Abstract
Aim
To study the role of gene promoter hypermethylation of the putative tumour suppressor genes involved in the death‐associated protein (DAP) kinase/p14/HDM2/p53/Apaf‐1 apoptosis pathway in multiple myeloma (MM).
Method
DNAs from 55 primary MM marrow samples and myeloma cell lines were analysed for aberrant promoter methylation of DAP kinase, p14 and Apaf‐1 genes by methylation‐specific polymerase chain reaction (MSP).
Result
In the methylated positive control, the sensitivity of M‐MSP for DAP kinase was 1×10−3. Aberrant hypermethylation of DAP kinase was found in 29/55 (52.7%) primary MM samples, whereas hypermethylation of p14 or Apaf‐1 was undetectable in any of the samples tested. 5‐Azacytidine treatment of two myeloma cell lines, WL2 and HS‐Sultan, led to de‐methylation and re‐expression of DAP kinase, thereby confirming gene silencing associated with promoter hypermethylation. Hypermethylation of DAP kinase did not correlate with age, sex, paraprotein subtype or Durie–Salmon stage, but negatively affected the overall survival.
Conclusion
Of the putative tumour suppressor genes in the DAP kinase/p14/HDM2/p53/Apaf‐1 apoptosis pathway, only DAP kinase is frequently methylated in MM, which is associated with gene silencing and might be of prognostic significance. p14 and Apaf‐1 were not methylated in MM.
Multiple myeloma (MM) is a disease with a low proliferative rate. Plasma cells actively synthesising DNA are typically <1%, and increase only in advanced disease.1,2 However, neoplastic plasma cells are protected from apoptosis by interacting with the marrow microenvironment.1,2
Death‐associated protein (DAP) kinase is a proapoptotic calcium/calmodulin‐regulated serine/threonine kinase with a multidomain structure. It participates in a wide array of apoptotic mechanisms initiated by interferon‐α, tumour necrosis factor‐α, activated Fas and detachment from the extracellular matrix.3 DAP kinase counteracts oncogene‐induced transformation by activating p53 in a p19ARF‐dependent manner, thereby providing an intrinsic p53‐dependent apoptotic checkpoint that is turned on by oncogenes at the initial stages of transformation.3
p14ARF is one of the two genes encoded by the INK4A/ARF locus at chromosome 9p21.4 Alternative first exons 1α and 1β, under the control of different promoters, specify the 5′ ends of p16INK4A and p14ARF, respectively. These alternative exons are spliced to the same splice acceptor site in exon 2, leading to translation in alternative reading frames. p14 physically associates with HDM2, which possesses E3 ubiquitin ligase activity. HDM2 is a negative regulator of p53, mediating its degradation in the proteosome. p14ARF binding to HMD2 leads to inhibition of the E3 ubiquitin ligase activity, and localisation of HDM2 to the nucleus, precluding its interaction with p53.4 Therefore, by stabilising p53, p14ARF acts as a tumour suppressor.
In normal cells, aberrant activation of a cellular oncogene may trigger an intrinsic tumour suppression mechanism, leading to apoptosis and thus eradication of the transformed cell.5 The DAP kinase/p14/HDM2/p53 pathway is one such mechanism, leading to the downstream activation of Apaf‐1 and formation of the apoptosome.6 In MM, p53 is functional in the majority of cases, and mutation or deletion is rarely demonstrated.7,8 Therefore, other tumour suppressor components of this pathway may be potentially affected in the pathogenesis of MM, in which oncogene activation is a frequent event.1,2
DNA methylation, catalysed by DNA methyltransferase, involves the addition of a methyl group to the carbon 5 position of the cytosine ring in the CpG dinucleotide, converting it to methylcytosine.9,10 In many cancers, the CpG islands of selected genes are aberrantly methylated (hypermethylated), resulting in transcriptional repression. This may serve as an alternative mechanism of gene inactivation.
We have demonstrated frequent epigenetic dysregulation of cell cycle control and Jak/STAT signalling in MM.11,12,13,14 However, as study of isolated genes or random assortment of genes precludes interpretation of the role of hypermethylation of putative tumour suppressor genes in a specific pathway, we investigated the role of aberrant gene promoter hypermethylation in disrupting the DAP kinase/p14/HDM2/p53/Apaf‐1 pathway in MM. p53 was not investigated, as it does not harbour a CpG island in the promoter region (accession number: J04238).
