During a 60-min exposure, H2O2 was depleted with first-order kinetics (Fig. 1). The rate constants for 100 and 200 μM H2O2 were 0.047 and 0.045 min−1, respectively, while the rate of depletion for 50 μM H2O2 was slightly faster at 0.067 min−1. In the absence of cells, H2O2 was very stable, indicating that depletion of H2O2 was the result of cellular metabolism and not the interaction with medium components.
Figure 1.
Kinetics of H2O2 depletion from the culture medium. SV40-transformed fibroblast monolayers (1 × 106 cells) were incubated at 37°C in 3 ml of serum-free MEM supplemented with either 50 μM (▴), 100 μM (▪), or 200 μM (•) H2O2. The H2O2 (200 μM) content in the medium in the absence of cells is represented by (▾). Medium aliquots were removed and the amount of H2O2 remaining was determined by spectrophotometric analysis and a standard curve of known H2O2 concentrations. Data are expressed as the mean ± SEM (n = 2 cell experiments in duplicate for each time point; error bars were omitted because the SEM were <1%). First-order rate constants of H2O2 depletion were 0.067 min−1 (▴), 0.047 min−1 (▪), and 0.045 min−1 (•) for 50, 100, and 200 μM, respectively.
DNA Isolation and Quantitative PCR (QPCR).
High molecular weight DNA was isolated with the QIAamp DNA isolation kit (Qiagen, Chatsworth, CA) as described by the manufacturer. DNA isolation by this technique has been shown to be suitable for long PCR (23). The concentration of total cellular DNA was determined by ethidium bromide fluorescence with an A4-filter fluorimeter with an excitation band pass filter at 365 nm and a emission cut-off filter at 600 nm (Optical Technology Devices, Elmsford, NY) using λ/HindIII DNA as a standard. QPCRs were performed in a GeneAmp PCR System 2400 with the GeneAmp XL PCR kit (Perkin–Elmer) as described (24). Reaction mixtures contained 15 ng of template DNA, 1.1 mM Mg(AOc)2, 100 μg/ml nonacetylated BSA, 0.2 mM deoxynucleotide triphosphates (Pharmacia), 0.2 μM primers, 0.2 μCi [α-32P]dATP (1 Ci = 37 GBq), and 1 unit of recombinant Thermus thermophilus DNA polymerase. The primer nucleotide sequences were as follows: for the 17.7-kb 5′ flanking region of the β-globin gene (GenBank data base accession number, J00179J00179), 5′-TTGAGACGCATGAGACGTGCAG-3′ (RH1024), and 5′-GCACTGGCTTAGGAGTTGGACT-3′ (RH1053); and for the 16.2-kb fragment of the mitochondrial genome (25), 5′-TGAGGCCAAATATCATTCTGAGGGGC-3′ (RH1065) and 5′-TTTCATCATGCGGAGATGTTGGATGG-3′ (RH1066).
The PCR was initiated with a 75°C hot-start addition of the polymerase and allowed to undergo the following thermocycler profile: an initial denaturation for 1 min at 94°C followed by 25 cycles of 94°C denaturation for 15 sec and 68°C primer extension for 12 min. A final extension at 72°C was performed for 10 min at the completion of the profile. To ensure quantitative conditions, a control reaction containing 7.5 ng of template DNA was included with each amplification. An aliquot of each PCR product was resolved on a 1% vertical agarose gel and electrophoresed in TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) at 80 V (5 V/cm) for 4 hr. The dried gels were then exposed to a phoshor screen for 12–18 hr and quantitated with imagequant (Molecular Dynamics).
DNA lesion frequencies were calculated as described (24, 26). Briefly, the amplification of damaged samples (AD) was normalized to the amplification of nondamaged controls (AO) resulting in a relative amplification ratio. Assuming a random distribution of lesions and using the Poisson equation [f(x) = e−λλx/x!, where λ = the average lesion frequency] for the nondamaged templates (i.e., the zero class; x = 0), the average lesion frequency per DNA strand was determined: λ = −ln AD/AO. Statistical analysis was performed with the unpaired Student’s t test.
