Abstract
Accurate localization of proteins within the substructure of cells and cellular organelles enables better understanding of structure–function relationships, including elucidation of protein–protein interactions. We describe the use of a near-field scanning optical microscope (NSOM) to simultaneously map and detect colocalized proteins within a cell, with superresolution. The system we elected to study was that of human red blood cells invaded by the human malaria parasite Plasmodium falciparum. During intraerythrocytic growth, the parasite expresses proteins that are transported to the erythrocyte cell membrane. Association of parasite proteins with host skeletal proteins leads to modification of the erythrocyte membrane. We report on colocalization studies of parasite proteins with an erythrocyte skeletal protein. Host and parasite proteins were selectively labeled in indirect immunofluorescence antibody assays. Simultaneous dual-color excitation and detection with NSOM provided fluorescence maps together with topography of the cell membrane with subwavelength (100 nm) resolution. Colocalization studies with laser scanning confocal microscopy provided lower resolution (310 nm) fluorescence maps of cross sections through the cell. Because the two excitation colors shared the exact same near-field aperture, the two fluorescence images were acquired in perfect, pixel-by-pixel registry, free from chromatic aberrations, which contaminate laser scanning confocal microscopy measurements. Colocalization studies of the protein pairs of mature parasite-infected erythrocyte surface antigen (MESA)(parasite)/protein4.1(host) and P. falciparum histidine rich protein (PfHRP1)(parasite)/protein4.1(host) showed good real-space correlation for the MESA/protein4.1 pair, but relatively poor correlation for the PfHRP1/protein4.1 pair. These data imply that NSOM provides high resolution information on in situ interactions between proteins in biological membranes. This method of detecting colocalization of proteins in cellular structures may have general applicability in many areas of current biological research.
One of the crucial aspects of current biological inquiry relates to the organization of cells and how interactions between proteins are involved in important cellular processes, such as signal transduction and receptor–ligand binding. Several experimental systems have been developed in recent years to try and identify such protein–protein interactions including the two hybrid and phage display systems (1, 2). All such attempts to identify interactions in disassembled cellular systems need to be confirmed in vivo, by approaches such as chemical cross-linking in cells or some form of colocalization study using microscopy. A common approach has been the use of confocal microscopy using fluorescent antibody probes. We report here the development of a technique based on dual-color immunofluorescence labeling together with near-field scanning optical microscopy (NSOM) (3, 4), which offers significant advances in the detection of colocalized proteins, particularly at the membrane of cells.
A near-field microscope is based on scanning a very small light source, with dimensions smaller than its wavelength, very close (in the near-field) to the specimen. This light source is constructed from a single-mode optical fiber, which is tapered at one of its ends and coated with aluminum on its side walls. Light that is coupled to the fiber is emanated only from the very end of the taper, through an aperture of 50–100 nm diameter and is exponentially attenuated away from the tip, but laterally confined to the aperture dimensions within the near-field range. Whereas previous NSOM studies were mainly performed on optically thin samples, here we show that the unique properties of the near-field light are suitable for selective excitation of fluorescence from the top surface of a thick sample as well. Therefore, fluorescence NSOM is a very useful tool for imaging cell membranes (5), unlike cross-sectioning confocal microscopy, which is suitable for imaging the inside of a cell. In addition, to maintain constant tip height over the specimen during scanning, a shear-force feedback scheme is used (6, 7). The error-signal of the feedback circuit contains topography information, and hence simultaneously acquired fluorescence and topography are mapped in perfect registry, with superresolution beyond the diffraction limit.
As a demonstration of the power of the application of NSOM to detecting colocalization of proteins in cells, we have studied malaria-infected red blood cells. Several proteins that are synthesized by intraerythrocytic Plasmodium falciparum malaria parasites and are transported through the erythrocyte cytosol (8–10) have been identified in association with the membrane of infected erythrocytes (9, 11–21). In many cases, the erythrocyte proteins with which they interact, the mechanism of association, and the consequences of association have not been fully defined. However, it is thought that parasite protein interactions with erythrocyte membrane proteins contribute to modifications in infected erythrocyte morphology, antigenicity, mechanical properties, and adhesive properties during infection of red blood cells (22–29).
Knob-associated P. falciparum histidine rich protein (PfHRP1) (15, 30) is the predominant parasite protein component of knobs, protrusions of ≈100 nm diameter on the infected erythrocyte membrane. Purified knob structures contain erythrocyte skeletal proteins spectrin, actin, dematin, and protein 4.1, as well as PfHRP1 (14, 31). A 30-kDa fragment of PfHRP1 forms stable complexes with spectrin and actin in vitro (32). Protein 4.1 also binds to spectrin and actin and provides mechanical stability to the red blood cell membrane (33). The mature parasite-infected erythrocyte surface antigen (MESA) (13), also called P. falciparum erythrocyte membrane protein 2 (PfEMP2) (9) associates with protein 4.1 (34, 35). MESA has been localized to the cytoplasmic face of knobs by immunoelectron microscopy (9) although analysis of purified knobs has failed to identify MESA (31), and MESA is not required for knob formation or cytoadherence (36).