Materials and methods
Patients, diagnosis and treatment
A total of 34 male and 21 female patients with MM, with a median age of 57 (25–87) years, were studied. The diagnosis of MM was based on standard criteria.15 Complete staging examination included bone marrow examination, skeletal survey, serum and urine protein electrophoresis, and serum immunoglobulin (IgG, IgA and IgM) levels. Monoclonal proteins were identified by electrophoresis, immunoelectrophoresis and immunofixation. There were 6 (10.9%) patients with Durie–Salmon stage I, 14 (25.5%) with stage II and 35 (63.6%) with stage III. Patients with stage I or asymptomatic disease did not receive chemotherapy until disease progression. Patients below 65 years of age were induced with three to six courses of vincristine, adriamycin and dexamethasone for initial myeloma cytoreduction. Those with a human leucocyte antigen‐identical donor and below 50 years of age received myeloablative allogeneic haematopoietic stem cell transplantation (HSCT; n = 8), and those between 50 and 65 years of age (n = 10) received non‐myeloablative HSCT. Patients without a suitable donor received autologous HSCT up to the age of 65 years (n = 11). Patients >65 years of age were given melphalan and prednisolone until they reached maximal response or the plateau phase. Patients progressing from the plateau phase received melphalan, cyclophosphamide, thalidomide, or vincristine, adriamycin and dexamethasone. Patients with local symptoms due to disease received local radiotherapy. The study was approved by the institutional review board of Queen Mary Hospital (Hong Kong, Hong Kong) and informed consent obtained.
Methylation‐specific polymerase chain reaction
DNA was extracted from 55 myeloma marrow samples at diagnosis and from myeloma cell lines (WL2 and HS‐Sultan) by the standard method. Methylation‐specific polymerase chain reaction (MSP) for aberrant gene promoter methylation was performed as described previously.16,17 Treatment of DNA with bisulphite for conversion of unmethylated cytosine to uracil (but not affecting methylated cytosine) was performed with a commercially available kit (CpGenome DNA modification kit, Intergen, New York, New York, USA). The genes p14, DAP kinase and Apaf‐1 were tested. Primers for the methylated (M‐MSP) and unmethylated (U‐MSP) promoters are shown in table 1.18,19 DNA from 19 normal bone marrow donors was used as negative control, while methylated control DNA (CpGenome Universal Methylated DNA, Intergen) was used as positive control in all experiments. MSP was performed in a thermal cycler (9700, ABI Biosystems, Foster City, California, USA) with the following cycling conditions: 95°C for 4 min, 35 cycles of 95°C for 45 s, specific annealing temperature for 30 s, 72°C for 30 s and a final extension of 10 min at 72°C. The MSP mixture contained 50 ng of bisulphite‐treated DNA, 0.2 mM dNTPs, 2 mM MgCl2, 10 pmol of each primer, 1× PCR buffer and 2.5 U of AmpliTaq Gold DNA Polymerase (ABI Biosystems) in a final volume of 50 μl. In all, 10 μl of PCR products were loaded onto 6% non‐denaturing polyacrylamide gels, electrophoresed and visualised under ultraviolet light after staining with ethidium bromide.
Table 1 Methylation‐specific polymerase chain reaction: primer sequences and reaction conditions.
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) | Tm/cycles | Reference |
|---|---|---|---|---|
| DAP kinase | ||||
| M‐MSP | GGATAGTCGGAT CGAGTTAACGTC | CCCTCCCAAA CGCCGA | 63°C/35 | Chan et al19 (2002) |
| U‐MSP | GGAGGATAGTTG GATTGAGTTAATGTT | CAAATCCCTCCC AAACACCAA | ||
| P14 | ||||
| M‐MSP | GTGTTAAAG GGCGGCGTAGC | AAAACCCTC ACTCGCGACGA | 65°C/35 | Esteller et al18 (2000) |
| U‐MSP | TTTTTGGTGT TAAAGGGTGGTGTAGT | CACAAAAAC CCTCACTCACAACAA | ||
| Apaf‐1 | ||||
| M‐MSP* | TATTGCGATATTGTT TTAAATTCGA | GAAACGTAACTAAA CCTCAAAAACG | 64°C/35 | Genebank |
| U‐MSP** | TATTGTGATATTGTT TTAAATTTGA | CAAAACATAACTAA ACCTCAAAAACAC | AB070 829 | |
DAP, death‐associated protein; M‐MSP, methylation‐specific polymerase chain reaction for the methylated allele; U‐MSP, MSP for the unmethylated allele; Tm, annealing temperature.
Primers for Apaf‐1: M‐MSP*, nucleotides 785–809; U‐MSP**, nucleotides 938–963 in Genbank accession number: AB070829.
Sensitivity of MSP
One microgram of DNA of the methylated positive control (Intergen) was diluted serially (10‐fold) in normal marrow DNA, modified by bisulphite and then amplified by MSP.
5‐Azacytidine treatment of the WL2 and HS‐Sultan cell lines
The human myeloma cell lines WL2 and HS‐Sultan were cultured in Rosewell Park Memorial Institute medium supplemented with 10% fetal calf serum. For treatment with 5‐azacytidine (5‐AC), cells from myeloma cell lines, WL2 (kindly donated by Dr Andrew Zannettino, Myeloma and Mesenchymal Research Laboratory, Division of Haematology, Institute of Medical and Veterinary Science, Adelaide, Australia) and HS‐Sultan (purchased from ATCC), were seeded in six‐well plates at a density of 106 cells/ml, and cultured with 5 and 0.5 μM of 5‐AC (Sigma, St Louis, Missouri, USA) for 5 days, with fresh medium containing 5‐AC replenished every 2 days. Cells on days 0 and 5 of 5‐AC treatment were harvested.
