Abstract
Background
Changes in epithelial cell interactions have been implicated in carcinogenesis, tumour invasion and metastasis.
Aim
To screen for altered expression of epithelial adhesion genes in lung cancer development.
Methods
Gene expression profiles were assessed with cDNA expression arrays in eight non‐small cell lung cancer (NSCLC) and eight normal bronchi obtained from the same patient. Immunohistochemistry (IHC) and RNA in situ hybridisation (ISH) were used to confirm the most prominently expressed adhesion molecules and to investigate their distribution at protein and mRNA levels.
Results
43 differentially expressed cancer‐related genes were identified in adenocarcinoma, squamous cell carcinoma (SCC) and normal bronchus. Five of these genes are related to epithelial adhesion—that is, integrin α3 (ITGA3), integrin β4 (ITGB4), desmoplakin I and II (DSP), plakoglobin, and desmocollin 3 (DSC3). ITGA3 and ITGB4, showing predominantly cell–matrix staining, were up regulated in adenocarcinoma and SCC, respectively. ITGB4 also showed strong staining in SCC with IHC and ISH. Components of the desmosome adhesion complex DSP, plakoglobin and DSC3 were strongly up regulated in SCC and showed a distinct cell–cell staining pattern. DSP and plakoglobin were predominantly present at central, more differentiated tumour cells, whereas DSC3 showed a stronger staining in the peripheral basal cells of SCC tumour areas.
Conclusions
Lack of cellular adhesion may have an important role in the metastatic potency of a primary tumour. A possible association of strong presence and normal‐distributed desmosomal molecules in SCC with the less frequent and late pattern of metastasis in SCC as compared with adenocarcinoma is suggested.
Non‐small cell lung cancer (NSCLC), which constitutes 80% of all lung cancers, encompasses several distinct subtypes, including the most common types of adenocarcinoma and squamous cell carcinoma (SCC). Gene expression profiles may help to better understand which pathogenetic mechanisms are associated with the development of lung cancer. Gene expression profiling has been applied to identify genes that are related to specific histological subtypes of lung cancer.1,2 In addition, several studies have focused on the relationship of gene expression profiles with clinical characteristics such as prediction of survival,1,3,4,5 chemosensitivity6,7 and metastatic behaviour,6,8,9 or to cellular processes such as apoptosis10 and cell cycle.11
Our aim was to compare gene expression profiles in patients with adenocarcinoma and SCC in comparison with normal bronchus obtained from the same patient. We have focused on expression of epithelial cell–cell and cell–matrix adhesion molecules, as they are known to correlate with tumour progression. We validated the expression levels and investigated the distribution of protein and mRNA from most prominently expressed epithelium‐related adhesion genes by immunohistochemistry (IHC) and RNA in situ hybridisation (ISH).
Methods
Tissue specimens
Tumour and normal bronchus tissue were obtained from well‐defined patients without chemotherapy and a smoking history of at least 45 packyears (table 112). Tissues were obtained from four patients with adenocarcinoma and four patients with SCC during surgery. The tissue was partially snap frozen for cDNA array and IHC, and partially fixed and embedded in paraffin wax for ISH. The tissues were stained with haematoxylin and eosin for histological confirmation of the diagnosis. Patients were only included when the bronchus showed normal bronchus epithelium without metaplasia or dysplasia. The study was approved by the local medical ethics committee of the University Medical Center Groningen (Groningen, The Netherlands) and written informed consent was obtained from all patients.
Table 1 Description of clinical features of patients with adenocarcinoma (AC) and squamous cell carcinoma (SCC), without prior treatment.
