Abstract
Background and objective
Until recently, there has been little agreement between conflicting results of osteoarthritis (OA) linkage. The purpose of this study was to conduct a whole‐genome linkage scan to identify susceptibility loci for idiopathic hand OA in a large, population‐based sample of females.
Methods
Two OA‐related radiographic phenotypes DIP (distal interphalangeal joints)‐OA and Tot‐KL (Kellgren‐Lawrence score for both hands) chosen a priori were examined on 538 (269 pairs) monozygous and 1256 (628 pairs) dizygous (DZ) females. A genome‐wide scan using microsatellite markers spaced 10 cM apart was performed on 1028 DZ twins. First, the heritability of the two OA phenotypes was estimated. Next, multipoint linkage analysis was conducted using a modified version of the Haseman–Elston method in a generalised linear model.
Results
Heritability for DIP‐OA and Tot‐KL was found to be 47.6% and 67.4%, respectively. A genome‐wide scan produced reliable evidence of significant linkage of DIP‐OA on chromosome 2 at 90 cM (logarithmic odds ratio (LOD) = 2.90) and for Tot‐KL on chromosome 19 at 65 cM (LOD = 4.26). These results are in agreement with data published previously. Several other significant linkage peaks were observed—for example, on chromosome 1 at 250 cM and on chromosome 3 at 30 cM—but were confirmed less reliably.
Conclusion
This is one of the largest OA linkage studies performed to date and provides clear evidence for linkage at two quantitative trait loci (on chromosome 2 at 90 cM and on chromosome 19 at 65 cM). As the results were robust and replicated in previous smaller studies, the fine mapping of these regions is a logical next step to pinpoint potential susceptibility gene(s) of interest.
Idiopathic osteoarthritis (OA), in particular hand OA, is a heterogeneous and multifactorial process of joint degeneration, which appears with exceptionally high prevalence in aged populations. A number of community‐based studies have shown that the majority of adults aged >55 years have radiographic evidence of hand OA.1 Determining definitive aetiological factors, however, poses methodological and other problems. Various factors are known to be associated with OA, including age, gender, biomechanics, ethnic background, genetics and others.2
After age, genetic factors play a major role and have explained 30–60% of the residual variation in most studies.2,3,4,5 Several genome‐wide scans with hand OA have been reported, but the results have been inconsistent, often because of small sample size and low power. Despite some studies having reasonable size, results of only marginal significance have been obtained.3,4,6 Such conflicting results might reflect differences in sample ascertainment, age and cohort effects, false‐positive results due to multiple‐testing problems, or methodological problems such as disease definitions. This is particularly complex in hand OA, where numerous and small joints are examined, which may have different age‐related rates of OA progression.7,8
In an attempt to clarify this situation, we used a large community‐based sample of dizygous (DZ) twin pairs to search for susceptibility loci by whole‐genome linkage screening.
Material and methods
Sample and phenotypes
The data examined in the present study were from the Twins UK Adult Twin Registry (described in detail elsewhere9). All participants gave written informed consent before entering the study, and the St Thomas' Hospital research ethics committee approved the project. Twins have been shown to be similar to age‐matched singletons for a range of health and lifestyle variables.10
Plain posterior–anterior hand radiographs with both hands placed flat were obtained from each study participant. The films from twins were not read paired, nor necessarily close in time. The radiographic features of hand OA, and the presence of osteophytes (OSP) and joint space narrowing (JSN), were examined for each of 15 joints, on each hand. OSP and JSN were separately evaluated and graded from 0 to 3 for increasing severity using a standardised atlas.11 In addition, the summary grade for each joint of the hand joints (30 in total) was evaluated according to the Kellgren and Lawrence (KL) grading system (0–4) and in accordance with the original atlas. The intraobserver reproducibility for each site/trait was estimated using a weighted κ statistic and found to be satisfactory (>0.78) for different joints and features. For the present genetic analysis, we selected a priori two clearly defined OA phenotypes used in other studies: (1) DIP‐OA, obtained as a sum of OSP and JSN scores of 10 DIP joints and (2) Tot‐KL, the sum of KL scores for all 30 joints, on both hands. The main reasons for selecting these phenotypes were that they had the highest reproducibility and highest prevalence in the sample, of all the OA phenotypes available. They showed the closest correlation with age, and after adjustment were the closest to a normal distribution. Finally, these traits had the strongest heritability of all hand–joint combinations studied.