Western Blot Analysis.
SV40-transformed fibroblasts were exposed to 200 μM H2O2 for either 15 or 60 min and then allowed to recover for 12, 24, or 36 hr in conditioned medium. Total cell extracts were prepared by lysis of pellets in 62 mM Tris (pH 6.8), 10% glycerol, 2% SDS, and 2.5% 2-mercaptoethanol and by boiling for 10 min. Equal amounts of proteins (20 μg) in the extracts were electrophoresed on a 14% SDS/polyacrylamide gel and electroblotted to a Hybond ECL nitrocellulose membrane (Amersham). The blot was probed with the anti-WAF1 antibody (Calbiochem) and immobilized horseradish peroxidase activity was detected by enhanced chemiluminescence (Amersham).
Reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) and Cell Survival.
SV40-transformed fibroblasts were treated with 200 μM H2O2 for either 15 or 60 min as described above and then allowed to recover in conditioned medium or harvested immediately. At the indicated times the cells were harvested and counted manually with a hemacytometer. For determination of mitochondrial function, cells were incubated during the last hour of the recovery period with MTT (Sigma) (27) at a final concentration of 2.0 μg/ml, lysed (20% SDS/50% dimethylformamide), and measured at an absorbance of 570 nm. Absorbance values were converted to MTT reduction using a standard curve generated with known numbers of viable cells. MTT reduction for treated samples was then normalized to nontreated control samples and is reported as a fraction of control.
RESULTS
H2O2-Induced DNA Damage Is Extensive in Mitochondria.
Our laboratory has developed conditions for quantitative amplification of long DNA fragments (24, 26). This QPCR assay was used to detect H2O2-induced DNA damage in both a 17.7-kb fragment from the β-globin loci and a 16.2-kb mtDNA fragment from SV40-transformed fibroblasts (Fig. 2). This technique is based on the premise that DNA lesions, including oxidative damage such as strand breaks, base modifications, and AP sites, will block the progression of the polymerase resulting in a decrease in amplification of a target sequence (24). Therefore, only those DNA templates that do not contain polymerase-blocking lesions will be amplified. H2O2, like ionizing radiation, has been shown to induce a wide variety of damaged bases (28, 29), several of which may not act as strong blocks to the thermostable polymerase. It has been suggested that both of these DNA-damaging agents induce an equal number of single-strand breaks and damaged bases; single-strand breaks are known to be absolute blocks to the polymerase. Hydrogen peroxide has been shown to induce at least 11 different base products (29), of which ≈35-50% of the total adducts are expected to result in strong blocks to the thermostable polymerase. It is interesting to note that 8-oxodeoxyguanine constituted 10% of the total base adducts. Based on these studies it is likely that the QPCR assay is detecting a majority of the lesions induced by hydrogen peroxide.
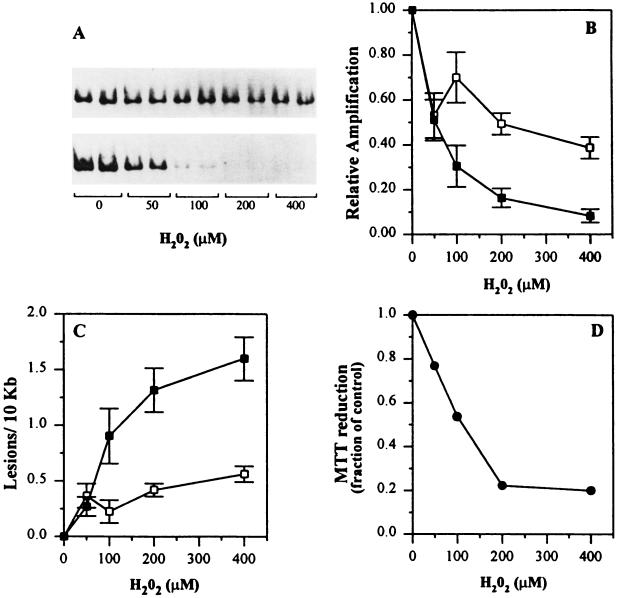
Figure 2.
