Abstract
Background
Tissue plasminogen activator (tPA) is the major activator of plasminogen in plasma. This serine protease is overexpressed by exocrine pancreas tumour cells, where it promotes tumour cell proliferation, growth, and invasion. Here we have explored the signalling pathways used by tPA to activate the proliferation of pancreatic cancer cells.
Methods
Transcriptional profiling on cDNA micro arrays was used to analyse the pattern of gene expression in response to tPA compared to the response to epidermal growth factor (EGF) and platelet derived growth factor (PDGF). Results were confirmed using different biochemical assays in which specific kinase inhibitors or RNA interference were used.
Results
Transcriptional profiling showed that tPA modulates the expression of a set of genes commonly regulated by EGF, but distinct from the major set of genes modulated by PDGF. This suggested that tPA and EGF share common signalling pathways, a conclusion supported by further experimental evidence. Firstly, we found that tPA induced a rapid and transient phosphorylation of the EGFR. Secondly, specific EGFR kinase inhibitors, but not PDGFR kinase inhibitors, abolished the tPA induced phosphorylation of the ERK1/2 kinases and cell proliferation. The mitogenic activity of tPA was also inhibited by siRNA depletion of EGFR, thus confirming the involvement of this receptor in tPA triggered signalling. Thirdly, we show that the signalling and mitogenic effects of tPA require its proteolytic activity, the activity of the metalloprotease‐9 and active hb‐EGF.
Conclusion
Our results suggest that tPA induces proliferation by triggering a proteolytic cascade that sequentially activates plasmin, metalloprotease‐9 (MMP‐9) and hb‐EGF. These events are required to activate the EGFR signalling pathway and cell proliferation.
Keywords: tissue plasminogen activator, pancreas cancer, EGF receptor, proliferation, Microarrays
Pancreatic carcinoma is a devastating disease. About 90% of patients present with locally advanced or metastatic disease. Although surgery remains the most successful therapy, less than 10% of patients diagnosed with pancreatic cancer actually have a curative resection.1 Improvements in therapy that can add to surgery and systemic treatments for the advanced disease provide the greatest hope of improving the clinical outcomes in this disease.
Tissue type plasminogen activator (tPA) is the major blood activator of plasminogen required for the degradation of fibrin clots.2 In addition to their well established fibrinolytic roles, tPA, plasmin, and urokinase type plasminogen activator (uPA), have been implicated in cancer growth and progression in vivo.3 tPA is overexpressed in pancreatic carcinoma, where it stimulates growth and angiogenesis.4,5,6 tPA activity correlates with poor prognosis in several other cancers, including melanomas and breast tumours.7,8
The mechanisms by which plasminogen activators (PAs) promote cancer cell proliferation are best known for uPA. uPA binds to a specific cell surface receptor (uPAR), that forms part of a multiprotein signalling‐receptor complex including integrins, FPR‐like receptor‐1/lipoxin A4 receptor, and epidermal growth factor receptor (EGFR).9,10,11 The binding of uPA to uPAR activates extracellular activated kinases 1 and 2 (ERK1, ERK2), among other kinases.12,13 This activation is blocked by EGFR tyrosine kinase antagonists.11,14 In addition, the activation of ERKs in response to uPA requires the activity of a metalloprotease (MMP), suggesting the implication of a released membrane bound EGFR ligand.14 For tPA, the signalling mechanisms leading to proliferation are currently unknown. In the brain, it is now becoming clear that tPA exerts both proteolytic and non‐proteolytic effects that contribute to various aspects of brain functioning at morphological, biochemical, and functional levels.15,16,17,18,19,20
EGFR, a member of the HER family of receptor tyrosine kinases, is a potent stimulator of cell growth upon binding by its high affinity ligands, EGF and transforming growth factor α (TGFα). The importance of EGFR in tumour progression has been extensively reviewed.21 Pancreatic carcinoma is one of many tumours overexpressing EGFR, thereby making it a rational target for antitumour therapy in this disease.22
Our previous work has shown that tPA, overexpressed in pancreatic carcinomas, promotes proliferation, angiogenesis, and invasion.4,5,6 tPA binds to the surface of pancreas cancer cells mostly through annexin II, also abundant in these tumours.23,24 This interaction maintains tPA active on the cell surface and is required for the cell's invasive capacity.23
Because tPA significantly stimulates pancreas cancer cell proliferation in vitro and in vivo,6 it is important to understand the mechanisms through which this protease induces tumour growth. Here we show that tPA elicits a transcriptional response that significantly overlaps that induced by EGF, which is clearly distinct from the response induced by PDGF. tPA stimulates a rapid phosphorylation and activation of the EGFR and downstream ERKs. By means of small interfering RNA duplexes (siRNA) knockdown and the use of chemical inhibitors we show that both the expression and the kinase activity of the EGFR are required for the transmission of the tPA promoted proliferation. Finally, we demonstrate that this process requires proteolytic active tPA, and the activation of plasmin, metalloprotease‐9 (MMP‐9), and heparin binding‐epidermal growth factor (hb‐EGF), that eventually lead to activation of the EGFR pathway and cell proliferation.
Experimental procedures
Cell culture and reagents
Cell lines obtained from the American Type Culture Collection (Rockville, MD, USA) were maintained in Dulbecco's Modified Eagle's Medium (DMEM) (Gibco‐brl, Gaithesburg, NY, USA) supplemented with 10% heat inactivated fetal bovine serum (FBS, Gibco‐Brl) at 37°C in an atmosphere of 5% CO2. Galardin (GM 6001), mutant [Glu52]diphtheria toxin CRM197, and active human recombinant MMP‐9 were from Calbiochem (Darmstadt, Germany); PD098059, LY294002, and tyrphostin AG1478 from Biomol (Butler Pike, PA, USA); recombinant human epidermal growth factor (EGF) from Invitrogen (Carlsbad, CA, USA); p‐aminobenzoyl‐gly‐pro‐d‐ala‐hydroxamic acid (AHA) from MP Biomedicals (Aurora, OH, USA); plasmin and bisindolylmaleimide (GF109203X) from Roche Diagnostics (Mannheim, Germany); recombinant tPA (Actilyse) from Boehringer Mannheim (Barcelona, Spain); Pefabloc/tPA [2,7‐bis‐(4‐amidinobenzylidene)‐cycloheptanone‐(1) dihydrochloride salt] from Pentapharm (Basel, Switzerland). Alpha 2 antiplasmin was from Athens Research and Technology (Athens, GA, USA). Catalytically inactive mutant tPA (S478A) was obtained from Molecular Innovations Inc (Southfield, MI, USA). Platelet derived growth factor (PDGF), sodium orthovanadate, β‐glycerophosphate, sodium fluoride, protease inhibitor cocktail, diisopropylfluorophosphate (DFP), and tyrphostin AG1296 were from Sigma (St Louis, MO, USA). Goat anti‐tPA neutralising antibodies were from American Diagnostica (Greenwich, CT, USA), mouse monoclonal anti‐phospho‐p44/42 MAPK (Thr 202/Tyr 204) and rabbit anti‐phospho‐Akt (Ser 473) from Cell Signaling (Beverly, MA, USA), rabbit anti‐EGF receptor and mouse monoclonal anti‐phospho‐EGF receptor (Y1173) from Upstate (Lake Placid, NY, USA), goat anti‐actin from Santa Cruz Biotechnology (Santa Cruz, CA, USA), rabbit anti‐Ki67 from Immunotek (Marseille, France), and peroxidase coupled anti‐goat antibodies from Dako (Glostrup, Denmark). Neutralising antibodies to hb‐EGF and antibody to PDGFR were from R&D Systems (Minneapolis, MN, USA). Mouse anti‐phospho‐p38 MAPK was a kind gift from S Ramon y Cajal (Hospital Vall d'Hebrón, Barcelona, Spain) and anti‐ERK5 was kindly provided by N Gómez (Universitat Autònoma, Barcelona, Spain). The EGFR‐GFP construct was kindly provided by A Sorkin (University of Colorado).