Genotyping
Genomewide linkage analysis used 737 highly polymorphic DNA markers spaced approximately 10 cM apart using standard fluorescence‐based genotyping methodologies, and have been described in full detail elsewhere.12 The estimated genotyping error rate was <1%.
Statistical analysis
First, 538 monozygous and 1256 DZ twins having complete radiological data available for all joints were included in analysis of the OA‐phenotype distribution, adjustment for age and model fitting analysis to estimate heritability of the chosen phenotypes.13 Next, 1028 DZ twins genotyped at each DNA marker were subjected to multipoint linkage (MPL) analysis. This was conducted using generalised linear modelling (GLM) in STATA V.9.0. This technique is based on optimal Haseman and Elston methods.14 This method is algebraically equivalent to other likelihood techniques, but has the advantage of being robust to deviations in multivariate normality by freely estimating the coefficient of variation (ie, the mean and variance‐corrected residual error) and by using a robust Huber estimate of variance.14 Regression diagnostics were used to check the reliability of model fit, including an iterative regression (IR) routine applied to GLM using the Huber estimate, followed by biweighted iterations that remove the gross outliers before calculating starting values and then performing the iterations. On a genomewide basis, any observed divergence between GLM and GLMIR results indicates potentially poor‐fitting models for specific regions. The prioritised regions were selected if the logarithmic odds ratio (LOD) score obtained was >2.5 and consistent between GLM and GLMIR results. The final confirmation of the positive results was made by computing the genome‐wide significance level (empirical p value) using a permutation approach, which is not affected by violation of the normality assumption, as implemented in the STATA package.
Results
Both OA phenotypes correlated significantly (p<0.001) with age and age squared. Some 12.0% and 26.4% of variation of DIP‐OA and Tot‐KL was attributable to age effects. Likelihood ratio test of the patterns of trait inheritance revealed that the most parsimonious model included contributions from additive genetic effects and random environment, with heritability estimates of 0.48 (95% CI 0.40 to 0.54) and 0.67 (95% CI 0.610 to 0.728) for DIP‐OA and Tot‐KL, respectively. In agreement with this were the intraclass correlation coefficients in the DZ cohort, for both DIP‐OA and Tot‐KL (0.24 and 0.25, respectively; p<0.001 for both variables). The phenotypic correlation between the adjusted Tot‐KL and DIP‐OA was 0.67 (p<0.001), suggesting that a substantial part of the variation (∼55%) of each variable may be governed by independent factors.
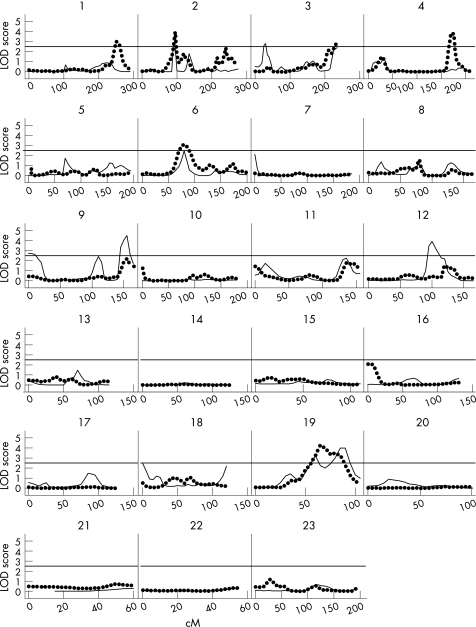
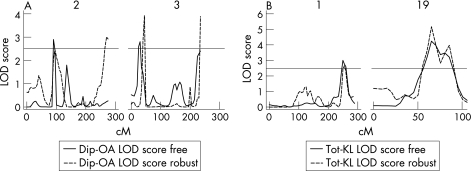
Figure 1 depicts the evidence for linkage for the entire genome scan, for DIP‐OA and Tot‐KL. The figure shows all nominal LOD scores estimated using the freely estimating coefficient of variation option, which is robust to phenotypic departure from normality assumption. By our rigid criteria of LOD>2.5, five chromosomal areas were identified both for DIP‐OA and for Tot‐KL. Table 1 summarises the corresponding LOD scores and their respective chromosomal locations. These results were checked using the robust iterative routine. For DIP‐OA, we obtained good confirmation of the results on chromosome 2, where both MPL peaks coincided (fig 2A). A marginal confirmation was also observed on chromosome 3, where the LOD score obtained in robust analysis reached 3.92; however, the corresponding linkage peak was located at the 45 cM position (fig 2A). For Tot‐KL, we obtained clear correspondence in the results on chromosome 1 (LODIR = 2.67) and chromosome 19, where the respective IR peak was even higher, achieving a value of 5.19 (fig 2B). The empirical p values for corresponding linkage peaks were estimated as 0.005 on chromosome 1 and 0.002 on chromosome 19 for Tot‐KL, and as 0.023 on chromosome 2 for DIP‐OA.