H2O2 dose response in human fibroblast cells. Human fibroblast cells were exposed to increasing concentrations of H2O2, in duplicate plates for each dose, for 1 hr at 37°C, and total cellular DNA was isolated. Control cultures were incubated in serum-free medium alone. (A) Representative autoradiogram depicting the decrease in amplification of the 17.7-kb β-globin fragment (Upper) and the 16.2-kb mitochondrial fragment (Lower). (B) The amount of radioactivity associated with each amplification product relative to the nondamaged controls was determined by PhosphorImager (Molecular Dynamics) analysis and is plotted as a function of H2O2 concentration: (▪), 16.2-kb mitochondrial fragment; (□), 17.7-kb β-globin fragment. (C) The decrease in relative amplification from B was then converted to lesion frequency using the Poisson equation as described. The data are expressed as the mean ± SEM from a minimum of two biological experiments in which 3–5 PCRs were performed per experiment. Statistically significant differences in the lesion frequency for both fragments at each dose were calculated using the unpaired Student’s t test (100 μM, P = 0.05; 200 μM, P = 0.004; 400 μM, P = 0.0001). (D) Mitochondrial function assayed by MTT reduction. Fibroblast cells were plated in 60-mm dishes and exposed to increasing concentrations of H2O2 for 60 min at 37°C. Following H2O2 exposure, the cells were rinsed with PBS and incubated for 60 min with conditioned medium containing 2.0 μg/ml MTT. Afterward the medium was removed, the cells lysed, and the absorbance measured at 570 nm (•). MTT reduction was determined with a standard curve and normalized to nontreated controls and is reported as a fraction of control. The data are expressed as the mean ± SD for triplicate plates (error bars were omitted because the SD were <1%).
Loss of amplification of the mitochondrial genome occurred at significantly lower doses of H2O2 than for the β-globin fragment (Fig. 2 A–C). Damage to both genomes appeared to plateau at higher concentrations of H2O2; however, mtDNA sustained 3-fold more DNA polymerase blocking adducts than the nuclear fragment at 200 μM H2O2. Time course experiments indicated that H2O2-induced damage to mtDNA occurred more rapidly than damage in the β-globin fragment (data not shown). Preferential mtDNA damage is not specific to H2O2 as other sources of ROS, such as asbestos and glucose oxidase, also induce significantly more mtDNA damage (F.M.Y., J. J. Salazar, and B.V.H., unpublished data).
Because damage to mtDNA was extensive (approximately four polymerase-blocking lesions per genome) it was important to also assess the physiological state of the mitochondrion. MTT reduction by succinate dehydrogenase, a component of complex II of the respiratory chain, is an indicator of mitochondrial function (27, 30). When cells were exposed to H2O2 for 60 min, a dose-dependent decrease in MTT reduction was observed (Fig. 2D). Therefore, these data demonstrate that a 60-min exposure to H2O2 not only induces extensive mtDNA damage, but also disrupts mitochondrial function.
Repair of mtDNA and Nuclear DNA.