For a better understanding of these associations and their relation to parasite gene expression and protein function, a technique is needed that can map the distribution of malarial proteins in the erythrocyte membrane and show their colocalization with host proteins. Fluorescence resonance energy transfer (37–39) can be used to determine intimate associations between macromolecules on a 1–7 nm scale. The technique was successfully applied to study membrane proteins and dynamic processes in living cells (for examples, see refs. 40–45). However, its application for static colocalization studies is problematical. Due to different binding affinities, it is very difficult to ensure that the sample is comprised only of donor–acceptor pairs when the two proteins are colocalized. Instead, we used nonresonant fluorophores to perform simultaneous dual-color excitation and dual-color detection with NSOM to image protein associations on a scale that is relevant to the PfHRP1 knob size (100 nm). We investigated the colocalization of parasite proteins MESA and PfHRP1 with erythrocyte skeletal protein 4.1. We find a high degree of physical correlation in the fluorescence maps of MESA and protein 4.1. Our data provide unequivocal support to data from previous biochemical and structural studies (34, 35), which suggested that MESA is anchored at the erythrocyte membrane through an association with protein 4.1. For the PfHRP1 and protein 4.1 pair, however, our measurements show poor correlation. Thus, our data indicate that whereas MESA, PfHRP1, and protein 4.1 are all present in the knob structures (14, 31), there is only a direct interaction between MESA and protein 4.1. PfHRP1 and protein 4.1 are not specifically associated with one another. We note that in our technique protein distributions are mapped in situ in the erythrocyte membrane, in registry with topography, whereas in the previous investigations associations were deduced from ex situ electrophoretic studies after the isolation of the proteins from the membrane.
These results show that (i) membrane proteins can be simultaneously mapped with membrane topography with resolution better than 100 nm; (ii) these fluorescence maps are generated by selective excitation of immunolabeled membranes proteins, while discriminating against fluorescence from the inside of the cell; and (iii) detection of colocalization of two (or more) different proteins can be performed with high accuracy, free from chromatic aberrations. This technique can be very useful for the studies of ligand–receptor binding, ion channels, and other signal transduction mechanisms through cell membranes.
MATERIALS AND METHODS
Cell Culture.
Parasites were grown in vitro as described (46) except that 10% human serum was replaced by 0.5% Albumax II (GIBCO) and 200 mM hypoxanthine (Sigma) was added as a supplement (52). Parasites used in these studies were ItG-P21 (18) and D63 (35). Parasites of both lines express PfHRP1 and MESA.
Antibodies.
mAb 89 (30) against knob-associated PfHRP1 was kindly provided by Diane Taylor (Georgetown University, Washington, DC). Mab Pf12.8B7.4 against MESA (9) was kindly provided by Jeffrey Lyon (Walter Reed Army Institute of Research, Washington, DC). Rabbit polyclonal antibody against protein 4.1 was kindly provided by Joel Chasis (Lawrence Berkeley National Laboratory).
Indirect Immunofluorescent Antibody Assay.
Infected erythrocytes from in vitro cultures were enriched to >50% trophozoites by gelatin flotation (47) and washed three times in phosphate buffered saline (PBS). Thin blood smears were air dried, and fixed with acetone/methanol. They were reacted at room temperature for 45 min with the appropriate primary antibody diluted 1:20 in PBS/1% bovine serum albumin (BSA), washed three times in PBS, and reacted for 45 min with fluorescein isothiocyanate (FITC), tetramethylrhodamine, or Texas Red (TR) conjugated goat anti-mouse or goat anti-rabbit IgG (Molecular Probes) diluted 1:100 in PBS/1% BSA. For two-color labeling, primary antibodies from mouse and rabbit were mixed, as were secondary antibodies against both species. In the two-color control, mAb against PfHRP1 was followed by goat anti-mouse conjugated to either TR or FITC. Slides were washed three times in PBS and stored on water-soaked filter paper at 4°C. For the measurements, the filter paper was removed and the samples were imaged in air, at room temperature, without further drying.
Near-Field Scanning Optical Microscopy.