Microarray hybridisation and analysis
cDNA arrays containing 15 360 cDNAs, of which 13 295 are known genes and 2257 control genes, were generated and processed as described.25 PANC1 cells were grown in low serum (0.1%) medium for 96 hours and treated with recombinant tPA (6 nM), EGF (3 nM) or PDGF (1 nM) for 15, 30, 60, 120, or 240 minutes. The concentration of ligands used are unsaturating with respect to each receptor in these cells.23,26,27. Each treatment and time point were done in duplicate. Dye swap hybridisations were performed with the same samples labelled with the reciprocal fluorophore. Background was subtracted from the signal, log2(signal) plotted versus log2(ratio), and a Lowess normalisation applied to adjust most spots to log ratio 0. This value was calculated for all replicates and results tabulated for signal, change (n‐fold), log ratio, standard deviation of the log ratio, and z score. Spots with an SD versus their dye swap replicate greater than 0.6 were filtered out. Normalised log2 ratios in gene expression were then used to further analyse and cluster the data.
Clustering of samples
Q‐mode factor analysis (FA) was applied on the normalised micro array data.28,29 Factor analysis uses the covariance in the transcript levels to group genes and samples, a better indicator than total variance of the occurrence of transcriptional profiles shared by different pathways.28,30,31 From the factor model, hierarchical trees were derived by means of UPGMA clustering, starting from the sample coordinates in loadings space. An estimation of the reliability of each branch was obtained by means of a jackknife bootstrap analysis, using 100 replicates and random subsets of 90% of the genes per sample. Genes were considered differentially expressed among groups when their associated two sided t test p value was below 10−5. This cut off considers a Bonferroni adjustment to take into account multiple testing.
Real time PCR
We used real time polymerase chain reaction (RT‐PCR) with SYBRGreen incorporation to determine the expression levels of selected genes. RNA was isolated from cells, and controlled for quality on a 2100 BioAnalyzer instrument (Agilent, Palo Alto, CA, USA). Total RNA, 2 μg, was reverse transcribed by priming with random hexamers at 37°C for 60 minutes, followed by RNase treatment at 37°C for 20 minutes. The resulting cDNAs were used as templates in PCR reactions with gene specific primers. Real time PCR was performed on Abi Prism 7700 instrument (Applied Biosystems, Foster City, CA, USA). Thermal cycler conditions were 95°C for 15 minutes, and 45 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. All determinations were performed in triplicate and in two independent experiments. Since the relative amplification efficiencies of target and reference samples were found to be approximately equal, the ΔΔCt method was applied to estimate relative transcript levels. Levels of RPS14 amplification were used for endogenous reference to normalise each sample Ct (threshold cycle) value and untreated cells were used as calibrators for growth factor treated cells in each case. The final results, expressed as n‐fold differences in target gene expression were calculated as follows:
nTARGET = 2–[(Ct target – Ct reference)TREATED – (Ct target – Ct reference)UNTREATED]
Western blotting, immunofluorescence, and immunoprecipitation
Western blotting, immunofluorescence, and immunoprecipitation were performed as described previously.23
Proliferation assays
For Ki67 proliferation assays, serum starved cells were incubated with the specified reagents for 6 hours, and fixed and processed for immunocytochemistry as described.23 Cells positive for nuclear Ki67 staining were scored as proliferating. At least 400 cells were counted for each condition, in at least two independent experiments. To assay the rate of DNA synthesis, cells grown in 0.1% FBS for 48 hours were exposed to factors for 16 hours and labelled with 1 μCi [3H]‐thymidine (Amersham Pharmacia Biotech)/well for 4 hours. DNA was precipitated with 5% trichloroacetic acid, washed, and incorporated radioactivity determined by scintillation counting. The effect of the addition of mitogens was expressed as a fold of incorporation of [3H]‐thymidine in cells grown at 0.1 % FBS in quadruplicate samples.
RNA interference
siRNA were synthesised by in vitro transcription using the Silencer RNA construction kit from Ambion (Austin, TX, USA) for the target sequence for EGFR (5′‐GAGCTGCCCATGAGAAAT‐3′). siRNAs for MMP‐9 were purchased from Ambion. Control (scrambled) siRNAs were synthesised corresponding to sequences that did not match any human transcripts or genes by BLAST searches in Genbank. PANC1 cells were transfected with siRNA duplexes using Lipofectamine Plus Reagent (Invitrogen). Co‐transfection with pEGFP vector (0.1 μg/well) (Clontech) monitored transfection efficiency in some experiments.
Statistics
Results are expressed as mean (SEM), and Student's t test was used for statistical analysis; p<0.05 was taken as level of significance.
Results
tPA and EGF elicit a shared transcriptional response in PANC1 pancreatic carcinoma cells
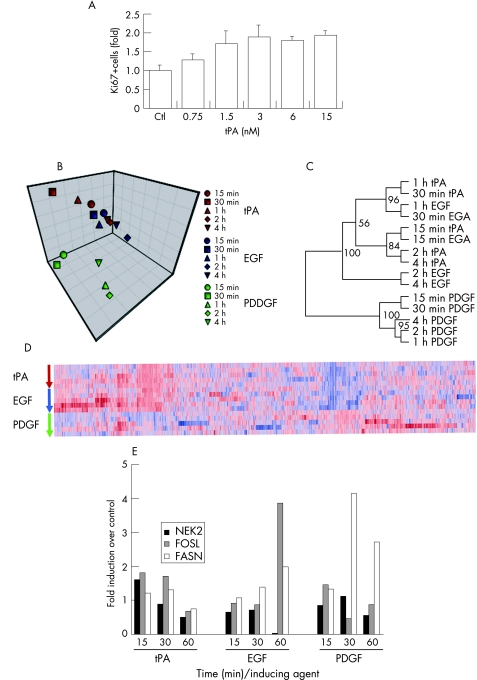
Many pancreatic carcinomas express high levels of tPA, and also EGF, PDGF, and their receptors, EGFR and PDGFR.5,32,33 In order to study the relationship of the proliferative signals induced by tPA on pancreas cancer cells with those triggered by the well studied growth factors EGF and PDGF, we performed a comparative analysis of transcriptional profiles induced by these factors.34 In a preliminary experiment, we established the optimal concentration of recombinant tPA (rtPA) in proliferation assays on serum starved PANC1 cells, which do not express the endogenous protease,6 showing that the mitogenic effect of rtPA is concentration dependent (fig 1A).
Figure 1 tPA and EGF elicit a common transcriptional response in PANC1 pancreas cancer cells. (A) PANC1 cells were serum starved for 96 hours and treated with increasing concentration of the recombinant active tPA for 6 hours. Induction of proliferation was analysed by scoring the number of cells with Ki67 positive nuclei compared to untreated serum starved control cells (Ctl). Shown is the fold induction of Ki67 positive cells for each treatment compared to untreated cells. At least 400 cells were counted for each condition in triplicate samples. Data from two independent experiments were combined. Activation of proliferation is significatively increased starting at 3 nM tPA. The error bars represent the SEM. (B–D) Serum starved PANC1 cells were treated with tPA (6 nM), EGF (3 nM), or PDGF (1 nM) for 15, 30, 60, 120 and 240 minutes. Total RNA was extracted and used for hybridisation on cDNA arrays. After normalisation, data were analysed by the factor analysis based package FADA (Materials and methods), and represented graphically (B) by using as coordinates the first three loading factors obtained for each sample (C), as a phylogenetic tree calculated from distances based on C‐index values (Materials and methods), and (D) as a heat map corresponding to normalised expression values of the genes selected by FADA as the most significant for each time series experiment. (E) Quantitative RT‐PCR to determine levels of NEK2, FOSL, and FASN in PANC1 cells after treatment with tPA, EGF, or PDGF at the indicated times, expressed as fold values over control untreated samples. Shown are average values from triplicate determinations and two independent experiments. Standard deviations for ΔΔCt values between replicate determinations were always less than 10%. Primers used were: NEK2, forward AGAACCTGAGAAACAGATGC, reverse TATTGGTCCGGTCAATAATC; FOSL, forward CAAGCATCAACACCATGAG, reverse GCTGTAGTGAGGGTAGGTCA; FASN, forward GACCTGTCTAGGTTTGATGC, reverse GCTTCATAGGTGACTTCCAG.