Figure 1 Multipoint linkage analysis results of two osteoarthritis (OA)‐related phenotypes adjusted for age for all chromosomes. Suggestive linkages at the logarithmic odds ratio score 2.5 shown by horizontal line. Solid line shows Tot‐KL (Kellgren–Lawrence score for both hands), interrupted line shows DIP (distal interphalangeal)‐OA.
Table 1 Significant multipoint logarithmic odds ratio scores obtained at the first stage of analysis.
| Chromosome | Location (cM) | Closest marker | Marker location (cM) | Nominal LOD |
|---|---|---|---|---|
| Phenotype: DIP‐OA | ||||
| 2p13.2–2p14* | 90 | D2S285 | 86.2–89.9 | 2.9 |
| 3p25.1–3p25.2* | 30 | D3S1263 | 30.4–36.1 | 2.8 |
| 9q34.2–9q34.3 | 155 | D9S164–D9S1826 | 147.9–148.1 | 4.5 |
| 159.6–160.2 | ||||
| 12q21.33–12q22 | 100 | D12S351–D12S346 | 95.6–97.1 | 3,93 |
| 104.6–106.1 | ||||
| 19q13.41 | 90 | D19S571–D19S418 | 84.1–87.7 | 3.99 |
| 92.6–97.5 | ||||
| Phenotype: Tot‐KL | ||||
| 1q42.12–1q42.13* | 250 | D1S425–D1S2842 | 231.1–235.3 | 3.02 |
| 273.5–277.3 | ||||
| 2p12–2p13.3 | 95 | D2S2110 | 90.82–95.1 | 3.97 |
| 4q32.3 | 170 | D4S1597 | 169.1–169.4 | 3.84 |
| 6q11.2–6q12 | 80 | D6S257 | 79.92–80.0 | 3.05 |
| 19q13.2* | 65 | D19S5420 | 70.14 | 4.26 |
DIP, distal interphalangeal; LOD, logarithmic odds ratio; OA, osteoarthritis; Tot‐KL, Kellgren–Lawrence score for both hands.
*Results confirmed by generalised linear modelling (iterative regression).
Figure 2 Significant multipoint linkage analysis results of two osteoarthritis (OA)‐related phenotypes adjusted for age. (A) shows results for DIP (distal interphalangeal)‐OA phenotype with chromosomes 2 and 3. (B) shows results for Tot‐KL (Kellgren–Lawrence score for both hands) phenotype with chromosomes 1 and 19. The figure clearly demonstrates overlapping of peaks obtained by two methods of generalised linear modelling technique observed on chromosomes 2 (A), and 1 and 19 (B). Suggestive linkages at the logarithmic odds ratio (LOD) score 2.5 shown by the horizontal line. The solid line shows LOD score estimate obtained with the total sample, the interrupted line shows LOD score obtained after removal of the outlying individuals.