| Patient no | Description (age, sex) | Location | Diagnosis | Packyears |
|---|---|---|---|---|
| 1 | 70 years, M | LUL | AC, T1N1M0 | 55 |
| Moderately differentiated | ||||
| 2 | 68 years, F | RLL | AC, T2N1M0 | 53 |
| Moderately differentiated | ||||
| 3 | 71 years, M | RUL | AC, T2N2M0 | 51 |
| Poorly differentiated | ||||
| 4 | 74 years, F | RUL | AC, T2N0M0 | 56 |
| Poorly differentiated | ||||
| 5 | 75 years, M | LUL | SCC, T1N0M0 | 62 |
| Moderately differentiated | ||||
| 6 | 72 years, M | LL | SCC, T2N1M0 | 52 |
| Poorly differentiated | ||||
| 7 | 66 years, M | LUL | SCC, T3N0M0 | 50 |
| Well differentiated | ||||
| 8 | 73 years, M | LUL | SCC, T2N1M0 | 49 |
| Poorly differentiated |
F, female; LL, left lung; LUL, left upper lobe; M, male; RLL, right lower lobe; RUL, right upper lobe.
Tumour–node–metastasis staging according to Mountain et al.12
cDNA expression array
Total RNA was isolated from frozen tissue sections using Strataprep total RNA miniprep kit (Stratagene, La Jolla, California, USA), according to the manufacturer's instructions, including DNase treatment.
Atlas Human Cancer 1.2 Arrays (Clontech, Palo Alto, California, USA) were hybridised according to the manufacturer's instructions. Briefly, 2–5 μg of RNA was converted into cDNA and labelled with [α‐33P]dATP (Amersham Biosciences, 3000 Ci/mmol, 10 mCi/ml, Piscataway, New Jersey, USA), purified and hybridised overnight at 68°C. Filters were washed and exposed to a phosphorimaging screen for 1–4 days and scanned with Storm (Amersham Biosciences).
Data analysis
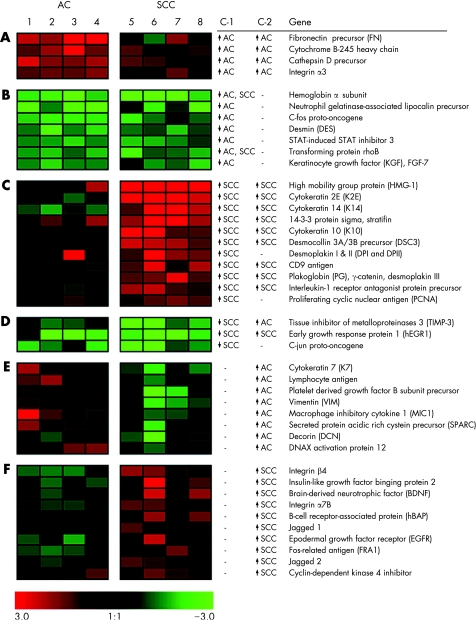
Images were analysed using Atlas Image software V.2.0 (Clontech). Natural logarithm (ln) was taken to improve a normal distribution of data. Data of the hybridisations were standardised to mean = 0 and standard deviation (SD) = 1. Subsequently, the principal components were calculated and the first principal component was deducted. Two independent selection criteria were applied to identify differentially expressed genes (fig 1).
Figure 1 Overview of differentially expressed genes in normal bronchus, adenocarcinoma (AC) and squamous cell carcinoma (SCC). Forty three gene names are listed at the right. The most differentially expressed genes are ranked per group from top to bottom. Selection criterion 1 (c‐1) was based on identifying differentially expressed genes in tumour versus normal bronchus in each patient. Ratios were determined by subtracting the standardised ln values of normal bronchus tissue from those of the tumour tissue from the same patient (T:B ratio). A consistent T:B ratio of >absolute 1 (which means a 2.3‐fold up regulation or down regulation) for each of the four individual samples per tumour type, was considered relevant. Selection criterion 2 (c‐2) was based on identifying differentially expressed genes between AC and SCC by using the sum of four standardised ln values of all AC (AC‐sum) or SCC (SCC‐sum) samples. Differences were considered relevant when the AC‐sum or SCC‐sum is >absolute 3 and the difference between the sums is at least 4. The brightness of colour correlates with the degree of “normalised expression difference” as shown at the bottom of image, where green represents low and red represents high level of expression. Groups A–D represent the T:B ratios used for c‐1, and groups E and F the tumour values used for c‐2. (A) Genes up regulated in AC in comparison with normal bronchus (c‐1) and SCC (c‐2). (B) Genes down regulated in AC in comparison with normal bronchus (c‐1). Two of the genes were also down regulated in SCC. (C) Genes up regulated in SCC in comparison with normal bronchus (c‐1) and AC (c‐2) except for two genes. (D) Genes down regulated in SCC in comparison with normal bronchus and in one case also in comparison with AC, whereas another case is down regulated in comparison with AC. (E) Genes with higher expression in AC in comparison with SCC. (F) Genes with higher expression in SCC in comparison with AC. c‐1, selection criterion 1; c‐2, selection criterion 2 (see Material and methods). ln, natural logarithm; ↑, up regulation; ↓ down regulation.