Next, PANC1 cells were treated with tPA (6 nM), EGF (3 nM), or PDGF (1 nM) for varying times, and transcriptional profiles analysed. The genes and the samples were clustered by applying an unsupervised approach, based on factor analysis.28,29 This analysis shows that the transcriptional responses to tPA closely group with those to EGF, and are clearly distinct from the responses to PDGF (fig 1B–D). The clustering of the transcriptional responses to tPA and EGF is particularly tight at short times after induction with either factor, followed by a divergence at later times (2 hours and 4 hours after stimulation). This is reflected by the occurrence of a large number of genes that show shared kinetics and intensity of induction in response to both tPA and EGF, compared to those genes that respond similarly to tPA and PDGF (fig 1C and supplementary table). The differential induction of several representative genes was further validated by quantitative PCR: NEK2 was predominantly induced by tPA (1.8‐fold at 15 minutes), FOSL at early times by tPA (1.7‐fold at 15 minutes) and at later times by EGF (3.8‐fold at 60 min), and FASN by PDGF (4‐fold at 30 minutes) (fig 1E). These results suggest that the signalling pathways induced by tPA in PANC1 cells significantly overlap those induced by EGF, but much less with those induced by PDGF. We thus hypothesised that tPA directly or indirectly activates one or more components of the EGF signalling pathway.
Signalling by tPA through the EGF receptor
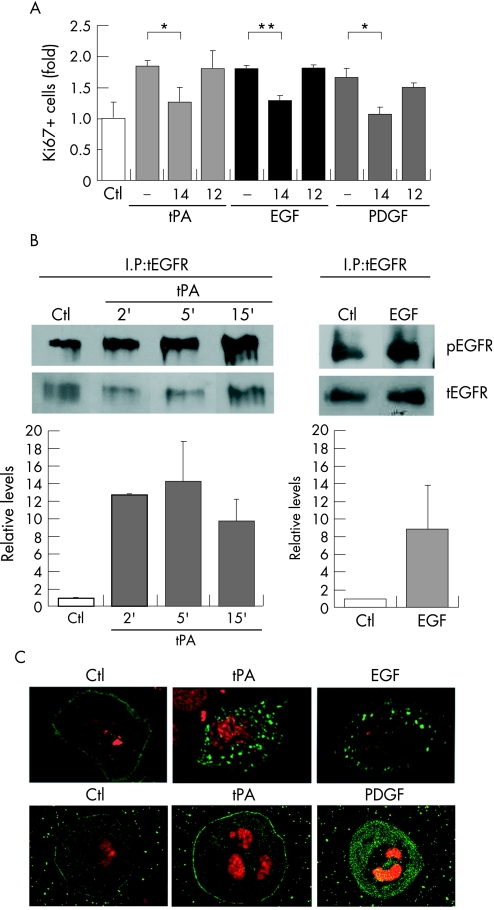
To test this hypothesis, PANC1 cells were assayed for the mitogenic activity of tPA, EGF or PDGF in the presence of selective EGFR or PDGFR inhibitors. As shown in figure 2A, all three factors were mitogenic at comparable levels for PANC1 cells, both by nuclear Ki67 reactivity assays and [3H]‐thymidine incorporation assays (not shown). The mitogenic activity of EGF was significantly reduced in the presence of the EGFR specific inhibitor AG1478, but not when pretreated with the PDGFR specific inhibitor AG1296. Correspondingly, the mitogenic activity of PDGF was greatly reduced by the PDGFR specific inhibitor AG1296, and to a lesser extent by the EGFR specific inhibitor AG1478, in agreement with the known requirement for EGFR for a full mitogenic activity of PDGFR.35 Relevant to the above hypothesis, the mitogenic activity of tPA on PANC1 cells was significantly reduced in the presence of the EGFR inhibitor AG1478, while it was unaffected by the PDGFR inhibitor AG1296 (fig 2A). This suggested that the mitogenic activity of tPA requires a catalytically active EGFR, but not PDGFR.
Figure 2 Mitogenic signalling by tPA requires active EGF receptor. (A) Mitogenic activity of tPA, EGF, and PDGF on PANC1 cells. Cells were treated with growth factors as described in figure 1, during 6 hours and analysed by scoring cells with nuclear staining for Ki67. Where indicated, cells were pre‐incubated with the EGFR specific inhibitor AG1478 (14) (2.5 μM) or the PDGFR specific inhibitor AG1296 (12) (2.5 μM) for 30 minutes before adding the growth factors, and the inhibitors maintained throughout stimulation. Shown is the fold of Ki67 positive cells for each treatment compared to untreated serum starved control cells (Ctl). At least 400 cells were counted for each condition in triplicate samples. Data from two independent experiments were combined. The error bars represent the SEM. (*p<0.03; **p<0.009). (B) Top: Induction of phosphorylation of EGFR by tPA. Cells were treated with tPA (6 nM) for 2, 5, and 15 minutes or EGF (3 nM), as a control. Lysates were immunoprecipitated with antibodies to EGFR. The phosphorylated receptor was detected by western blotting with an antibody specific for pTyr 1173 in EGFR. Bottom: The bar graphs show the average from three independent experiments where the densitometric quantitation of the signals corresponding to the phosphorylated receptor were normalised with respect to total EGFR. The error bars represent the SEM. (C) Top images: tPA stimulates EGFR internalisation. PANC1 cells were transiently transfected with GFP‐EGFR and serum starved for 48 hours before the addition of tPA (6 nM) or EGF (3 nM). Bottom images: tPA does not induce PDGFR internalisation. As a control, cells were serum starved for 48 hours and treated with tPA (6 nM) or PDGF (1 nM) for 30 minutes before staining with anti‐PDGFR antibody. Nuclei were counterstained with propidium iodide (red). Fluorescent images were obtained after 30 minutes of treatment with growth factors.
We then determined if tPA could activate EGFR in PANC1 cells. Figure 2B shows that treatment of PANC1 cells with tPA induced a rapid and transient tyrosine phosphorylation of EGFR reaching maximal levels at 5 minutes after the induction, and decreased thereafter. The amount of phosphorylated EGFR induced by tPA was comparable to that induced by EGF (fig 2B). In A431 cells, which express high numbers of EGFR molecules on their surface, tPA was also able to induce phosphorylation of EGFR with identical kinetics (see Gut website (http://gut.bmj.com/supplemental) fig 1A), although activation of this pathway by either tPA or EGF in these cells did not result in stimulation of proliferation.