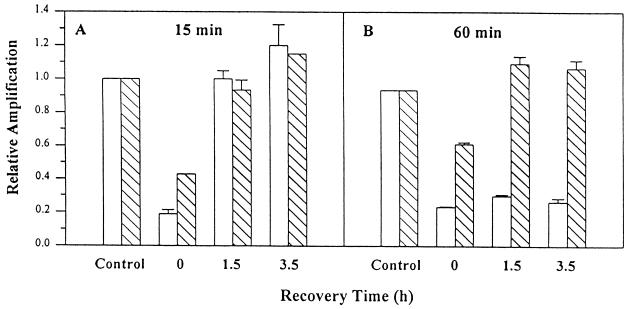
When cells were treated with 200 μM H2O2 for 60 min and then allowed to recover in conditioned medium, DNA repair in the β-globin fragment occurred within 1.5 hr (Fig. 3B). In contrast, no repair in mtDNA was observed, even after 3.5 hr. Similar results were observed when cells were exposed to 400 μM H2O2 (data not shown). When the period of H2O2 exposure was reduced to 15 min, repair of mtDNA was observed within 1.5 hr (Fig. 3A), which was similar to the repair kinetics observed for the nuclear β-globin fragment. Together these data demonstrate that mitochondria are proficient in the repair of H2O2-induced DNA damage following short exposures; however, longer periods of H2O2 treatment lead to persistent DNA damage.
Figure 3.
DNA repair activity in mtDNA and nuclear DNA. Fibroblast cells were exposed to 200 μM H2O2 for either 15 (A) or 60 (B) min as described in Fig. 2 and either harvested immediately or allowed to recover in conditioned medium for the indicated times. QPCR was performed for both the mitochondrial (open bars) and β-globin fragments (hatched bars). The data are expressed as the mean ± SD (n = 3).
Persistent mtDNA Damage Correlates with the Loss of Mitochondrial Function, Permanent Growth Arrest, and Apoptosis.
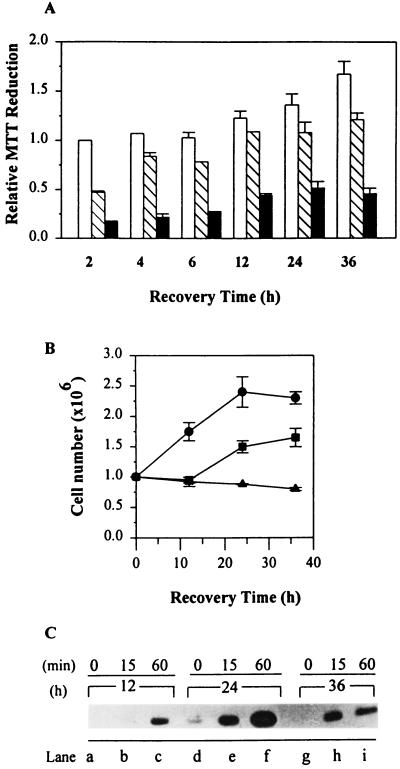
Because persistent mtDNA damage might be an early indicator of a decline of mitochondrial physiology, we next sought to correlate persistent mtDNA damage with mitochondrial function and cell growth. Recovery of MTT reduction was observed for cells exposed to H2O2 for 15 min; however, this activity was compromised in cells treated for 60 min (Fig. 4A). Fibroblasts exposed for 15 min to H2O2 experienced a transient growth arrest, whereas those exposed for 60 min entered a permanent growth arrest (Fig. 4B). No cell loss was apparent when the cells were counted at 12 and 24 hr of recovery, although morphological changes associated with apoptotic cell death were present within the population at 36 hr (data not shown). The inability to reduce MTT and the lack of recovery therefore was not due to a loss in cells but rather a consequence of prolonged H2O2 exposure.
Figure 4.
Effect of H2O2 on mitochondrial function and cell growth. Fibroblast cells were treated with 200 μM H2O2 for either 15 or 60 min as described in Fig. 2 and then allowed to recover in conditioned medium for the indicated times. (A) Mitochondrial function for control cells (open bars) or cells treated with H2O2 for 15 min (hatched bars) or 60 min (filled bars) was determined after incubating the cells with 2.0 μg/ml MTT for 60 min at 37°C during the final hour of the indicated recovery time. The data are expressed as the mean ± SEM (n = 2 biological experiments, where each point was performed in duplicate) relative to nontreated controls at time zero. (B) The total number of cells was determined, following a 15-min (▪) or 60-min (▴) exposure to H2O2, by manual counting at 12, 24, and 36 hr posttreatment, and treated cells were compared with nontreated cells (•). These measurements were performed simultaneously with the MTT assay (A). The data are expressed as the mean ± SEM (n = 2 biological experiments). (C) Increased expression of WAF1/CIP1 in response to H2O2. WAF1/CIP1 levels were analyzed in control cells (lanes a, d, and g), and cells were exposed to 200 μM H2O2 for either 15 min (lanes b, e, and h) or 60 min (lanes c, f, and i) at 12 hr (lanes a–c), 24 h (lanes d–f), and 36 h (lanes g–i) of recovery.