Discussion
There are no clear‐cut or universally accepted criteria for the selection of the primary OA phenotype, which has resulted in substantial heterogeneity in the epidemiological and genetic literature. However, regardless of the method employed, all previous studies have invariably reported a significant familial component in variation of OA, after adjusting for age.2,3,4,5,6
In the present linkage study, we used two phenotypes selected a priori on the basis of their distribution, sample size, substantial interindividual variation, significant familial aggregation and similarity to phenotypes examined in previous studies. The estimates of heritability obtained in this study were highly significant and were in accordance with previously published data. The major findings were the three reliable MPL signals observed for our two OA phenotypes. The genomic regions with multipoint LOD scores >2.5 were observed on chromosome 2 at 90 cM for DIP‐OA, and on chromosome 1 at 250 cM and chromosome 19 at 65 cM for Tot‐KL. These relatively strong linkage signals were obtained by both linkage methods (fig 2). The reliability and significance of all three peaks were confirmed by the estimated empirical p values. The additional significant linkage (LOD = 2.80) for DIP‐OA on chromosome 3 (30 cM from p terminus) was not fully supported by the robust routine. Despite a high LOD score (3.92), the peak of the MPL plots from the free and robust analyses did not overlap (30 vs 45 cM). The positive linkage data for this chromosome are therefore less reliable than the data generated for chromosomes 1, 2 and 19. The same is true with respect to chromosome 6 at 80 cM, where LOD = 3.05 was initially observed with Tot‐KL, and also coincided with a lower peak for DIP. Note also that DIP‐OA and Tot‐KL linkage peaks overlapped substantially on some chromosomes—namely, 2, 6 and 19 (fig 1), although this was not confirmed by our strict statistical criteria. These regions are nevertheless of interest, as they may reflect the common genetic effects shared by two phenotypes, and thus at least partly explain the significant phenotypic correlation between them. Of particular interest is chromosome 19q13 (table 1), in which both peaks were reliably significant: LOD = 3.99 and 4.46 for DIP‐OA and TOT‐KL, respectively. Although the regions did not exactly coincide (90 vs 65 cM respectively) the 95% CIs for the locations of quantitative trait loci obtained in model free linkage analyses are very wide and are often defined as 1.5–2 LOD score drop.15 Even with the more conservative estimate recently proposed by Manichaikul et al,16 if the spacing between markers is 10 cM (as was the case in our study), a drop of ∼1.2 in LOD is needed. This produces a 95% CI of approximately 30 cM, which overlaps with the empirical MPL location of the peaks, suggesting possible common quantitative trait loci.
Our three main linkage regions on chromosomes 1, 2 and 19 have been reported in previous linkage scans for OA (table 2). Thus, for example, Hunter et al,4 using the Framingham sample, recently reported significant linkage of the first principal component derived from 96 variables (KL scores, OSP scores and JSN scores) for 32 joints with chromosome 1 at 218 cM. Their signal was relatively close to our result for Tot‐KL at 250 cM. Interestingly, their original LOD of 2.02 increased to 3.03 when males were excluded from the analysis, indicating that the specific‐linkage signals may be sex‐specific. This is a particular advantage of the present study, as we used a single‐sex sample. Hunter et al also noted that most of the variation of their first principal component was caused by DIP scores, suggesting a more joint‐specific consideration of the observed linkage. Table 2 shows that our highest LOD score of 4.26 was found at 65 cM on chromosome 19 for Tot‐KL, and concurs with the results of Demissie et al,6 who recorded a significant association of the Tot‐KL score in the Framingham cohort to D19S178 marker, located in almost precisely the same area, 68 cM.
Table 2 Summary of linkage literature on hand osteoarthritis replicated in this study.
| Chromosome number, map position (cM) | Nearest marker | LOD score | Phenotype | Sample | Reference |
|---|---|---|---|---|---|
| 1, 250 | D1S425–D2S2842 | 3.02 | Tot‐KL | Community, UK | Present data |
| 1, 218 | Not available | 2.02 | DIP‐OA | Community, US | 4 |
| 1, 202 | Not available | 3.03 | DIP‐OA | Community, US | 4 (only females) |
| 1, 102 | D1S1665 | 2.96 | Total JSN score | Community, US | 6 |
| 19, 65 | D19S414–D19S418 | 4.26 | Tot‐KL | Community, UK | Present data |
| 19, 52 | D19S433 | 1.82 | Total JSN score | Community, US | 6 |
| 19, 68 | D19S178 | 1.83 | Tot‐KL | Community, US | 6 |
| 2, 90 | D2S136–D2S139 | 2.87 | DIP‐OA | Community, UK | Present data |
| 2, 94.8 | D2S1566 | 2.20 | DIP‐OA | Affected families; original mapping; Iceland | 3 |
| 2, 48 | D2S2168 | 2.44 | DIP‐OA | Affected families; additional mapping; Iceland | 3 (additional markers) |
| 2, 48 | D2S2168 | 4.44 | DIP‐OA and CMC‐1 | Affected families; additional mapping; Iceland | 3 (additional markers) |
| 2, 51.5 | D2S405 | 2.23 | Total JSN score | Community, US | 6 |
| 2, 103.7 | Not available | 1.04 (p = 0.014) | DIP‐OA | Affected families, UK | 18 |
| 2, 116 | IL1R1 (q shoulder) | p = 0.0001 | DIP‐OA | Affected families, Finland | 17 |
CMC, carpometacarpal; DIP, distal interphalangeal; IL1R1, interleukin 1 receptor, type 1; JSN, joint space narrowing; LOD, logarithmic odds ratio; OA, osteoarthritis; Tot‐KL, Kellgren–Lawrence score for both hands.