A NSOM scan head was constructed as an add-on to a conventional upright fluorescence microscope (Zeiss Axioskop), which also had confocal and bright-field capabilities. Fluorescence was collected with a 100× 1.4NA oil immersion objective (Zeiss Apoplan) and detected with an Avalanche photodiode (SPCM-200, EG & G, Quebec). This scheme enabled us to explore a large field-of-view by brightfield, zoom-in by confocal, and final zoom-in by near-field microscopy. Using this system we routinely obtained fluorescence maps with sub-100 nm resolution and with single-molecule sensitivity (48, 49). Such exquisite sensitivity suggests that immunofluorescence studies can be performed with low copy number membrane proteins.
Dual-Color Imaging.
For colocalization studies, proteins of interest were labeled with antibodies conjugated to two different fluorophores: FITC and TR. FITC was excited by a 488-nm line of an air-cooled Ar+ laser (532-AP, Omnichrome, Chino, CA) and TR was excited by a 568-nm line of a Krypton-Ar+ laser (Innova 70 Spectrum, Coherent Laser Group, Santa Clara, CA). The two excitation beams were colinearly combined and simultaneously coupled to the NSOM fiber probe. Two detectors were used to simultaneously acquire the two emissions (48). Both excitation colors were single-mode guided in the fiber (SpecTran, single mode 515 nm FS SMC-A0515B, Sturbridge, MA). Far-field inspection of the light emanating the near-filed aperture revealed diffraction-limited spots for both excitation wavelengths. Dipole moment effects that alter the point-spread-function of the microscope for single molecules (50) were averaged out for a large number of fluorophores present in this study. Because the two excitation wavelengths shared the near-field aperture with the same point-spread-function, fluorescence images were in perfect, pixel-by-pixel, registry. This is in contrast to confocal and wide-field fluorescence microscopy, where chromatic aberrations cause misalignment of the two-color images acquired on the two detectors. The excitation was blocked with a color glass long-pass filter (515 nm, LP515; Oriel, Stratford, CT) and a Raman notch filter (568 nm, HSPF-568-1.0; Kaiser Optical Systems, Ann Arbor, MI). The collected light was split into two channels by a dichroic mirror (558 nm, 558DRLP; Omega Optical, Brattleboro, VT). To eliminate cross-talk between channels, band pass filters 535 nm center, 25 nm full-width half-maximum (FWHM) (535DF25, Omega Optical) and 635 nm center, 25 nm FWHM (635DF25, Omega Optical) were placed in front of the short wavelength (FITC) and long wavelength (TR) detectors, respectively. With the band pass filters in place, the cross-talk was measured by exciting the dual-color-labeled erythrocyte with either of the two single laser lines. With 568 nm excitation alone, the signal in the long wavelength channel was 90% of the signal obtained with dual-color excitation, and the signal in the short wavelength channel was at the dark count level of the detector, indicating no cross-talk from TR to FITC. With 488 nm excitation alone, the signal in the long wavelength channel was 10% of the signal obtained with dual-color excitation. In the short wavelength channel, on the other hand, we measured the same signal level as with dual-color excitation. The 10% cross-talk on the long wavelength channel is due to the absorption tail of TR at 488 nm and the emission tail of FITC, and was subtracted for all images.
A commercial data acquisition software (SPM32, RHK Technology, Rochester, MI) was used to simultaneously acquire data from the two Avalanche photodiodes (the fluorescence channels) and the voltage on the piezo scanner (topography). Distribution of proteins in the erythrocyte membrane was presented as a color-coded image overlaid on topography, which was displayed as height in a three-dimensional plot. For detection of protein colocalization, FITC and TR images were pseudo-colored with red and green, respectively, and then overlaid through pixel-by-pixel addition of their red-green-blue values. Signals coming from areas labeled by FITC appeared green; signals coming from areas labeled by TR appeared red; signals coming from areas labeled with both FITC and TR appeared yellow.
A quantitative analysis of the colocalization was carried out through pixel-by-pixel correlation of red and green images: the intensity in each channel was analyzed for every pixel and displayed as a single point in a two-dimensional histogram. The x,y coordinates of each point presented the intensities in red and green channels, respectively. The number of occurrences of each x,y pair was presented by a gray scale value (brighter color corresponds to more occurrences). In such two-dimensional histograms, two perfectly correlated images show up as a straight line along the diagonal; anti-correlated images result in two lines along the two axes and un-correlated images exhibit a broad, unstructured distribution. We note that such analyses can be performed only when the two images are in perfect registry.
RESULTS AND DISCUSSION
Mapping of Malaria Protein PfHRP1 in the Infected Erythrocyte Membrane.