WAF1/CIP1 is known to be expressed in cells damaged by a number of different agents (16). Both H2O2 treatment protocols resulted in an increase in WAF1/CIP1 protein expression (Fig. 4C). However, the 60-min treatment induced a rapid increase within 12 hr and an additional 4-fold induction at 24 hr (Fig. 4C, lanes c and f).
DISCUSSION
The purpose of this study was to examine the role of nuclear DNA and mtDNA damage and repair in the sensitivity of human cells to H2O2. In this study we found that (i) H2O2-induced mtDNA damage occurred rapidly and was more extensive than damage to a 17.7-kb fragment 5′ to the β-globin gene; (ii) a 60-min exposure to H2O2 resulted in mtDNA damage that was not repaired, whereas damage to the β-globin fragment was completely removed within 1.5 hr; (iii) a 15-min exposure to H2O2 resulted in DNA damage that was efficiently repaired in both the β-globin fragment and mtDNA; and (iv) the persistence of mtDNA damage correlated with the loss of mitochondrial function, the induction of WAF1/CIPI, a permanent growth arrest, and apoptosis.
Analysis of H2O2-induced mtDNA damage by alkaline gel electrophoresis and Southern hybridization indicated that the damage measured by the QPCR assay included both strand breaks and base modification(s) (unpublished data). The increased susceptibility of the mitochondrial genome to DNA damage could be due to several factors, including: (i) the absence of complex chromatin organization, which may serve as a protective barrier against ROS; (ii) alterations in DNA repair activity; (iii) the presence of localized metal ions that may function as catalysts in the generation of ROS; and (iv) the stimulation of secondary ROS reactions due to damage to the electron transport chain and/or through lipid peroxidation.
The organization of chromatin by histones has been shown to result in less ROS-induced DNA strand breaks (31) and base damage when compared with naked DNA (32). It is possible that the genomic region used in the present studies may not be damaged as much as transcriptionally active regions. Further analysis has revealed that H2O2-induced damage in the hprt gene was identical to the β-globin region (data not shown). Because mtDNA is deficient in histones, and therefore lacks the complexity associated with nuclear DNA, mtDNA may be more prone to attack by oxidants. The plateau of nuclear damage following H2O2 treatment has been reported (33). For example, in mammalian cells, single-strand breaks plateau at concentrations >250 μM H2O2 (33). Because H2O2 is unreactive unless transition metals or superoxide radicals are present, it is possible that metal ions, such as Fe2+, may be limiting in the nucleus and an increase in H2O2 may not necessarily show increasing amounts of DNA damage.
Removal of oxidative stress-induced DNA strand breaks (formamidopyrimidine DNA glycosylase- and endonuclease III-sensitive lesions in mtDNA), presumably through base-excision repair, has been examined (8, 20). However, these studies did not examine damage and repair in a nuclear gene and thus no direct comparison between the two genomes could be made. Our observation of persistent mtDNA damage during the same period when repair to the β-globin fragment was >100% suggests that either mitochondrial repair was inhibited by H2O2 or that the steady-state level of damage increased even in the presence of an active repair system.