We also expressed a green fluorescent protein tagged form of EGFR in PANC1 cells. This chimeric protein behaves as a fully functional receptor, and responds to binding by its ligand by endocytosis and stimulation of the EGFR pathway.36 In serum starved cells, EGFR‐GFP showed a uniform distribution on the plasma membrane, with little or no intracellular localisation (fig 2C). In contrast, after 30 minutes of exposure to either tPA or EGF, the fluorescent receptor underwent a marked intracellular redistribution, showing a similar pattern of intracellular localisation in both treatments. For EGF treated cells, this distribution has been shown to represent early endosomes.37 As control, the addition of tPA did not induce the internalisation of PDGF receptors (fig 2C). Therefore, tPA stimulates the autophosphorylation and internalisation of EGFR in PANC1 cells, with kinetics and effects closely resembling those caused by EGF.
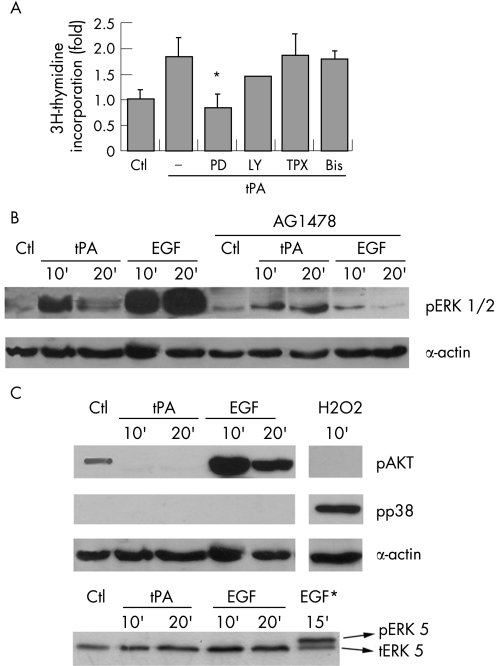
Phosphorylation and activation of the ERK1/2 is part of a cascade that relays signals triggered by activated EGFR.38 Figure 3A shows that MEK activity is required for proliferation by tPA. In contrast, the poor inhibition of the tPA mediated proliferation by LY294002, pertussis toxin, or GF109203x, indicate that PI3‐K, G proteins, and PKC activities, respectively, are not required for this activity of tPA. In addition, tPA can phosphorylate MEK (not shown) and ERK 1/2 in PANC1 and A431 cells, with maximal induction of the phosphorylated forms 10 minutes after the addition of tPA (fig 3B and Gut website (http://gut.bmj.com/supplemental) fig 1B). This induction is significantly reduced by incubation with AG1478, indicating that the phosphorylation of ERK1/2 induced by tPA is mostly dependent on the kinase activity of EGFR.
Figure 3 Active ERK1/2 are required for tPA effects on proliferation. (A) Induction of proliferation was analysed by the incorporation of [3H]‐thymidine and expressed as fold over unstimulated serum starved controls (Ctl). Serum starved PANC1 cells were treated with tPA (6 nM) in the presence of PD098059 (PD) 50 μM, LY294002 (LY) 50 μM, pertussis toxin (TPX) 100 μg/ml, and bisindolylmaleimide (Bis) 5 μM. Each treatment is the mean of quadruplicate measurements. At least three independent experiments were performed. The error bars represent the SEM. (*p<0.05). (B) tPA induces phosphorylation of ERK 1/2. Cells were serum starved and treated with tPA (6 nM) or EGF (3 nM) for the indicated times, without or with AG1478. Cell lysates (50 μg) were analysed for ERKs phosphorylation by western blotting with a phospho‐ERK specific antibody. Equal protein loading was verified by re‐analysing the membranes for actin expression levels. A representative blot of three performed is shown. (C) The ability of tPA to phosphorylate AKT (pAKT), p38 MAP kinase (pp38), or ERK5 (pERK5) was analysed by western blotting using specific antibodies to the phosphorylated proteins. The following treatments were performed as controls: H2O2 (1 mM), as a control for p38 MAP kinase phosphorylation; EGF (3 nM), as control for AKT phosphorylation; and HeLa cells treated with 3 nM EGF (EGF*), as a control for ERK5 activation.
The state of activation of other signalling kinases in response to tPA was also tested. The phosphoinositide 3′‐phosphate activated kinase AKT (PKB) was activated by EGF but not by tPA (fig 3C). Neither tPA nor EGF were able to promote the phosphorylation of the stress induced kinase p38 MAPK or ERK5 in PANC1 cells (fig 3C). These results suggest that the signalling pathways used by tPA, although significantly overlapping, are not identical to the EGF signalling pathway.
Knockdown of EGFR by RNAi prevents tPA induced mitogenesis
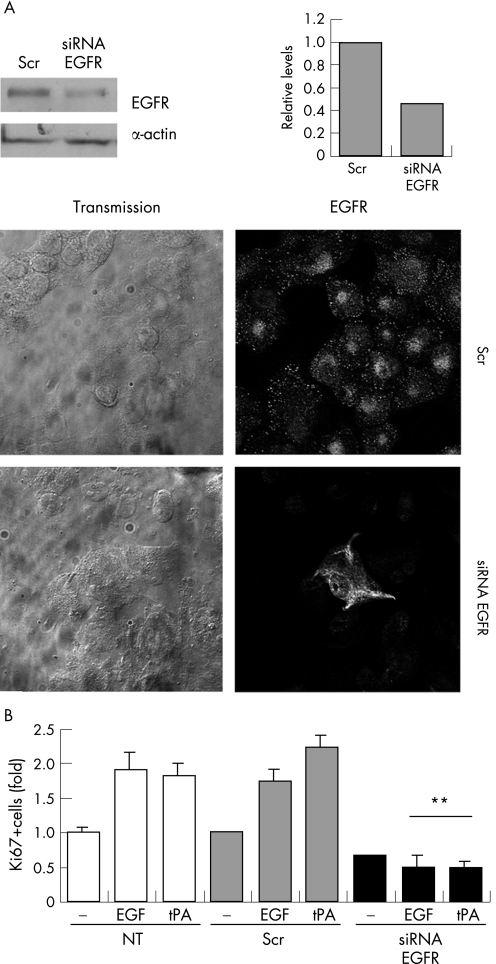
Next, PANC1 cells were depleted of EGFR by RNAi, and assayed for tPA induced proliferation (nuclear Ki67 reactivity). Seventy‐two hours after transfection of 80 nM EGFR specific siRNA duplexes, the protein levels for the receptor decreased by 60% compared with a control siRNA (fig 4A). Both tPA and EGF produced a marked mitogenic effect (Ki67 positive fraction) on cells expressing the EGFR that either were not transfected, or were transfected with a control siRNA (fig 4B). After treatment of PANC1 cells with EGFR specific siRNAs, but not with control siRNAs, the mitogenic effects of tPA and EGF were significantly reduced (fig 4B). Therefore, the presence of EGFR is required for the mitogenic effect of tPA on PANC1 cells.