Immunohistochemistry (IHC)
Mouse monoclonal antibodies directed against ITGA3 (1:1000, Telios, San Diego, California, USA), ITGB4 (1: 500 Telios), DSP (1:200, Chemicon, Temecula, California, USA), plakoglobin (γ‐catenin, 1:1000, BD‐Biosciences, San Jose, California, USA) and DSC3 (undiluted, Research Diagnostics, Flanders, New Jersey, USA) were used to stain the tissue sections according to standard laboratory procedures. Frozen sections were fixed in acetone for 10 min and incubated with the primary monoclonal antibodies for 1 h. Endogenous peroxidase activity was blocked with 0.075% hydrogen peroxide for 1 h. Positive staining was visualised after the second and third steps using peroxidase rabbit‐α‐mouse and goat‐α‐rabbit antibodies (Dako, Glostrup, Denmark) using 3‐amino‐9‐ethyl‐carbazole (AEC).
RNA in situ hybridisation
Total RNA of SCC was primed with oligo (dT) for cDNA synthesis according to the manufacturer's instructions (Invitrogen, Carlsbad, California, USA). Primers were selected from sequence files of the corresponding genes to obtain polymerase chain reaction (PCR) products of about 500 bp (table 2). After amplification, the PCR products were subcloned in the pCRII‐TOPO vector (Invitrogen) according to the manufacturer's instructions. SP6 and T7 RNA polymerases were used to synthesise anti‐sense and sense dioxigenin (DIG)‐labelled RNA probes using the DIG RNA labelling kit (Sp6/T7; Roche, Basel, Switzerland). The sense probe was used as a negative control in the in situ hybridisation (ISH) experiments.
Table 2 Primer sequences and Genbank accession number used for amplification of integrin α3, integrin β4, desmoplakin I and II, plakoglobin and desmocollin 3.
| Gene | Forward primer 5′‐3′ | Reverse primer 5′‐3′ | Genbank reference | PCR product (bp) |
|---|---|---|---|---|
| Integrin α3 | TCCTGCCTAAGGTTGTCTACG | TTGGAACTAGCCTCAGCTTCTC | NM_002204 | 546 |
| Integrin β4 | CTGTGAGATGGCCCAAGGAG | GGGTGCAAGGATGGAGTAGC | NM_000213 | 528 |
| Desmoplakin I and II | CCTGCACAGGTGGCATCATC | CCGGAGCCGAAGACATGTTG | NM_004415 | 507 |
| Plakoglobin | CAGGTGCGGTTCCTCATCTG | AAGCACCCTGGAGAGAGAAGC | NM_002230 | 531 |
| Desmocollin 3 | ACTGTGGAGCCTCACCAAAG | CCGGCAGGATTCCAAGGTCT | NM_024423 | 517 |
PCR, polymerase chain reaction.
ISH was carried out on paraffin wax sections using standard laboratory protocols. Deparaffinised tissue sections were treated with 5, 10, 15 and 20 μg/ml proteinase K (Roche) at 37°C for 1 h and washed with Tris‐buffered saline. Slides were incubated with 1 ng/μl DIG‐labelled probe in a hybridisation solution consisting of 5× Denhardt's solution, 2× SSC, 10% dextran, 30% formamide, 1 mg/ml t‐RNA and 2 mg/ml fish sperm DNA overnight at 55°C. Slides were washed and treated for 30 min with 10 mg/ml RNase at 37°C. Anti‐DIG‐labelled alkaline phosphatase (Roche) in 0.1 M maleic acid buffer containing 0.15 M sodium chloride, 2% blocking buffer and 0.1% Triton X‐100, was incubated on the slides for 1 h at room temperature. Positive cells were visualised by overnight staining with nitroblue tetrazolium and BCIP (Roche) in TBS (pH 9.0) containing 50 mM magnesium chloride.