One potential source of continued DNA damage is through the generation of secondary ROS, such as lipid peroxidation products (34). Lipids within the inner mitochondrial membrane contain components of the electron transport chain, many of which contain transition metal ions. Stimulation of both radical and nonradical species through metal-catalyzed lipid peroxidation reactions have been shown to damage DNA (35–39). Therefore, due to the proximity of mtDNA to the inner mitochondrial membrane, ROS-induced chain-propagation reactions have the potential for contributing to the overall increase in the steady-state level of mtDNA damage. Secondary reactions that overwhelm the repair capacity of the mitochondrion would be observed as persistent DNA damage. This hypothesis is supported by our inability to monitor repair to mtDNA even in the context of efficient repair in the β-globin fragment. Furthermore, repair to mtDNA was not observed at 6, 12, and 24 hr following a 60-min exposure to 200 μM H2O2 (data not shown).
It might be expected that ROS-mediated damage to mitochondria may inactivate electron transport complexes or inhibit mtDNA transcription thereby altering normal mitochondrial function. Because 1–2% of all oxygen consumed within the cell is thought to be released from mitochondria as ROS (40), any cellular insult that leads to disruption of the electron transport chain could lead to an increase in mitochondrial-generated ROS. It is known that components of the electron transport chain and ATPase are damaged by ROS (41). The loss of succinate dehydrogenase activity observed in our experiments clearly indicates the detrimental effect of H2O2 on normal mitochondrial function. In addition, the increased expression of the cyclin-dependent kinase inhibitor WAF1/CIP1 and subsequent induction of a permanent growth arrest following a 60-min exposure to H2O2 correlated with those cells in which mtDNA repair was undetectable. Thus, continuous ROS exposure contributes to a decrease in overall mitochondrial function and to permanent growth arrest (13).
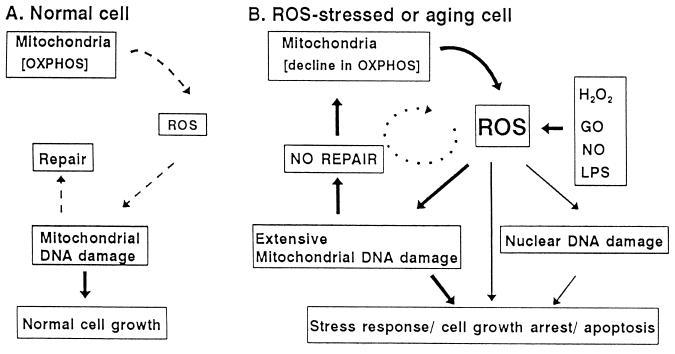
Our results provide support for the role of mitochondrial-derived ROS and oxidative stress (Fig. 5). Under normal growth conditions, ROS generated within the mitochondrion leads to a low level of mtDNA damage, which is rapidly repaired (Fig. 5A). However, during periods of chronic ROS exposure, which may include degenerative diseases associated with aging and ischemia/reperfusion injury, mitochondrial macromolecules including DNA are extensively damaged (Fig. 5B). This mtDNA damage is more extensive and persistent than nuclear DNA damage. Persistence of mtDNA damage could be due to continued ROS production by lipid peroxidation and/or damage within the electron transport system. These alterations in mitochondrial physiology may contribute to a cellular stress response, cell growth arrest, and subsequent apoptosis. This model is supported by the finding that steady-state level of oxidative damage to rat liver mtDNA has been shown to be elevated when compared with nuclear DNA (7). This difference is more dramatic when DNA damage was analyzed as a function of age; mtDNA from older rats contained more damage than mtDNA from younger rats (10, 11, 42). The results presented in this study demonstrate and further support the sensitivity of mtDNA to ROS.
Figure 5.
Schematic representation of the role of mitochondria in the generation of ROS and oxidative stress. The mitochondrion is responsible for oxidative phosphorylation, the biochemical pathway that generates ATP via the respiratory chain. During this process, 1–2% of the oxygen that is consumed is released as ROS, which can damage mtDNA and subsequently be repaired. However, under ROS-stressed conditions, the generation of ROS leads to persistent mtDNA damage. The propagation of mitochondrial damage through the generation of secondary ROS could lead to a decline in oxidative phosphorylation and of mitochondrial physiology. In response to changes in mitochondrial physiology, stress response genes are activated and cells undergo growth arrest, which may be followed by apoptosis.