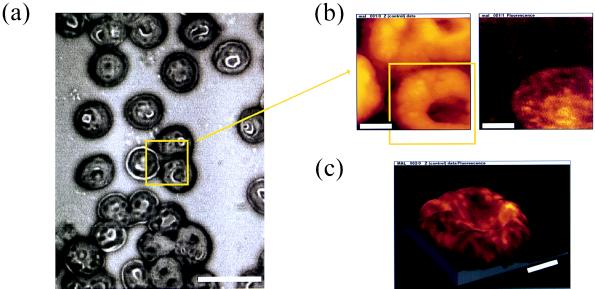
Fig. 1a shows a bright-field image of red blood cells labeled with antibody against PfHRP1 and immunolabeled with tetramethylrhodamine-conjugated secondary antibody. The NSOM tip was positioned at the center of the field-of-view and a zoomed-in scan was performed (area shown by yellow rectangle). Fig. 1b shows the acquired topography signal (Left) and fluorescence signal (Right). As can be seen, the three cells that are enclosed by the yellow box in Fig. 1a are visible in the topography image, but only the infected erythrocyte, recognized by mAb to PfHRP1, in the lower right corner of the rectangle, is detected in the fluorescence image. In addition, the topography image of the infected cell shows “grainy” features and the membrane surface is deformed compared with neighboring, uninfected cells. Examination of a large number of cells confirmed the finding that infected cells were consistently more deformed in topography maps. The strength of the fluorescence signal from the immunolabeled cells was typically 300 kHz. The background, dominated by the fiber tip autofluorescence, was about 10 kHz. Autofluorescence of erythrocytes was negligible compared with the fiber background, most probably due to the membrane specificity of the technique. The excitation intensity drops exponentially away from the tip and hence fluorescence from the inside of the cell is orders of magnitude weaker than fluorescence excited at the membrane. In addition, as shown below by cross-section confocal imaging (Fig. 2b), PfHRP1 mostly resides in the membrane for this late stage of infection. A further zoom-in around the infected cell in Fig. 1b (yellow box) is shown in Fig. 1c. Here the fluorescence signal is superimposed as a color value (brightness scale goes from black via red to yellow) on the topography to show the distribution of PfHRP1 in the erythrocyte membrane. Fluorescence spots with typical sizes of 100–150 nm are clearly resolved. Their average density is found to be 10 spots per μm2, which corresponds to the surface density of knobs deduced from electron microscopy images of isolated infected erythrocyte membranes (51). We conclude that the fluorescence spots in Fig. 1 originate from PfHRP1 in individual electron-dense knobs on the infected erythrocyte membrane. We cannot, however, resolve the fluorescence distribution within individual knobs, since the average knob size is 105 nm (31, 51), comparable to the resolution of the technique.
Figure 1.
Mapping of malarial protein in the erythrocyte membrane. A thin blood smear of the trophozoite infected erythrocytes was fixed, then reacted with antibodies against PfHRP1, and labeled with tetramethylrhodamine. (a) Bright-field image of the blood smear. The NSOM tip was positioned above the three cells in the center of the field-of-view and a scan was taken within the yellow framed area. (Scale bar = 10 μm.) (b) Result of the NSOM imaging in topography (Left) and fluorescence (Right) (128 × 128 pixels, 60 nm/pixel). While all three cells are imaged in topography, only the one in the lower right corner was infected and is visible in the fluorescence image. (c) Zoom around the infected cell, which was seen in the lower right corner of b (128 × 128 pixels, 52 nm/pixel). The fluorescence signal is superimposed as a color value (brightness scale goes from black via red to yellow) to the topography to show the distribution of PfHRP1 in the erythrocyte membrane. (Scale bar for b and c = 2 μm.) Z range for c is 1 μm.
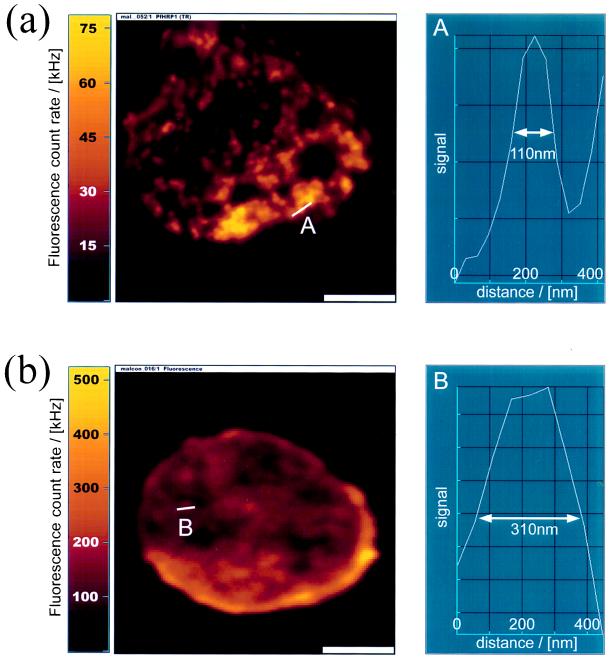
Figure 2.