Results
Gene expression in normal bronchus, adenocarcinoma and SCC
Comparison of tumour tissue with normal bronchus (c‐1) and of adenocarcinoma with SCC (c‐2) showed 43 differentially expressed genes, including 5 epithelium‐related adhesion genes (fig 1). Two were integrins, ITGA3 being up regulated in adenocarcinoma in comparison with normal bronchus and SCC, and ITGB4 being up regulated in SCC compared with adenocarcinoma. The other three genes, all components of the desmosome adhesion complex, DSP, plakoglobin and DSC3, were up regulated in SCC in comparison with normal bronchus. Plakoglobin and DSC3 were also up regulated when compared with adenocarcinoma.
Distribution of integrins
Immunohistochemistry
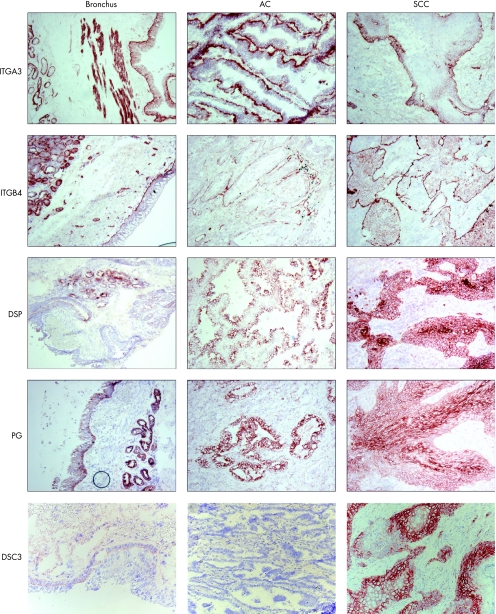
Strong cell–matrix and weak cell–cell staining of ITGA3‐protein was observed in normal bronchial epithelium, and additionally in bronchial glands, muscular layer and small blood vessels (fig 2). In adenocarcinoma and SCC, strong cell–matrix and weak cell–cell staining of ITGA3 was observed at the borders of tumour areas. ITGB4 showed a staining pattern comparable with ITGA3 staining, except in the bronchial muscular layer, where no ITGB4 was detected. Additionally ITGB4 was more restricted to basal cell–matrix borders in the normal bronchus and in adenocarcinomas. In SCC tumour cells, ITGB4 staining was stronger compared with bronchial epithelium, whereas in adenocarcinoma tumour cells, staining was less prominent than in bronchial epithelium.
Figure 2 Immunohistochemistry on normal bronchus, adenocarcinoma (AC) and squamous cell carcinoma (SCC). Integrin α3 (ITGA3) and integrin β4 (ITGB4) show predominantly cell–matrix staining. The desmosomal proteins desmoplakin I and II (DSP) and plakoglobin (PG) show cell–cell staining, with strongest staining between more central localised cells in SCC. Desmocollin 3 (DSC3) shows strong basal staining in SCC.