Finally, the sensitivity of the QPCR assay and the ability to simultaneously measure oxidant-induced mitochondrial and nuclear DNA damage may be a useful biomarker for chronic oxidative stress associated with many degenerative diseases.
Acknowledgments
We would like to acknowledge Drs. Samuel Wilson and Sankar Mitra for the critical reading of the manuscript and Dr. Judah Rosenblatt for advice concerning statistical analysis. We would also like to thank Dr. Suzanne Cheng (Roche Molecular Systems) for her generous support throughout these studies. This work was supported in part by grants from the National Institutes of Health (ES07281 and ES07038) to B.V.H.
Footnotes
Abbreviations: ROS, reactive oxygen species; H2O2, hydrogen peroxide; SV40, simian virus 40; QPCR, quantitative PCR; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; AP, abasic site.
References
- 1.Janssen Y M, Van Houten B, Borm P J, Mossman B T. Lab Invest. 1993;69:261–274. [PubMed] [Google Scholar]
- 2.Breen A P, Murphy J A. Free Radical Biol Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 3.Clark J M, Pattabiraman N, Jarvis W, Beardsley G P. Biochemistry. 1987;26:5404–5409. doi: 10.1021/bi00391a028. [DOI] [PubMed] [Google Scholar]
- 4.Teoule R, Bert C, Bonicel A. Radiat Res. 1977;72:190–200. [PubMed] [Google Scholar]
- 5.Demple B, Harrison L. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 6.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 7.Richter C, Park J-W, Ames B. Proc Natl Acad Sci USA. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Driggers W J, LeDoux S P, Wilson G L. J Biol Chem. 1993;268:22042–22045. [PubMed] [Google Scholar]
- 9.Tritschler H, Medori R. Neurology. 1993;43:280–288. doi: 10.1212/wnl.43.2.280. [DOI] [PubMed] [Google Scholar]
- 10.Ames B N, Shigenaga M K, Hagen T M. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shigenaga M, Hagen T M, Ames B N. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallace D C, Shoffner J M, Trounce I, Brown M D, Ballinger S, Corral-Debrinski M, Horton T, Jun A S, Lott M T. Biochim Biophys Acta. 1995;1271:141–151. doi: 10.1016/0925-4439(95)00021-u. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q, Ames B. Proc Natl Acad Sci USA. 1994;91:4130–4134. doi: 10.1073/pnas.91.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clopton D A, Saltman P. Biochem Biophys Res Commun. 1995;210:189–196. doi: 10.1006/bbrc.1995.1645. [DOI] [PubMed] [Google Scholar]
- 15.Corcaran G B, Fix L, Jones D P, Moslen M T, Nicotera P, Oberhammer F A, Buttyan R. Toxicol Appl Pharmacol. 1994;128:169–181. doi: 10.1006/taap.1994.1195. [DOI] [PubMed] [Google Scholar]
- 16.de Bono D P, Yang W D. Atherosclerosis (Ireland) 1995;114:235–245. doi: 10.1016/0021-9150(94)05488-5. [DOI] [PubMed] [Google Scholar]
- 17.Whittemore E R, Loo D T, Cotman C W. NeuroReport. 1994;5:1485–1488. doi: 10.1097/00001756-199407000-00019. [DOI] [PubMed] [Google Scholar]
- 18.El-Deiry W S, Harper J W, O’Connor P M, Velculescu V E, Canman C, Jackman E J, Pietenpol J, Burrell A M, Hill D E, Wang Y, Wiman K G, Mercer W E, Kastan M B, Kohn K W, Elledge S J, Kinzler K W, Vogelstein B. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 19.Russo T, Zambrano N, Esposito F, Ammendola R, Cimino F, Fiscella M, Jackman J, O’Connor P M, Anderson C W, Appella E. J Biol Chem. 1995;270:29386–29391. doi: 10.1074/jbc.270.49.29386. [DOI] [PubMed] [Google Scholar]