NSOM (a) (256 × 256 pixels, 30 nm/pixel) and confocal (b) (128 × 128 pixels, 70 nm/pixel) imaging of infected erythrocytes labeled with antibodies against PfHRP1. The figures show the fluorescence image (Left) and a cross section through the smallest feature (marked with a line) found in the image (Right). The fluorescence signal is given as a color value (brightness scale goes from black via red to yellow). (a) Individual knobs are resolved by NSOM imaging. The resolution is about 100 nm. (b) Confocal cross section through the cell. The membrane is seen as a bright ring. The resolution is about 310 nm. (Scale bars = 2 μm.)
Comparison of NSOM and Confocal Membrane Imaging.
Fig. 2 compares NSOM imaging (Fig. 2a) with confocal imaging (Fig. 2b) of infected erythrocytes. These images were separately taken on two different infected red blood cells from the same culture, which were fixed and dried in parallel and labeled against PfHRP1. The NSOM image (Fig. 2a Left) shows an infected red blood cell reacted with anti-PfHRP1. A cross section taken along the line “A” is shown in Fig. 2a (Right). It shows the smallest feature in the image, with a FWHM of about 100 nm. A confocal image of a similar anti-PfHRP1-labeled infected erythrocyte is shown in Fig. 2b. Here again the smallest feature in the image is shown in a cross section “B” (Fig. 2b Right), with a FWHM of 310 nm. In contrast to the selective imaging of the membrane proteins by NSOM, the confocal image shows a cross section through the cell. A bright ring of fluorescence in the confocal image indicates that most of the PfHRP1 resides in the membrane as expected in the mature trophozoite stage of the intraerythrocytic cycle. However, the confocal image is a cross section of the cell and cannot reveal the distribution of PfHRP1 on the membrane surface, or the membrane topography. Furthermore, the resolution of the confocal image is three times lower compared with the NSOM image.
Colocalization of Malarial and Host Membrane Proteins.
Dual-color labeling and dual-color detection, together with pixel-by-pixel analysis described above, were used to study colocalization of two host–parasite protein pairs. A thin blood smear of erythrocytes infected with trophozoite stage parasites was fixed and then reacted with either mouse monoclonal or rabbit polyclonal antibodies against the proteins under study. Secondary antibodies (anti-mouse or anti-rabbit) labeled with either FITC or TR were then used to stain the proteins.
The results for the control experiment are shown in Fig. 3a, and for the two host–parasite protein pairs in Fig. 3 b and c. The left column of the figure shows the two simultaneously acquired and pseudo-colored images for each case. The red and green images represent raw data acquired on long wavelength (TR) and short wavelength (FITC) channels, respectively. The center column shows an overlay of the two images, and the right column shows pixel-by-pixel correlation of the red and green channels. All images were taken with pixel size (30–86 nm) smaller than aperture size (110 nm).
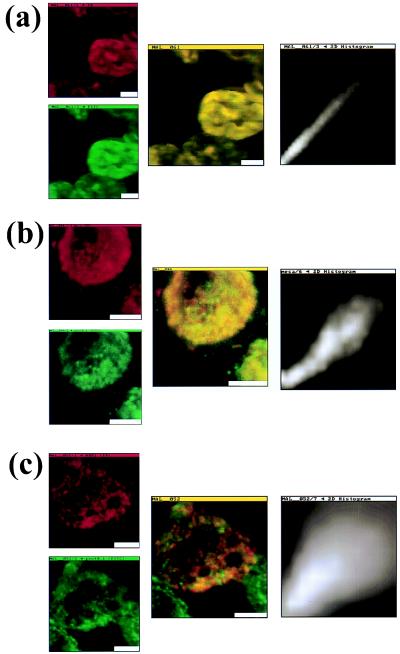
Figure 3.
Colocalization of malarial and host proteins in the erythrocyte membrane. A blood smear of erythrocytes infected with the trophozoite stage parasites was fixed and then reacted with two different antibodies (from rabbit and from mouse) against the proteins under study. Secondary antibodies labeled with FITC (anti-rabbit) and TR (anti-mouse) were used, respectively. The left column shows the two simultaneously acquired fluorescence channels by NSOM dual-color imaging. The center column presents the corresponding overlay of the fluorescence images and the right column is pixel-by-pixel correlation between the red and green images. (a) Control experiment: primary antibodies against PfHRP1 (128 × 128 pixels, 86 nm/pixel). (b) Colocalization of MESA and protein 4.1 (128 × 128 pixels, 48 nm/pixel). (c) Colocalization of PfHRP1 and protein 4.1 (256 × 256 pixels, 30 nm/pixel). (Scale bars = 2 μm.)