Figure 4 Proliferation induced by tPA requires the expression of the EGFR protein. (A) Specific depletion of EGFR protein attained by transfecting PANC1 cells with the indicated double stranded siRNAs. Seventy‐two hours post‐transfection, EGFR levels were analysed by western blotting and quantitated by densitometric scanning of the signal (right graphs). Normalisation was done against signals for actin. As controls, non‐specific scrambled (Scr) siRNAs were transfected in parallel at the same concentrations. Bottom panels: immunocytochemistry with anti‐EGFR antibodies showing the depletion of EGFR protein by the specific siRNA compared with control scrambled siRNA (Scr). (B) Effect of the depletion of EGFR on the proliferative response to tPA. Untransfected or siRNA transfected cells were serum starved for 48 hours and then treated with tPA (6 nM) or EGF (3 nM). Cells were double immunostained for Ki67 and EGFR and the effect of RNAi was analysed by scoring the number of Ki67 positive cells in the EGFR negative population. The percentage of positive cells in each treatment with respect to untreated control cells (Ctl), is shown. For proliferation assays, the error bars represent the SEM. At least 400 cells were counted for each condition in two independent experiments (**p<0.001).
tPA induced activation of ERK 1/2 requires enzymatically active tPA, plasmin, metalloproteases, and the activity of hb‐EGF
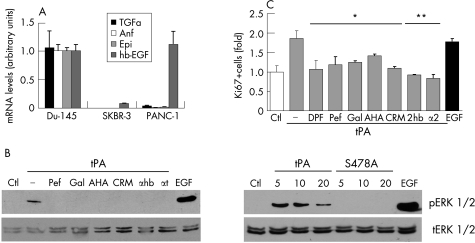
It has been reported that EGFR is transactivated by MMP dependent mechanisms,11 which involve the proteolytic release of membrane anchored EGF ligands.39 To test the possibility that tPA transactivates EGFR through the release of ligands for this receptor, we first analysed the expression of endogenous EGFR ligands in PANC1 cells by quantitative RT‐PCR. Figure 5A shows that PANC1 cells express high levels of hb‐EGF, but have low or undetectable levels of TGF‐α, amphiregulin, and epiregulin, suggesting that hb‐EGF might be responsible for the tPA mediated activation of EGFR in these cells.
Figure 5 Activation of proliferation and ERK 1/2 phosphorylation by tPA require plasmin, metalloproteases and hb‐EGF activities. (A) Expression levels for EGFR ligands TGFα, amphiregulin (Amp), epiregulin (Epi), and hb‐EGF were analysed in PANC1 cells by real time RT‐PCR. Data were normalised to actin and are expressed as mean (SEM) (bars). Du‐145 prostate cancer cells, known to secrete these ligands,64 were used as the calibrator. For comparison, SKBR‐3 breast cancer cells with very low expression of EGFR ligands are shown. Primers used were: amphiregulin, forward 5′‐ATATAGAGCACCTGGAAGCA‐3′, reverse 5′‐TGTCAATCATGCTGTGAGTT‐3′; epiregulin, forward 5′‐TTCTACAGGCAGTCCTCAGT‐3′, reverse 5′‐ACGTGGATTGTCTTCTGTCT‐3′; hb‐egf, forward 5′‐TTCCACATCATAACCTCCTC‐3′, reverse 5′‐ACCTATGACCACACAACCAT‐3′; TGF‐α, forward 5′‐CATTAAAATGGGACACCACT‐3′, reverse 5′‐TCGCTCTGGGTATTGTGT‐3′. (B) tPA proteolytic activity is required for ERKs phosphorylation. Cells were treated for 10 minutes with tPA (6 nM) or EGF (3 nM) as described in figure 1; specific inhibitors were added 30 min before stimulating with growth factors. Lysates (50 μg) were analysed by western blotting with antibodies to phosphorylated ERKs. Ctl, control untreated cells; Pef, pefabloc/tPA (30 μM); Gal, galardin (10 μM); AHA, p‐aminobenzoyl‐gly‐pro‐d‐alahydroxamic acid (1 μM); CRM, CRM 197 (5 μg/ml); αhb, neutralising antibody to hb‐EGF (10 μg/ml); αt, neutralising antibody to tPA (10 μg/ml). Activated ERK1/2 are detected with active tPA (5–20 nM), or EGF (3 nM) but not with the catalytically inactive mutant tPA (tPAS478A) (5–20 nM). Total ERK1/2 signals from the same blots are also shown. (C) Requirements for the mitogenic effects of tPA. Serum starved cells were treated with tPA (6 nM) or EGF (3 nM), without or with specific inhibitors, and analysed after 6 hours by fluorescent staining with anti‐Ki67 antibodies. At least 400 cells were scored for each condition. Results are the mean from at least two independent experiments. DFP, diisopropylfluorophosphate treated tPA (6 nM); Pef, pefabloc/tPA (30 μM); Gal, galardin (10 μM); AHA, p‐aminobenzoyl‐gly‐pro‐d‐ala‐hydroxamic acid (1 μM); CRM, CRM 197 (5 μg/ml); αhb, neutralising antibody to hb‐EGF (10 μg/ml); α2, alpha 2 antiplasmin, 2 μg/ml. Control isotype matched antibody used at the same concentration had no effects in these assays (not shown) (*p<0.04; **p<0.004).
To characterise the proteolytic cascade that leads to the release of hb‐EGF and the activation of EGFR, specific protease inhibitors were used. Blocking the proteolytic activity of tPA by pretreatment with the specific inhibitor Pefabloc/tPA, by a neutralising antibody or irreversibly with DFP (fig 5B and data not shown), markedly reduced the phosphorylation of ERK1/2 by tPA, indicating the requirement for active tPA, as also previously shown.6 These results were paralleled by those obtained in mitogenic assays. The fraction of proliferating cells (Ki67 positive) induced by tPA was significantly reduced when its proteolytic activity was inhibited by Pefabloc or DFP treatment (fig 5C). In other cell systems, the effects of tPA on cell signalling appear to also require metalloproteases activities.40,41,42 Different metalloprotease inhibitors, including galardin and the gelatinase specific inhibitor aminobenzoyl‐gly‐pro‐d‐ala‐hydroxamic acid (AHA), prevented both the activation of ERK1/2 and proliferation by tPA (fig 5B and 5C). The neutralising hb‐EGF antibody (αhb) and CRM197, a non‐toxic diphtheria toxin analogue that selectively binds to hb‐EGF and inhibits strongly the mitogenic activity of the secreted form of human hb‐EGF,43 also inhibited ERK1/2 phosphorylation and proliferation by tPA, indicating that hb‐EGF is required for the tPA promoted activation of the EGFR pathway. In addition, the plasmin specific inhibitor α2‐antiplasmin also significantly inhibited the tPA promoted proliferation of PANC1 cells (fig 5C), but not the mitogenic activity of EGF (see Gut website (http://gut.bmj.com/supplemental) fig 2). Finally, a catalytically inactive tPA mutant (tPAS478A) was unable to stimulate activation of ERK1/2, demonstrating the requirement for the proteolytic active tPA in the activation of the pathway (fig 5B). These results suggest that tPA, plasmin, metalloproteases, and hb‐EGF activities are required for the tPA promoted cell proliferation and the activation of the EGFR pathway.
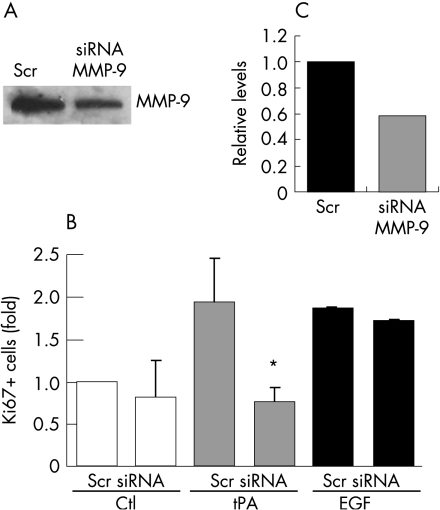
tPA induced proliferation requires MMP‐9
Plasmin has been shown to be epistatic to gelatinases and to activate directly MMP‐9 in neutrophils.41,42 Thus, we tested whether MMP‐9 is the gelatinase activated by the tPA/plasmin system, by using PANC1 cells depleted of MMP‐9 by RNAi, and assayed for tPA induced proliferation (nuclear Ki67 reactivity). Transfection of siRNA for MMP‐9 caused a specific and significant decrease (48% reduction for 80 nM siRNA) of secreted protein levels after 48 hours (fig 6A). This level of depletion of MMP‐9 in PANC1 cells was sufficient to prevent proliferation by tPA, but not by EGF (fig 6B). Transfection with control siRNA duplexes did not cause any effects on tPA induced proliferation (fig 6B). As shown in figure 2 on the Gut website (http://gut.bmj.com/supplemental), the proliferation of PANC1 cells in response to recombinant plasmin requires the activity of metalloproteases, and the proliferation induced by MMP‐9 requires active hb‐EGF. These results confirm that plasmin is epistatic to metalloproteases while MMP‐9 is epistatic to hb‐EGF in the activation of cell proliferation (see Gut website (http://gut.bmj.com/supplemental) fig 2).