RNA‐in situ hybridisation
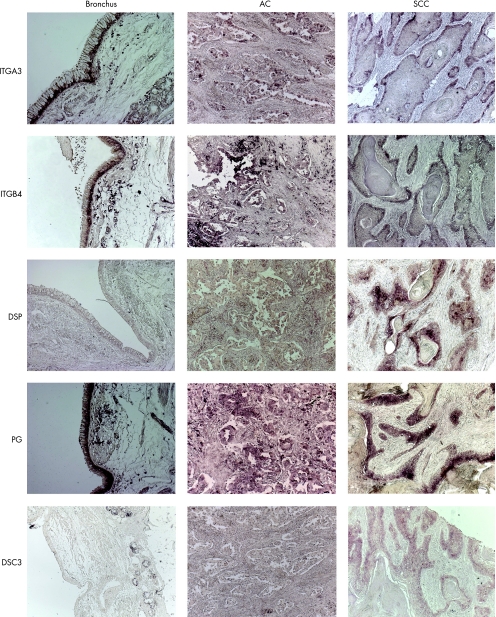
ISH showed a positive staining for ITGA3 and ITGB4 in the basal cells of the normal bronchial epithelium (fig 3). Adenocarcinoma tumour cells and predominantly basal cells of SCC tumour areas showed a positive staining for ITGA3 and ITGB4. Overall, ISH showed a staining pattern comparable to IHC for ITGA3 and ITGB4.
Figure 3 RNA in situ hybridisation staining of normal bronchus, adenocarcinoma (AC) and squamous cell carcinoma (SCC). Integrin α3 (ITGA3) and integrin β4 (ITGB4) show a predominant staining in basal cells. The desmosomal genes desmoplakin I and II (DSP) and plakoglobin (PG) show evident staining of SCC tumour cells, most pronounced in central localised cells of the tumour areas in SCC. For desmocollin 3 (DSC3) strong basal staining was observed in SCC.
Distribution of desmosomal adhesion molecules
Immunohistochemistry
In the normal bronchus, DSP showed weak staining in bronchial epithelial cells, at the basolateral borders of suprabasal cells and at the apical side of ciliated cells. Strong positive staining was observed in epithelial cells of bronchial glands (fig 2). In adenocarcinoma and SCC, positive cell–cell staining was clearly visible at the borders between tumour cells. The strongest staining was shown in SCC, especially between tumour cells in the centre of SCC tumour areas.
Plakoglobin showed strong staining in the normal bronchus at apical and lateral borders of the basal bronchial epithelial cells and at cells of the bronchial glands. Adenocarcinoma and SCC showed strong tumour cell–cell staining. In SCC, cell–cell staining of tumour cells located in the centre of tumour areas was stronger than the surrounding basal tumour cells. Often, plakoglobin also showed cytoplasmic staining in bronchial epithelial and tumour cells.
DSC3 showed weak staining in some normal bronchi, and if so, only at apical borders of basal bronchial epithelial cells. DSC3 was not detected in adenocarcinoma. By contrast, SCC showed strong cell–cell staining of tumour cells. The strongest staining was observed in the basal cells of SCC.
RNA‐in situ hybridisation
ISH for DSP showed a strong staining in SCC especially in the central SCC tumour areas (fig 3). Plakoglobin showed a similar pattern in SCC. Plakoglobin staining in the normal bronchus and adenocarcinoma was less apparent compared with SCC. DSC3 staining was only visible in bronchial glands and not in bronchial epithelial cells of normal bronchus. Clear DSC3 staining was shown in SCC, with the strongest intensity in basal tumour cells, whereas no staining was observed in adenocarcinoma. Overall, ISH showed a pattern comparable to IHC for DSP, plakoglobin and DSC3.
Discussion
Epithelial adhesion is a fundamental pathway in tissue architecture and epithelial differentiation. Changes in epithelial cell–cell and cell–matrix interactions have been implicated in carcinogenesis, tumour invasion and metastasis. Thus, there is a putative role of these adhesion genes in cancer susceptibility and metastasis. This study explored differential expression of cancer‐related, inclusive adhesion‐related, genes in NSCLC and normal bronchi from patients with a long smoking history. We identified 43 differentially expressed genes representing in part key regulators of biological functions including five epithelium‐related adhesion genes, two subunits of integrin receptors, ITGA3 and ITGB4, and three genes involved in the desmosome complex, DSP, plakoglobin and DSC3. These genes were further investigated with IHC and RNA ISH and showed consistent results for all three techniques for three of the five genes (table 3).