Fig. 3a shows the results of the control experiment. Immunofluorescence staining against PfHRP1 was performed with mAb 89. This mAb maps to a repeated epitope on the HRP1 protein. Secondary antibodies (goat anti-mouse) were labeled with FITC and TR. The overlay image (center column) shows only yellow areas, indicating the expected perfect overlap between red and green channels, since the same protein moiety has been labeled with two different fluorophores. This overlap is confirmed by the pixel-by-pixel correlation, which shows a straight line along the diagonal (right column). This result confirms the validity of the technique, since no off-diagonal red–green pairs could be found.
Fig. 3b shows colocalization results for the host–parasite pair protein 4.1/MESA. Protein 4.1 was labeled with FITC-conjugated, and MESA with TR-conjugated, secondary antibodies. The overlay image (center column) predominantly shows yellow areas of overlap between the two fluorophores. There are very few regions where green only or red only fluorescence can be found. The correlation analysis (right column) shows linear distribution, indicating colocalization of MESA with protein 4.1. This result indicates that MESA and erythrocyte protein 4.1 interact in situ in the membranes of infected red blood cells.
Fig. 3c shows colocalization results for the host–parasite pair protein 4.1/PfHRP1. Secondary antibodies conjugated with FITC or TR were used to localize protein 4.1 and PfHRP1, respectively, in the infected erythrocyte membrane. The overlay image (center column) clearly shows that PfHRP1 and protein 4.1 are only poorly colocalized, as confirmed by pixel-by-pixel correlation, which exhibits a very broad distribution around the diagonal (right column). Thus, we conclude that there is no association between protein 4.1 and PfHRP1 in the membrane of infected red blood cells. While both protein 4.1 and PfHRP1 are present in knob structures, these two proteins do not directly interact with each other as do MESA and protein 4.1.
CONCLUSIONS
NSOM was used to perform membrane oriented colocalization measurements, together with topography mapping, of parasite and host proteins in malaria infected erythrocytes. Our results imply that the malarial protein MESA interacts with erythrocyte protein 4.1 in membranes of infected red blood cells, whereas PfHRP1 does not interact with protein 4.1.
Extrapolating from previous biochemical analyses that indicate the presence of protein 4.1 in knobs (14, 31), and the evidence that PfHRP1 and protein 4.1 both bind to spectrin and actin in the red blood cell skeleton (32, 33), it has been tacitly assumed that PfHRP1 and protein 4.1 may interact in infected red blood cell membranes. Our results unequivocally show that this is not the case. These data extend our previous finding that protein 4.1 is not required for PfHRP1 to localize to the infected erythrocyte membrane, whereas protein 4.1 is necessary for localizing MESA at the membrane surface (35).
We have shown that near-field scanning optical microscopy can be specifically applied to cell membrane immunofluorescence studies. We applied NSOM to study membrane oriented colocalization, together with topography measurements, of parasite and host proteins in malaria infected red blood cells. The technique exhibits several advantages compared with confocal microscopy: (i) NSOM imaging exhibits superresolution; (ii) The near-field source excites fluorescence only from the outermost layer (50–100 nm) of the cell—i.e., the cell membrane; (iii) As a result, very good signal-to-background ratio is obtained, because autofluorescence is not integrated across the whole cell thickness; (iv) Topography information from the shear-force feedback allows real space mapping of proteins with subwavelength resolution (100 nm) in the cell membrane; (v) Dual-color excitation via shared aperture eliminates chromatic aberration, and hence permits superresolution colocalization studies; (vi) Mapping and colocalization studies can be performed with only few copies of proteins due to the exquisite sensitivity of the instrument.
Acknowledgments
We thank M. Narla and R. L. Coppel for critical reading and useful comments. We gratefully acknowledge the loan of the Innova 70 Spectrum laser from Coherent Laser Group (Santa Clara, CA). This work was supported by the Laboratory Directed Research and Development Program of Lawrence Berkeley National Laboratory under U.S. Department of Energy Contract DE-AC03-76SF00098 and Office of Naval Research Contract N00014-95-F-0099. C.M. is supported by National Institutes of Health Grant DK 32094.
Footnotes
Abbreviations: NSOM, near-field scanning optical microscopy; PfHRP1, P. falciparum histidine rich protein; MESA, mature parasite-infected erythrocyte surface antigen; FITC, fluorescein isothiocyanate; TR, Texas Red; FWHM, full-width half-maximum.