- 20.LeDoux S, Wilson G L, Beecham E J, Stevnsner T, Wassermann K, Bohr V. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 21.Shull S N, Heintz H, Periasamy M, Manohar M, Janssen Y M, Marsh J P, Mossman B T. J Biol Chem. 1991;266:24398–24403. [PubMed] [Google Scholar]
- 22.Nowak D. Biomed Biochim Acta. 1990;49:353–362. [PubMed] [Google Scholar]
- 23.Cheng S, Chen Y, Monforte J A, Higuchi R, Van Houten B. PCR Methods Appl. 1995;4:294–298. doi: 10.1101/gr.4.5.294. [DOI] [PubMed] [Google Scholar]
- 24.Yakes F M, Chen Y, Van Houten B. In: Technologies for Detection of DNA Damage and Mutations. Pfeifer G P, editor. New York: Plenum; 1996. pp. 169–182. [Google Scholar]
- 25.Cheng S, Higuchi R, Stoneking M. Nat Genet. 1994;7:350–351. doi: 10.1038/ng0794-350. [DOI] [PubMed] [Google Scholar]
- 26.Van Houten B, Chandrasekhar D, Huang W, Katz E. Amplifications Forum PCR Users. 1993;10:10–17. [Google Scholar]
- 27.Hansen M B, Nielsen S E, Berg K. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 28.Ward J F. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 29.Jaruga P, Dizdaroglu M. Nucleic Acids Res. 1996;24:1389–1394. doi: 10.1093/nar/24.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Behl C, Davis J B, Lesley R, Schubert D. Cell. 1994;77:817–827. doi: 10.1016/0092-8674(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 31.Ljungman M, Hanawalt P C. Mol Carcinogen. 1992;5:264–269. doi: 10.1002/mc.2940050406. [DOI] [PubMed] [Google Scholar]
- 32.Dizdaroglu M, Rao G, Halliwell B, Gajewski E. Arch Biochem Biophys. 1991;285:317–324. doi: 10.1016/0003-9861(91)90366-q. [DOI] [PubMed] [Google Scholar]
- 33.Hoffman M E, Meneghini R. Photochem Photobiol. 1979;29:301–305. doi: 10.1111/j.1751-1097.1979.tb07052.x. [DOI] [PubMed] [Google Scholar]
- 34.Bindoli A. Free Radical Biol Med. 1988;5:247–261. doi: 10.1016/0891-5849(88)90018-4. [DOI] [PubMed] [Google Scholar]
- 35.Hruazkewycz A M. Biochem Biophys Res Commun. 1988;153:191–197. doi: 10.1016/s0006-291x(88)81207-5. [DOI] [PubMed] [Google Scholar]
- 36.Hruazkewycz A M. Mutat Res. 1992;275:243–248. doi: 10.1016/0921-8734(92)90028-n. [DOI] [PubMed] [Google Scholar]
- 37.Stohs S J, Bagghi D. Free Radical Biol Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 38.Vaca C E, Wilhelm J, Harms-Ringdahl M. Mutat Res. 1988;195:137–149. doi: 10.1016/0165-1110(88)90022-x. [DOI] [PubMed] [Google Scholar]
- 39.Zastawny T H, Altman S A, Randers-Eichhorn L, Madurawe R, Lumpkin J A, Dizdaroglu M, Rao G. Free Radical Biol Med. 1995;18:1013–1022. doi: 10.1016/0891-5849(94)00241-b. [DOI] [PubMed] [Google Scholar]
- 40.Chance B, Sies H, Boveris A. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Marcillat O, Giulivi C, Ernster L, Davies K J. J Biol Chem. 1990;265:16330–16336. [PubMed] [Google Scholar]
- 42.Ames B N, Shigenaga M K, Hagen T M. Biochim Biophys Acta. 1995;1271:165–170. doi: 10.1016/0925-4439(95)00024-x. [DOI] [PubMed] [Google Scholar]