Table 3 Overview of the results of cDNA expression array (Array), immunohistochemistry (IHC) and RNA in situ hybridisation (ISH).
| Gene | Tissue | Array | IHC | ISH |
|---|---|---|---|---|
| Integrin α3 | Bronchus | w | s | s |
| AC | s | s | s | |
| SCC | m | s | s | |
| Integrin β4* | Bronchus | m | m | m |
| AC | w | w | w | |
| SCC | s | s | s | |
| Desmoplakin I and II* | Bronchus | w | w | w |
| AC | m | m | w | |
| SCC | s | s | s | |
| Plakoglobin | Bronchus | w | s | m |
| AC | w | s | w | |
| SCC | s | s | s | |
| Desmocollin 3* | Bronchus | w | w | w |
| AC | w | w | w | |
| SCC | s | s | m |
AC, adenocarcinoma; IHC, immunohistochemistry; ISH, in situ hybridisation; m, moderate expression or staining; s, strong expression or staining; SCC, squamous cell carcinoma; w, weak, or no expression or staining.
*Genes with consistent expression levels using the three different techniques.
The advantage of this study is that representative normal bronchus tissue were selected, containing the putative lung cancer precursor cells. We analysed both normal bronchus and NSCLC tissue from the same patient; moreover here, it is assumed that the greatest source of variation in gene expression profiles originates from interindividual biological variation.13 Another advantage of this approach is that genetic variance due to smoke exposure, which indeed can induce changes in the resulting gene expression profiles, will be excluded.14,15
Integrins are a family of heterodimeric transmembrane glycoproteins, consisting of α and β subunits, which serve primarily as cell–matrix adhesion molecules but which can also be involved in cell–cell adhesion. Integrins have an important role in the regulation of cellular morphology, differentiation and proliferation. Of the seven existing integrins expressed on airway epithelial cells of healthy people, integrins α3β1 and α6β4 recognise components of the basement membrane (laminins 5, 10 and 11) and function therefore as true cell–matrix adhesion molecules.16 Remarkably, integrin subunits α3 and β4 were up regulated in our adenocarcinoma and SCC samples, respectively. On the basis of lower levels of integrin β1 and α6, we could not access differential expression pattern for these two subunits. Differential expression for IGTA3 and ITGB4 were confirmed by IHC and ISH. This confirmation was most pronounced for ITGB4, which showed the strongest expression of both protein and mRNA in SCC, in contrast with the weakest expression in adenocarcinoma. ITGA3 and ITGB4 showed prominently cell–matrix staining in normal bronchus, adenocarcinoma and SCC. Previous immunohistochemical studies have shown comparable frequency, intensity and distribution of those integrins in normal bronchial epithelium and NSCLC.16,17,18,19,20,21
Increasing evidence suggests modulation of desmosomes is an important step in the initiation and invasiveness of human malignancies. Desmosomes are adhesive intercellular junctions, made up of heterodimers of different isoforms of the transmembrane desmosomal cadherins, desmogleins and desmocollins, which are linked to cytokeratin intermediate filaments in epithelial cells through interactions with desmoplakins, plakoglobin and plakophilins (for a review, see Wheelock and Johnson 22). Little is known about the colocalisation of the desmosome components and their distribution in normal bronchial epithelium and NSCLC. So far, a tissue‐dependent and differentiation‐specific distribution of individual desmosomal components has only been described in squamous‐like epithelial tissues, such as epidermis, hair follicles, mammary glands and oral mucosa,23,24,25,26,27,28,29 suggesting that desmosomal cadherins have a role in directing differentiation in epithelial tissues.26,30 This is the first study illustrating the distribution of differentially expressed desmosomal molecules in SCC.
We observed strong enhancement of RNA and protein expression in SCC for three desmosomal components: DSP, plakoglobin and DSC3. Normal bronchus and adenocarcinoma showed a relatively low or no expression of desmosome molecules. However, plakoglobin protein expression was observed in normal bronchus and adenocarcinoma at levels similar to SCC, which might be related to its role in adherens junction, attaching to actin filaments.31 Strongest expression of DSP and plakoglobin was observed in more differentiated cells centrally located in tumour areas, whereas DSC3 showed stronger expression in basal cells. Remarkably, this differentiation‐specific distribution of the desmosomal molecules in SCC is very similar to the pattern in normal squamous epithelium of the skin. This strongly suggests a normal expression regulation of these molecules in malignant transformed SCC and may have implications for the biological behaviour of tumour cells. Lack of cellular adhesion has an important role in the metastatic potency of a primary tumour. Although additional studies are required to elucidate the exact relationship of desmosomal molecules with metastatic behaviour in lung cancer, the strong presence of these molecules in SCC and its apparently normal distribution may be associated with the less frequent and late metastasis pattern of SCC as compared with adenocarcinoma.