The association of DIP‐OA to chromosome region 2 at 90 cM, observed in this linkage analysis, also seems to be a promising finding. We found a LOD of 3.95 close by at 95 cM for Tot‐KL, although not confirmed in the robust method. Several independent genome‐wide scans have reported the linkage of this region to total hand scores and to a variety of DIP‐OA phenotypes (eg, Stefansson et al,3 table 2). The latter study used a large sample of Icelandic families with probands having severely affected DIP joints. The group initially estimated a LOD of 2.2, which almost completely coincided with our peak, at 94.8 cM. However, when they added additional markers and altered the phenotype to include the first carpometacarpal joints, their linkage peak increased to 4.4, but moved to 48 cM. Demissie et al6 reported their total hand sum of JSN to be associated with this area, 51.5 cM. In contrast, at least two other studies suggested that DIP‐OA could be linked to the chromosomal region observed in the present study (table 2). Leppavuori's team,17 in a small family‐based sample, found a significant association of DIP to 116 cM near the IL1R1gene cluster. Greig et al,18 using a modest sample of affected families from a similar ethnic background to ours, found significant linkage to markers mapped to the 103.7 cM position on this chromosome. As the CIs are wide for model‐free linkage methods,16 it is possible that these areas coincide and represent a replication.
A potential limitation of this study is the simple structure of the twin families, which has some ambiguity in identical by descent status definition. This would have had the effect of reducing the chances of finding significant linkage. Having said this, it should be noted that the single‐sex twin pair design does have a number of advantages in genetic analysis of common age‐related and sex‐related traits. To strengthen the reliability of linkage results, we used a large, community‐based sample, one of the largest that has been examined to date. We examined well‐defined phenotypes, with very high intraobserver consistency of assessment and strong familial aggregation, and used techniques of linkage analysis robust to deviations from normality.14 In addition, to avoid possible false‐positive linkage signals, we validated our results with additional statistical tests. It should also be mentioned that the results of our analysis are unaffected by the potential sex‐specific metabolic pathways and genetic influences that have been observed in a number of genetic epidemiological studies, both in animal models19 and in humans.20,21,22 The endocrine milieus and physiological background differ between males and females throughout life. Therefore, even in the absence of the sex‐specific genes, sex could act as an environmental factor that can influence disease risk and severity through sex‐specific genotype interaction. We were unable to look separately for symptomatic disease due to small numbers, but believe the results would be equally applicable to the correlations with severe disease and pain.
In summary, significant linkage peaks were observed on chromosomes 2 and 3 for DIP‐OA, and on chromosomes 1 and 19 for Tot‐KL. We found some overlap (chromosomes 2, 6 and 19) between the linkage peaks for the two phenotypes, which are certainly of interest but clearly require confirmation. However, our major peaks showed little overlap, which may suggest the potential influence of different metabolic pathways, governed by different genes, on different OA‐related phenotypes, as was recently suggested by Hunter et al.4 Of our four significant linkage results, two (on chromosome 2 for DIP‐OA, and on chromosome 19 for Tot‐KL) are supported further by results obtained by other teams using other samples. The fine mapping of these regions is now warranted and may well reveal gene(s) of interest in OA.
Acknowledgements
The Twin Research and Genetic Epidemiology Unit received support from the Welcome Trust and Arthritis Research Campaign. The funding for the Genome scan came from Gemini/Sequenom Inc. We thank Yulia Vistoropsky (TAU) and Sergey Ermakov (TAU) for their assistance in manuscript preparation.
Abbreviations
DIP - distal interphalangeal
DZ - dizygous
GLM - generalised linear modelling
IR - iterative regression
JSN - joint space narrowing
KL - Kellgren and Lawrence
LOD - logarithmic odds ratio
MPL - multipoint linkage
OA - osteoarthritis
OSP - osteophyte
Tot‐KL - Kellgren–Lawrence score for both hands
Footnotes
Competing interests: None.