Figure 6 Depletion of MMP‐9 inhibits the proliferative response to tPA. (A) Cells were transfected with pEGFP and siRNA duplexes specifically targeting MMP‐9 (siRNA, 80 nM), or control siRNA duplexes with no known targets (Scr, 80 nM). Seventy‐two hours post‐transfection, culture supernatants were concentrated, and equal amounts of proteins were analysed by western blotting. Secreted MMP‐9 levels were quantitated by densitometric scanning of the signals (C). (B) Effects of the depletion of MMP‐9 on the proliferative status of PANC1 cells. Cells were transfected with pEGFP and the specific MMP‐9 siRNA (siRNA) or a control siRNA (Scr), serum deprived for 48 hours and either not treated (Ctl) or treated with tPA (6 nM) or EGF (3 nM). Double fluorescence counting for Ki67 and GFP was used to score the number of proliferating cells in the transfected population. At least 400 cells were counted for each condition in two independent experiments. The error bars represent the SEM (*p = 0.04).
Collectively, our observations suggest that the effects of tPA on cell proliferation and signalling are mediated by the sequential activation of plasmin and MMP‐9. In addition, hb‐EGF, expressed at high levels in PANC1 cells, is also required to engage the EGFR signalling pathway.
Discussion
Tumours of the exocrine pancreas overexpress tPA and EGFR.4,5,22,32,44 tPA acts by stimulating cell proliferation in these tumours, promoting their growth and the associated angiogenesis in vitro and in vivo.6 The mechanism by which tPA promotes cell proliferation has remained elusive so far. In this study we provide evidence that tPA induces the activation of the EGFR and the transmission of the proliferative signal to the downstream ERK1/2 kinases through the proteolytically activation of plasmin, MMP‐9, and hb‐EGF. These are new findings, with potentially important therapeutic implications in pancreatic cancer.
The activation of individual biochemical pathways may induce overlapping alterations in the pattern of expressed genes, forming networks of interactions.45 However, activation of specific pathways can elicit specific transcriptional profiles, such that a given profile corresponds to a phenotypic window that may be used to infer the active pathways and the triggering signals.46,47,48 The use of time series is particularly useful in these reverse biochemistry approaches, especially when parallel comparative experiments are performed.34 Our comparative transcriptional profiling of responses to tPA, EGF, and PDGF on PANC1 pancreas cancer cells shows a significant overlap between responses to tPA and EGF, and also a clear distinction of these two responses from that elicited by PDGF. The similarities in the transcriptional responses to tPA and EGF were more evident at early times of exposure to either factor (from 15 minutes to 1 hour), followed by a relative divergence at later times (2 hours and 4 hours). This indicates that, despite the use by tPA of pathways common to EGF, additional factors modulate the response of PANC1 cells to tPA. The physiological relevance of these differences was also supported experimentally. Indeed, activation of ERKs by tPA was less vigorous compared to EGF, and AKT (PKB) was activated by EGF but not by tPA. Nevertheless, the transcriptional profiling analysis was highly suggestive of a significant overlap in the signalling pathways elicited by tPA and EGF, which was confirmed by further experiments. Firstly, chemical inactivation of the catalytic activity of EGFR, but not PDGF, blocked the tPA promoted proliferation of PANC1 cells. Secondly, tPA induced a rapid phosphorylation of EGFR, MEK and ERK1/2, with kinetics similar to the phosphorylation induced by EGF, and inhibitors of EGFR decreased this induction. Thirdly, tPA promoted specifically the endocytosis of EGFR, also with kinetics comparable to endocytosis induced by EGF. And fourthly, knockdown of EGFR with siRNA prevented the mitogenic effect of tPA on PANC1 cells. Collectively, these observations led us to conclude that tPA activates the EGF signalling pathway through EGFR.
Although tPA contains an EGF‐like domain, our results demonstrate that its catalytic activity is required for activation of the EGFR pathway, and therefore a direct activation of EGFR by tPA as a potential ligand for this receptor seems unlikely. Part of the mitogenic stimulus of some receptors can be produced by the transactivation of EGFR by other tyrosine kinase receptors (like PDGFR, IGF1R),27,49 protease receptors (uPAR, PAR‐1),11,14,50 CAM and integrins,13,51,52 or hormones acting via heterotrimeric G‐proteins.54,55 The latter is promoted by intracellular signalling components that promote a metalloprotease dependent proteolytic activation of EGFR ligands.53,54,55 Our results show that the proliferative signals generated by tPA, that activate EGFR and ERK1/2, require active tPA, the activity of plasmin, and MMP‐9. In addition, the use of mutant [Glu52]diphtheria toxin (CRM197) and of a specific neutralising antibody to hb‐EGF, indicate that active hb‐EGF is also required. Based on our results, that describe for the first time the signalling pathway for tPA in cancer cell proliferation, we suggest a mechanism by which tPA induces a rapid sequential activation of plasmin and MMP‐9. MMP‐9 activity is required for the activation of hb‐EGF that binds to the EGFR, induces its autophosphorylation,53,54 and initiates a signalling cascade with phosphorylation and activation of MEK and ERK1/2. Recently, a different pathway of transduction of the tPA signal to the nucleus has been reported for the tPA dependent activation of gene expression and protein secretion in renal interstitial fibroblasts.56 The mechanism that we propose for tPA in cancer cell proliferation is supported by several findings. Firstly, proteolytic activity of tPA and plasmin are necessary for the tPA promoted EGFR activation and proliferation: both proteases required MMPs for these actions as shown by the use of several inhibitors. Secondly, MMP‐9 expression and activity are required for the tPA promoted EGFR pathway activation and proliferation, as demonstrated by specific inhibitors and RNAi assays. Thirdly, PANC1 cells express considerably higher levels of hb‐EGF compared to other EGFR ligands, and active hb‐EGF is required for the tPA promoted EGFR pathway activation and proliferation. Additional evidence by others also lends support to the proposed mechanism. (i) Plasmin can directly activate pro‐MMP‐9 in infiltrating neutrophils41; (ii) the plasminogen/plasmin system is epistatic to MMP‐9 and MMP‐2 activation in different cell systems41,42; and (iii) MMP‐2 and MMP‐9 may mediate the transactivation of EGFR by G protein coupled receptors activation of hb‐EGF.51 In addition, MMP‐9 is strongly expressed in aggressive pancreatic cancer.57 Furthermore, serine proteases like plasmin, trypsin, or kallikrein are able to release membrane anchored pro‐EGF in vivo and in vitro.58,59
Recently, tPA has been shown to proteolyse and activate PDGF‐CC, generating an efficient PDGFR‐α ligand that induces proliferation of COS‐7 fibroblasts.60 However, the divergent transcriptional responses induced by tPA and PDGF and the lack of inhibition of the tPA proliferative signal by AG1296, argue against the involvement of this receptor in the tPA mitogenic activity on PANC1 cells.
Because human pancreatic carcinomas overexpress tPA, MMP‐9, hb‐EGF, and EGFR, autocrine loops can generate signals that stimulate cell proliferation and anchorage independent growth.4,5,6,22,32,44,61,62 We have shown that tPA provides an additional autocrine mode of activation of the EGFR pathway that leads to growth stimulation. Our observations provide additional support for targeting the EGFR in antitumour therapy in pancreatic cancer.63 Conceivably, future combination therapies including selective inhibitors or tPA blocking agents could further improve EGFR targeted therapies.