References
- 1.Fields S. Methods: Comp Methods Enzymol. 1993;5:116–124. [Google Scholar]
- 2.Smith G P, Scott J K. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 3.Pohl D W, Denk W, Lanz M. Appl Phys Lett. 1984;44:651–653. [Google Scholar]
- 4.Lewis A, Isaacson M, Harootunian A, Murray A. Ultramicroscopy. 1984;13:227–231. [Google Scholar]
- 5.Kirsch A, Meyer C, Jovin T M. In: Proceedings of NATO Advanced Research Workshop: Analytical Use of Fluorescent Probes in Oncology. Kohen E, Hirschberg J G, editors. New York: Plenum; 1996. in press. [Google Scholar]
- 6.Toledo-Crow R, Yang P C, Chen Y, Vaez-Iravani M. Appl Phys Lett. 1992;60:2957–2959. [Google Scholar]
- 7.Betzig E, Finn P L, Weiner J S. Appl Phys Lett. 1992;60:2484–2486. [Google Scholar]
- 8.Elford B C, Cowan G M, Ferguson D J P. Biochem J. 1995;308:361–374. doi: 10.1042/bj3080361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howard R J, Lyon J A, Uni S, Saul A J, Aley S B, Klotz F, Patton L J, Sherwood J A, Marsh K, Aikawa M, Rock E P. J Cell Biol. 1987;104:1269–1280. doi: 10.1083/jcb.104.5.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gormley J A, Howard R J, Taraschi T F. J Cell Biol. 1992;119:1481–1495. doi: 10.1083/jcb.119.6.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perlmann H, Berzins K, Wahlgren M J, Carlsson J, Bjorkman A, Patarroyo M E, Perlmann P. J Exp Med. 1984;159:1686–1704. doi: 10.1084/jem.159.6.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown G V, Culvenor J G, Crewther P E, Bianco A E, Coppel R L, Saint R B, Stahl H D, Kemp D J, Anders R F. J Exp Med. 1985;162:774–779. doi: 10.1084/jem.162.2.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coppel R L, Culvenor J G, Bianco E, Crewther P E, Stahl H-D, Brown G V, Anders R F, Kemp D J. Mol Biochem Parasitol. 1986;20:265–277. doi: 10.1016/0166-6851(86)90107-6. [DOI] [PubMed] [Google Scholar]
- 14.Leech J H, Barnwell J W, Aikawa M, Miller L H, Howard R J. J Cell Biol. 1984;98:1256–1264. doi: 10.1083/jcb.98.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilejian A. Proc Natl Acad Sci USA. 1979;76:4650–4653. doi: 10.1073/pnas.76.9.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howard R J. Prog Allergy. 1988;41:98–147. doi: 10.1159/000415221. [DOI] [PubMed] [Google Scholar]
- 17.Leech J H, Barnwell J W, Miller L H, Howard R J. J Exp Med. 1984;159:1567–1575. doi: 10.1084/jem.159.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magowan C, Wollish W, Anderson L, Leech J. J Exp Med. 1988;168:1307–1320. doi: 10.1084/jem.168.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baruch D I, Pasloske B L, Singh H B, Bi X H, Ma X C, Feldman M, Taraschi T F, Howard R J. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 20.Su X Z, Heatwole V M, Wertheimer S P, Guinet F, Herrfeldt J A, Peterson D S, Ravetch J A, Wellems T E. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 21.Smith J D, Chitnis C E, Craig A G, Roberts D J, Hudsontaylor D E, Peterson D S, Pinches R, Newbold C I, Miller L H. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trager W, Rudzinska M A, Bradbury P C. Bull WHO. 1966;35:883–885. [PMC free article] [PubMed] [Google Scholar]
- 23.Kilejian A, Abati A, Trager W. Exp Parasitol. 1977;42:157–164. doi: 10.1016/0014-4894(77)90073-x. [DOI] [PubMed] [Google Scholar]
- 24.Aikawa M. Bull WHO. 1977;55:139–156. [PMC free article] [PubMed] [Google Scholar]
- 25.Luse S A, Miller L H. Am J Trop Med Hyg. 1971;20:655–660. [PubMed] [Google Scholar]
- 26.Lee M V, Ambrus J L, DeSouza J M, Lee R V. J Med (Westbury, NY) 1982;13:479–485. [PubMed] [Google Scholar]