References
- 1.Hart D J, Spector T D. Definition and epidemiology of osteoarthritis of the hand: a review. Osteoarthritis Cartilage 20008(Suppl A)S2–S7. [DOI] [PubMed] [Google Scholar]
- 2.Spector T D, MacGregor A J. Risk factors for osteoarthritis: genetics. Osteoarthritis Cartilage 200412(Suppl A)S39–S44. [DOI] [PubMed] [Google Scholar]
- 3.Stefansson S E, Jonsson H, Ingvarsson T, Manolescu I, Jonsson H H, Olafsdottir G.et al Genomewide scan for hand osteoarthritis: a novel mutation in matrilin‐3. Am J Hum Genet 2003721448–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter D J, Demissie S, Cupples L A, Aliabadi P, Felson D T. A genome scan for joint‐specific hand osteoarthritis susceptibility: the Framingham Study. Arthritis Rheum 2004502489–2496. [DOI] [PubMed] [Google Scholar]
- 5.Suk E K, Malkin I, Dahm S, Kalichman L, Ruf N, Kobyliansky E.et al Association of ENPP1 gene polymorphisms with hand osteoarthritis in a Chuvasha population. Arthritis Res Ther 200571082–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demissie S, Cupples L A, Myers R, Aliabadi P, Levy D, Felson D T. Genome scan for quantity of hand osteoarthritis: the Framingham Study. Arthritis Rheum 200246946–952. [DOI] [PubMed] [Google Scholar]
- 7.Egger P, Cooper C, Hart D J, Doyle D V, Coggon D, Spector T D. Patterns of joint involvement in osteoarthritis of the hand: the Chingford Study. J Rheumatol 1995221509–1513. [PubMed] [Google Scholar]
- 8.Kalichman L, Cohen Z, Kobyliansky E, Livshits G. Patterns of joint distribution in hand osteoarthritis: contribution of sex, age and handedness. Am J Hum Biol 200416125–134. [DOI] [PubMed] [Google Scholar]
- 9.Spector T D, MacGregor A. The St Thomas' UK Adult Twin Registry. Twin Res 20025440–443. [DOI] [PubMed] [Google Scholar]
- 10.Andrew T, Hart D J, Snieder H, de Lange M, Spector T D, MacGregor A J. Are twins and singletons comparable? A study of disease‐related and lifestyle characteristics in adult women. Twin Res 20014464–477. [DOI] [PubMed] [Google Scholar]
- 11.Burnett S, Hart D J, Cooper C, Spector T D.A radiographic atlas of osteoarthritis. London: Springer‐Verlag, 1994
- 12.Wilson S G, Reed P W, Andrew T, Barber M J, Lindersson M, Langdown M.et al A genome‐screen of a large twin cohort reveals linkage for quantitative ultrasound of the calcaneus to 2q33–37 and 4q12–21. J Bone Miner Res 200419270–277. [DOI] [PubMed] [Google Scholar]
- 13.Snieder H, MacGregor A J, Spector T D. Genes control the cessation of a woman's reproductive life: a twin study of hysterectomy and age at menopause. J Clin Endocrinol Metab 1998831875–1880. [DOI] [PubMed] [Google Scholar]
- 14.Barber M J, Cordell H J, MacGregor A J, Andrew T. Gamma regression improves Haseman‐Elston and variance components linkage analysis for sib‐pairs. Genet Epidemiol 20042697–107. [DOI] [PubMed] [Google Scholar]
- 15.Dupuis J, Siegmund D. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics 1999151373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manichaikul A, Dupuis J, Sen S, Broman K W. Poor performance of bootstrap confidence intervals for the location of a quantitative trait locus. Genetics 2006174481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leppavuori J, Kujala U, Kinnunen J, Kaprio J, Nissila M, Heliovaara M.et al Genome scan for predisposing loci for distal interphalangeal joint osteoarthritis: evidence for a locus on 2q. Am J Hum Genet 1999651060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greig C, Spreckley K, Aspinwall R, Gillaspy E, Grant M, Ollier W.et al Linkage to nodal osteoarthritis: quantitative and qualitative analyses of data from a whole genome screen identifies trait dependent susceptibility loci. Ann Rheum Dis 2006651131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orwoll E S, Belknap J K, Klein R F. Gender specificity in the genetic determinants of peak bone mass. J Bone Miner Res 2001161962–1971. [DOI] [PubMed] [Google Scholar]
- 20.Ralston S H, Galwey N, MacKay I, Albagha O M E, Cardon L, Compston J E.et al Loci for regulation of bone mineral density in men and women identified by genome wide linkage scan: the FAMOS study. Hum Mol Genet 200514943–951. [DOI] [PubMed] [Google Scholar]
- 21.Peacok M, Koller D L, Lai D, Hui S, Foroud T, Econs M J. Sex‐specific quantitative trait loci contribute to normal variation in bone structure at the proximal femur in men. Bone 200537467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss L A, Pan L, Abney M, Ober C. The sex specific genetic architecture of quantitative traits in humans. Nat Genet 200638218–222. [DOI] [PubMed] [Google Scholar]