Two further figures and a table are available on Gut website (http://gut.bmj.com/supplemental)
Copyright © 2007 BMJ Publishing Group & British Society of Gastroenterology
Supplementary Material
Acknowledgements
We thank S Vilá de Muga and M Muñoz for technical help, and J Reventós for support and advices.
Abbreviations
DFP - diisopropyl fluorophosphate
DMEM - Dulbecco's Modified Eagle's Medium
EGFR - epidermal growth factor receptor
ERK 1/2 - extracellular activated kinases 1 and 2
FA - factor analysis
FBS - fetal bovine serum
hb‐EGF - heparin binding‐epidermal growth factor
MMP‐9 - metalloprotease‐9
PAs - plasminogen activators
PCR - polymerase chain reaction
PDGF - platelet derived growth factor
rtPA - recombinant tPA
siRNA - small interfering RNA duplexes
TGFα - transforming growth factor α
tPA - tissue type plasminogen activator
uPA - urokinase type plasminogen activator
Footnotes
Funding: This work was supported by grants from former Ministerio de Educación (SAF2000‐0195), Fondos de Investigación Sanitaria (PI020764), Fundació Marató‐TV3 and Hospital Vall d'Hebrón (to RP), grant GEN2001‐4856‐C13‐07 and GEN2001‐4856‐C13‐10 from the former Ministerio de Ciencias y Tecnología (to TMT and ARO, respectively). MH was a recipient of a fellowship from the Institut de Recerca Hospital Vall d'Hebrón.
Competing interests: none.
Two further figures and a table are available on Gut website (http://gut.bmj.com/supplemental)
References
- 1.Lockhart A C, Rothenberg M, Berlin J D. Treatment for pancreatic cancer: current therapy and continued progress. Gastroenterology 20051281642–1654. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P, Schoonjans L, Kieckens L.et al Physiological consequences of loss of plasminogen activator gene function in mice. Nature 1994368419–424. [DOI] [PubMed] [Google Scholar]
- 3.Duffy M J. The urokinase plasminogen activator system: role in malignancy. Curr Pharm Des 20041039–49. [DOI] [PubMed] [Google Scholar]
- 4.Paciucci R, Berrozpe G, Tora M.et al Isolation of tissue‐type plasminogen activator, cathepsin H, and non‐specific cross‐reacting antigen from SK‐PC‐1 pancreas cancer cells using subtractive hybridization. FEBS Lett 199638572–76. [DOI] [PubMed] [Google Scholar]
- 5.Paciucci R, Tora M, Díaz V M.et al The plasminogen activator system in pancreas cancer: role of tPA in the invasive potential in vitro. Oncogene 199816625–633. [DOI] [PubMed] [Google Scholar]
- 6.Díaz V M, Planagumá J, Thomson T M.et al Tissue plasminogen activator is required for the growth, invasion, and angiogenesis of pancreatic tumor cells. Gastroenterology 2002122806–819. [DOI] [PubMed] [Google Scholar]
- 7.Stack M S, Gray R D, Pizzo S V. Modulation of murine B16F10 melanoma plasminogen activator production by a synthetic peptide derived from the laminin A chain. Cancer Res 1993531998–2004. [PubMed] [Google Scholar]
- 8.Jessani N, Humphrey M, McDonald W H.et al Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci USA 200410113756–13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei Y, Lukashev M, Simon D I.et al Regulation of integrin function by the urokinase receptor. Science 19962731551–1555. [DOI] [PubMed] [Google Scholar]
- 10.Resnati M, Pallavicini I, Wang J M.et al The fibrinolytic receptor for urokinase activates the G protein‐coupled chemotactic receptor FPRL1/LXA4R. Proc Natl Acad Sci USA 2002991359–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu D, Aguirre Ghiso J, Estrada Y.et al EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer Cell 20021445–457. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen D H, Hussaini I M, Gonias S L. Binding of urokinase‐type plasminogen activator to its receptor in MCF‐7 cells activates extracellular signal‐regulated kinase 1 and 2 which is required for increased cellular motility. J Biol Chem 19982738502–8507. [DOI] [PubMed] [Google Scholar]
- 13.Aguirre Ghiso J A, Kovalski K, Ossowski L. Tumor dormancy induced by downregulation of urokinase receptor in human carcinoma involves integrin and MAPK signaling. J Cell Biol 199914789–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jo M, Thomas K S, Marozkina N.et al Dynamic assembly of the urokinase‐type plasminogen activator signaling receptor complex determines the mitogenic activity of urokinase‐type plasminogen activator. J Biol Chem 200528017449–17457. [DOI] [PubMed] [Google Scholar]
- 15.Akassoglou K, Kombrinck K W, Degen J L.et al Tissue plasminogen activator‐mediated fibrinolysis protects against axonal degeneration and demyelination after sciatic nerve injury. J Cell Biol 20001491157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicole O, Docagne F, Ali C.et al The proteolytic activity of tissue‐plasminogen activator enhances NMDA receptor‐mediated signaling. Nat Med 2001759–64. [DOI] [PubMed] [Google Scholar]
- 17.Pawlak R, Rao B S, Melchor J P.et al Tissue plasminogen activator and plasminogen mediate stress‐induced decline of neuronal and cognitive functions in the mouse hippocampus. Proc Natl Acad Sci USA 200510218201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yepes M, Sandkvist M, Coleman T A.et al Regulation of seizure spreading by neuroserpin and tissue‐type plasminogen activator is plasminogen‐independent. J Clin Invest 20021091571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matys T, Pawlak R, Matys E.et al Tissue plasminogen activator promotes the effects of corticotropin‐releasing factor on the amygdala and anxiety‐like behavior. Proc Natl Acad Sci USA 200410116345–16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medina M G, Ledesma M D, Domínguez J E.et al Tissue plasminogen activator mediates amyloid‐induced neurotoxicity via ERK1/2 activation. EMBO J 2005241706–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendelsohn J, Baselga J. Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol 2003212787–2799. [DOI] [PubMed] [Google Scholar]
- 22.Yamanaka Y, Friess H, Kobrin M S, Buchler M, Beger H G, Korc M. Coexpression of epidermal growth factor receptor and ligands in human pancreatic cancer is associated with enhanced tumor aggressiveness. Anticancer Res 199313565–569. [PubMed] [Google Scholar]
- 23.Díaz V M, Hurtado M, Thomson T.et al Specific interaction of tissue‐type plasminogen activator (tPA) with annexin II on the membrane of pancreatic cancer cells activates plasminogen and promotes invasion in vitro. Gut 200453993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kassam G, Choi K S, Ghuman J.et al The role of annexin II tetramer in the activation of plasminogen. J Biol Chem 19982734790–4799. [DOI] [PubMed] [Google Scholar]
- 25.Guerra S, López‐Fernandez L A, Pascual‐Montano A.et al Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J Virol 2003776493–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korc M, Magun B E. Recycling of epidermal growth factor in a human pancreatic carcinoma cell line. Proc Natl Acad Sci USA 1985826172–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowen‐Pope D F, Ross R. Platelet‐derived growth factor. II. Specific binding to cultured cells. J Biol Chem 19822575161–5171. [PubMed] [Google Scholar]
- 28.Lozano J J, Soler M, Bermudo R.et al Dual activation of pathways regulated by steroid receptors and peptide growth factors in primary prostate cancer revealed by Factor Analysis of microarray data. BMC Genomics 20056109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez‐Carbayo M, Socci N D, Lozano J J.et al Gene discovery in bladder cancer progression using cDNA microarrays. Am J Pathol 2003163505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liebermeister W. Linear Modes of gene expression determined by independent component analysis. Bioinformatics 20021851–60. [DOI] [PubMed] [Google Scholar]