In summary, we reported a detailed description of distribution of differentially expressed adhesion molecules on protein and mRNA levels in NSCLC and normal bronchi. A remarkably high expression of desmosomal molecules was observed in SCC, with a specific central or peripheral localisation. This may signify a role of these molecules in tumour metastasis.
Acknowledgements
This study was supported by the Spinoza award of DS Postma.
Abbreviations
DSC3 - desmocollin 3
DSP - desmoplakin I and II
IHC - immunohistochemistry
ISH - in situ hybridisation
ITGA3 - integrin α3
ITGB4 - integrin β4
NSCLC - non‐small cell lung cancer
PCR - polymerase chain reaction
SCC - squamous cell carcinoma
Footnotes
Competing interests: None declared.
References
- 1.Bhattacharjee A, Richards W G, Staunton J.et al Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA 20019813790–13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garber M E, Troyanskaya O G, Schluens K.et al Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA 20019813784–13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wigle D A, Jurisica I, Radulovich N.et al Molecular profiling of non‐small cell lung cancer and correlation with disease‐free survival. Cancer Res 2002623005–3008. [PubMed] [Google Scholar]
- 4.Beer D G, Kardia S L, Huang C C.et al Gene‐expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 20028816–824. [DOI] [PubMed] [Google Scholar]
- 5.Ikehara M, Oshita F, Sekiyama A.et al Genome‐wide cDNA microarray screening to correlate gene expression profile with survival in patients with advanced lung cancer. Oncol Rep 2004111041–1044. [PubMed] [Google Scholar]
- 6.Kikuchi T, Daigo Y, Katagiri T.et al Expression profiles of non‐small cell lung cancers on cDNA microarrays: identification of genes for prediction of lymph‐node metastasis and sensitivity to anti‐cancer drugs. Oncogene 2003222192–2205. [DOI] [PubMed] [Google Scholar]
- 7.Oshita F, Ikehara M, Sekiyama A.et al Genomic‐wide cDNA microarray screening to correlate gene expression profile with chemoresistance in patients with advanced lung cancer. J Exp Ther Oncol 20044155–160. [PubMed] [Google Scholar]
- 8.Hoang C D, D'Cunha J, Tawfic S H.et al Expression profiling of non‐small cell lung carcinoma identifies metastatic genotypes based on lymph node tumor burden. J Thorac Cardiovasc Surg 20041271332–1341. [DOI] [PubMed] [Google Scholar]
- 9.Takada M, Tada M, Tamoto E.et al Prediction of lymph node metastasis by analysis of gene expression profiles in non‐small cell lung cancer. J Surg Res 200412261–69. [DOI] [PubMed] [Google Scholar]
- 10.Singhal S, Amin K M, Kruklitis R.et al Differentially expressed apoptotic genes in early stage lung adenocarcinoma predicted by expression profiling. Cancer Biol Ther 20032566–571. [DOI] [PubMed] [Google Scholar]
- 11.Singhal S, Amin K M, Kruklitis R.et al Alterations in cell cycle genes in early stage lung adenocarcinoma identified by expression profiling. Cancer Biol Ther 20032291–298. [DOI] [PubMed] [Google Scholar]
- 12.Mountain C F. Revisions in the International System for Staging Lung Cancer. Chest 19971111710–1717. [DOI] [PubMed] [Google Scholar]
- 13.Simon R. Bioinformatics and whole genome technologies. Technical Report 005 2002. ftp://linus.nci.nih.gov/pub/techreport/TechReport005.pdf (accessed 7 Sept 2006)