- 27.Cranston H A, Boylan C W, Carroll G L, Sutera S P, Williamson J R. Science. 1984;223:400–403. doi: 10.1126/science.6362007. [DOI] [PubMed] [Google Scholar]
- 28.Nash G B, O’Brien E, Gordon S E, Dormandy J A. Blood. 1989;74:855–861. [PubMed] [Google Scholar]
- 29.Paulitschke M, Nash G B. J Lab Clin Med. 1993;122:581–589. [PubMed] [Google Scholar]
- 30.Taylor D W, Parra M, Chapman G B, Stearns M E, Rener J, Aikawa M, Uni S, Aley S B, Panton L J, Howard R J. Mol Biochem Parasitol. 1987;25:165–174. doi: 10.1016/0166-6851(87)90005-3. [DOI] [PubMed] [Google Scholar]
- 31.Chishti A H, Andrabi K I, Derick L H, Palek J, Liu S. Mol Biochem Parasitol. 1992;52:283–288. doi: 10.1016/0166-6851(92)90062-o. [DOI] [PubMed] [Google Scholar]
- 32.Kilejian A, Rashid M A, Aikawa M, Aji T, Yang Y F. Mol Biochem Parasitol. 1991;44:175–182. doi: 10.1016/0166-6851(91)90003-o. [DOI] [PubMed] [Google Scholar]
- 33.Discher D E, Winardi R, Schischmanoff P O, Pavra M, Conboy J G, Mohandas N. J Cell Biol. 1995;130:897–907. doi: 10.1083/jcb.130.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lustigman S, Anders R F, Brown G V, Coppel R L. Mol Biochem Parasitol. 1990;38:261–270. doi: 10.1016/0166-6851(90)90029-l. [DOI] [PubMed] [Google Scholar]
- 35.Magowan C, Coppel R L, Lau A O T, Moronne M M, Tchernia G, Narla M. Blood. 1995;86:3196–3204. [PubMed] [Google Scholar]
- 36.Petersen C, Nelson R, Magowan C, Wollish W, Jensen J, Leech J. Mol Biochem Parasitol. 1989;36:61–65. doi: 10.1016/0166-6851(89)90200-4. [DOI] [PubMed] [Google Scholar]
- 37.Förster T. In: Modern Quantum Chemistry Lectures at the Istanbul International Summer School. Sinanoglu O, editor. New York: Academic; 1965. pp. 93–137. [Google Scholar]
- 38.Stryer L, Haugland R P. Proc Natl Acad Sci USA. 1967;58:719–730. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantor C R, Schimmel P R. Biophysical Chemistry. Vol. 2. San Francisco: Freeman; 1980. [Google Scholar]
- 40.Bastiaens P I, Jovin T M. Proc Natl Acad Sci USA. 1996;93:8407–8412. doi: 10.1073/pnas.93.16.8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jurgens L, Arndt-Jovin D, Pecht I, Jovin T M. Eur J Immunol. 1996;26:84–91. doi: 10.1002/eji.1830260113. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez J E, Tsien R Y. Biophys J. 1995;69:1272–1280. doi: 10.1016/S0006-3495(95)80029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo C, Dower S K, Holowka D, Baird B. J Biol Chem. 1995;270:27562–27568. doi: 10.1074/jbc.270.46.27562. [DOI] [PubMed] [Google Scholar]
- 44.Kam Z, Volberg T, Geiger B. J Cell Sci. 1995;108:1051–1062. doi: 10.1242/jcs.108.3.1051. [DOI] [PubMed] [Google Scholar]
- 45.Young R M, Arnette J K, Roess D A, Barisas B G. Biophys J. 1994;67:881–888. doi: 10.1016/S0006-3495(94)80549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trager W, Jensen J B. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 47.Pasvol G, Wilson R J M, Smalley M E, Brown J. Ann Trop Med Parasitol. 1978;72:87–88. doi: 10.1080/00034983.1978.11719283. [DOI] [PubMed] [Google Scholar]
- 48.Ha T, Enderle Th, Ogletree D F, Chemla D S, Selvin P R, Weiss S. Proc Natl Acad Sci USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ha T, Enderle Th, Chemla D S, Selvin P R, Weiss S. Phys Rev Lett. 1996;71:3979–3982. doi: 10.1103/PhysRevLett.77.3979. [DOI] [PubMed] [Google Scholar]
- 50.Betzig E, Chichester R J. Science. 1993;262:1422–1424. doi: 10.1126/science.262.5138.1422. [DOI] [PubMed] [Google Scholar]
- 51.Gruenberg J, Allred D R, Sherman I W. J Cell Biol. 1983;97:795–802. doi: 10.1083/jcb.97.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cranmer, S. L., Magowan, C., Liang, J., Coppel, R. & Cooke, B. M. (1997) Trans. R. Soc. Trop. Med. Hyg., in press. [DOI] [PubMed]