- 31.Lee S I, Batzoglou S. Application of independent component analysis to microarrays. Genome Biol 20034R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korc M, Chandrasekar B, Yamanaka Y.et al Overexpression of the epidermal growth factor receptor in human pancreatic cancer is associated with concomitant increases in the levels of epidermal growth factor and transforming growth factor alpha. J Clin Invest 1992901352–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebert M, Yokoyama M, Friess H.et al Induction of platelet‐derived growth factor A and B chains and over‐expression of their receptors in human pancreatic cancer. Int J Cancer 199562529–535. [DOI] [PubMed] [Google Scholar]
- 34.Fambrough D, McClure K, Kazlauskas A.et al Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell 199997727–741. [DOI] [PubMed] [Google Scholar]
- 35.Saito Y, Haendeler J, Hojo Y.et al Receptor heterodimerization: essential mechanism for platelet‐derived growth factor‐induced epidermal growth factor receptor transactivation. Mol Cell Biol 2001216387–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter R E, Sorkin A. Endocytosis of functional epidermal growth factor receptor‐green fluorescent protein chimera. J Biol Chem 199827335000–35007. [DOI] [PubMed] [Google Scholar]
- 37.Hunyady L, Baukal A J, Gaborik Z.et al Differential PI 3‐kinase dependence of early and late phases of recycling of the internalized AT1 angiotensin receptor. J Cell Biol 20021571211–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fantl W J, Johnson D E, Williams L T. Signalling by receptor tyrosine kinases. Annu Rev Biochem 199362453–481. [DOI] [PubMed] [Google Scholar]
- 39.Peschon J J, Slack J L, Reddy P.et al An essential role for ectodomain shedding in mammalian development. Science 19982821281–1284. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Lee S R, Arai K.et al Lipoprotein receptor‐mediated induction of matrix metalloproteinase by tissue plasminogen activator. Nat Med 200391313–1317. [DOI] [PubMed] [Google Scholar]
- 41.Zhao B Q, Ikeda Y, Ihara H.et al Essential role of endogenous tissue plasminogen activator through matrix metalloproteinase 9 induction and expression on heparin‐produced cerebral hemorrhage after cerebral ischemia in mice. Blood 20041032610–2616. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z, Li N, Díaz L A.et al Synergy between a plasminogen cascade and MMP‐9 in autoimmune disease. J Clin Invest 2005115879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitamura T, Higashiyama S, Taniguchi N.et al Diphtheria toxin binds to the epidermal growth factor (EGF)‐like domain of human heparin‐binding EGF‐like growth factor/diphtheria toxin receptor and inhibits specifically its mitogenic activity. J Biol Chem 19952701015–1019. [DOI] [PubMed] [Google Scholar]
- 44.Lemoine N R, Hughes C M, Barton C M.et al The epidermal growth factor receptor in human pancreatic cancer. J Pathol 19921667–12. [DOI] [PubMed] [Google Scholar]
- 45.Pawson T, Saxton T M. Signaling networks—do all roads lead to the same genes? Cell 199997675–678. [DOI] [PubMed] [Google Scholar]
- 46.Hughes T R, Marton M J, Jones A R.et al Functional discovery via a compendium of expression profiles. Cell 2000102109–126. [DOI] [PubMed] [Google Scholar]
- 47.Slonim D K. From patterns to pathways: gene expression data analysis comes of age. Nat Genet 200232(Suppl)502–508. [DOI] [PubMed] [Google Scholar]
- 48.Jenner R G, Young R A. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol 20053281–294. [DOI] [PubMed] [Google Scholar]
- 49.El‐Shewy H M, Kelly F L, Barki‐Harrington L.et al Ectodomain shedding‐dependent transactivation of epidermal growth factor receptors in response to insulin‐like growth factor type I. Mol Endocrinol 2004112727–2739. [DOI] [PubMed] [Google Scholar]
- 50.Darmoul D, Gratio V, Devaud H, Peiretti F, Laburthe M. Activation of proteinase‐activated receptor 1 promotes human colon cancer cell proliferation through epidermal growth factor receptor transactivation. Mol Cancer Res 20042514–522. [PubMed] [Google Scholar]
- 51.Islam R, Kristiansen L V, Romani S.et al Activation of EGF receptor kinase by L1‐mediated homophilic cell interactions. Mol Biol Cell 2004152003–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sieg D J, Christof R. Hauck CR, Ilic D, et al. FAK integrates growth‐factor and integrin signals to promote cell migration. Nature Cell Biol 20002249–256. [DOI] [PubMed] [Google Scholar]
- 53.Roelle S, Grosse R, Aigner A.et al Matrix metalloproteinases 2 and 9 mediate epidermal growth factor receptor transactivation by gonadotropin‐releasing hormone. J Biol Chem 200327847307–47318. [DOI] [PubMed] [Google Scholar]
- 54.Prenzel N, Zwick E, Daub H.et al EGF receptor transactivation by G‐protein‐coupled receptors requires metalloproteinase cleavage of pro hb‐EGF. Nature 1999402884–888. [DOI] [PubMed] [Google Scholar]
- 55.Luttrell L M, Della Rocca G J, van Biesen T.et al G‐betagamma subunits mediate Src‐dependent phosphorylation of the epidermal growth factor receptor. A scaffold for G proteincoupled receptor‐mediated Ras activation. J Biol Chem 19972724637–4644. [DOI] [PubMed] [Google Scholar]
- 56.Hu K, Yang J, Tanaka S.et al Tissue‐type plasminogen activator acts as a cytokine that triggers intracellular signal transduction and induces matrix metalloproteinase‐9 gene expression. J Biol Chem 20062812120–2127. [DOI] [PubMed] [Google Scholar]
- 57.Takada M, Hirata K, Ajiki T.et al Expression of receptor for advanced glycation end products (RAGE) and MMP‐9 in human pancreatic cancer cells. Hepatogastroenterology 200451928–930. [PubMed] [Google Scholar]
- 58.Jorgensen E, Nexo E, Poulsen S S. The membrane fraction of homogenized rat kidney contains an enzyme that releases epidermal growth factor from the kidney membranes. Biochem Biophys Acta 19911074284–288. [DOI] [PubMed] [Google Scholar]
- 59.Schaudies R P, Johnson J P. Increased soluble EGF after ischemia is accompanied by a decrease in membrane‐associated precursors. Am J Physiol 1993264F523–F531. [DOI] [PubMed] [Google Scholar]
- 60.Fredriksson L, Li H, Fieber C.et al Tissue plasminogen activator is a potent activator of PDGF‐CC. EMBO J 2004233793–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith J, Dryneck R, Korc M. Production of transforming growth factor α in human pancreatic cancer cells: evidence for a superagonist autocrine cycle. Proc Natl Acad Sci USA 1987847567–7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kobrin M S, Funatomi H, Friess H.et al Induction and expression of heparin‐binding EGF‐like growth factor in human pancreatic cancer. Biochem Biophys Res Commun 19942021705–1709. [DOI] [PubMed] [Google Scholar]
- 63.Yokoi K, Sasaki T, Bucana C D.et al Simultaneous inhibition of EGFR, VEGFR, and platelet‐derived growth factor receptor signaling combined with gemcitabine produces therapy of human pancreatic carcinoma and prolongs survival in an orthotopic nude mouse model. Cancer Res 20056510371–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torring N, Jorgensen P E, Sorensen B S, Nexo E. Increased expression of heparin binding EGF (hb‐EGF), amphiregulin, TGF alpha and epiregulin in androgen‐independent prostate cancer cell lines. Anticancer Res 20002091–95. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.