- 14.Miura K, Bowman E D, Simon R.et al Laser capture microdissection and microarray expression analysis of lung adenocarcinoma reveals tobacco smoking‐ and prognosis‐related molecular profiles. Cancer Res 2002623244–3250. [PubMed] [Google Scholar]
- 15.Spira A, Beane J, Shah V.et al Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA 200410110143–10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damjanovich L, Albelda S M, Mette S A.et al Distribution of integrin cell adhesion receptors in normal and malignant lung tissue. Am J Respir Cell Mol Biol 19926197–206. [DOI] [PubMed] [Google Scholar]
- 17.Koukoulis G K, Warren W H, Virtanen I.et al Immunolocalization of integrins in the normal lung and in pulmonary carcinomas. Hum Pathol 1997281018–1025. [DOI] [PubMed] [Google Scholar]
- 18.Bartolazzi A, Cerboni C, Flamini G.et al Expression of alpha 3 beta 1 integrin receptor and its ligands in human lung tumors. Int J Cancer 199564248–252. [DOI] [PubMed] [Google Scholar]
- 19.Adachi M, Taki T, Huang C.et al Reduced integrin alpha3 expression as a factor of poor prognosis of patients with adenocarcinoma of the lung. J Clin Oncol 1998161060–1067. [DOI] [PubMed] [Google Scholar]
- 20.Smythe W R, LeBel E, Bavaria J E.et al Integrin expression in non‐small cell carcinoma of the lung. Cancer Metastasis Rev 199514229–239. [DOI] [PubMed] [Google Scholar]
- 21.Patriarca C, Alfano R M, Sonnenberg A.et al Integrin laminin receptor profile of pulmonary squamous cell and adenocarcinomas. Hum Pathol 1998291208–1215. [DOI] [PubMed] [Google Scholar]
- 22.Wheelock M J, Johnson K R. Cadherins as modulators of cellular phenotype. Annu Rev Cell Dev Biol 200319207–235. [DOI] [PubMed] [Google Scholar]
- 23.Getsios S, Huen A C, Green K J. Working out the strength and flexibility of desmosomes. Nat Rev Mol Cell Biol 20045271–281. [DOI] [PubMed] [Google Scholar]
- 24.Garrod D R, Merritt A J, Nie Z. Desmosomal cadherins. Curr Opin Cell Biol 200214537–545. [DOI] [PubMed] [Google Scholar]
- 25.Garrod D R, Merritt A J, Nie Z. Desmosomal adhesion: structural basis, molecular mechanism and regulation [review]. Mol Membr Biol 20021981–94. [DOI] [PubMed] [Google Scholar]
- 26.Borrmann C M, Mertens C, Schmidt A.et al Molecular diversity of plaques of epithelial‐adhering junctions. Ann N Y Acad Sci 2000915144–150. [DOI] [PubMed] [Google Scholar]
- 27.Kurzen H, Moll I, Moll R.et al Compositionally different desmosomes in the various compartments of the human hair follicle. Differentiation 199863295–304. [DOI] [PubMed] [Google Scholar]
- 28.Runswick S K, O'Hare M J, Jones L.et al Desmosomal adhesion regulates epithelial morphogenesis and cell positioning. Nat Cell Biol 20013823–830. [DOI] [PubMed] [Google Scholar]
- 29.Shinohara M, Hiraki A, Ikebe T.et al Immunohistochemical study of desmosomes in oral squamous cell carcinoma: correlation with cytokeratin and E‐cadherin staining, and with tumour behaviour. J Pathol 1998184369–381. [DOI] [PubMed] [Google Scholar]
- 30.North A J, Chidgey M A, Clarke J P.et al Distinct desmocollin isoforms occur in the same desmosomes and show reciprocally graded distributions in bovine nasal epidermis. Proc Natl Acad Sci USA 1996937701–7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowin P, Kapprell H P, Franke W W.et al Plakoglobin: a protein common to different kinds of intercellular adhering junctions. Cell 1986461063–1073. [DOI] [PubMed] [Google Scholar]