Reverse transcription‐PCR
One microgram of total cellular RNA was reverse transcribed with M‐MLV reverse transcriptase (Life Technologies, Rockville, Maryland, USA). cDNA was amplified by PCR with DAP kinase‐specific primers (forward: 5′‐GATAGAAATGTCCCCAAACCTCG‐3′ and reverse: 5′‐TCTTCTTGGATCCTTGACCAGAA‐3′, which spanned nucleotides 781–1124, Genbank accession number: X76104). RNA that was not reverse transcribed served as control. Reverse transcription‐PCR for glyceraldehyde 3‐phosphate dehydrogenase (forward: 5′‐GAAGGTGAAGGTCGGAGTC‐3′ and reverse: 5′‐GAAGATGGTGATGGGATTC‐3′) was performed to confirm RNA integrity. PCR conditions for DAP kinase were 35 cycles of 95°C for 30 s, 58°C for 60 s and 72°C for 60 s, and those for glyceraldehyde 3‐phosphate dehydrogenase 30 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s. PCR products were electrophoresed on 6% non‐denaturing polyacrylamide gels, stained with ethidium bromide and visualised under UV illumination. The identity of the PCR products was confirmed by DNA sequencing.
Statistical analysis
Association between DAP kinase hypermethylation and other clinical parameters (age, sex, Durie–Salmon stage, paraprotein subtypes) was studied by the χ2 test (for categorical variables) or Student's t test (for continuous variables). Overall survival (OS) was measured from the date of diagnosis to the date of last follow‐up or death. Survival was estimated by the Kaplan–Meier method and compared by the log‐rank test. All p values were two‐sided.
Results
Methylation‐specific polymerase chain reaction
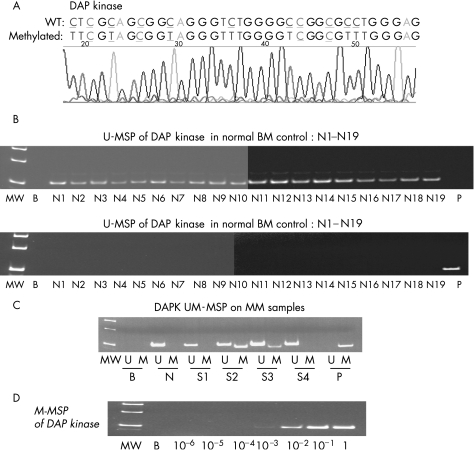
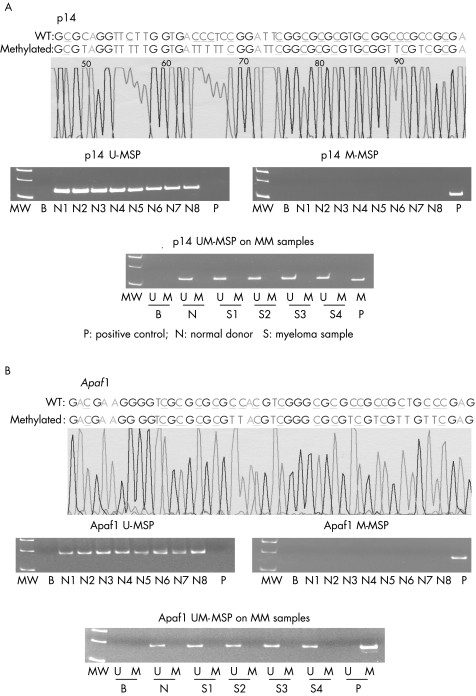
None of the 19 normal control marrows showed aberrant methylation of p14, DAP kinase and Apaf‐1 (fig 1B, 2A,B). The positive and negative controls showed the expected MSP results (normal DNA, U‐MSP positive/M‐MSP negative; methylated DNA: U‐MSP negative/M‐MSP positive). Sequencing of the MSP products showed the expected nucleotide changes after bisulphite treatment (fig 1A, 2A,B). For the primary MM samples, DAP kinase was methylated in 29 cases (52.7%; fig 1C), whereas p14 and Apaf‐1 were not methylated in any of the samples (fig 2A,B).
Figure 1 Methylation‐specific polymerase chain reaction (MSP) of death‐associated protein (DAP) kinase. (A) Sequence analysis of the methylated MSP (M‐MSP) product showed the expected changes after bisulphite treatment. Wild‐type (WT) cytosine (C) residues that remained unchanged in methylated CpG were coloured blue and underlined, while those that were changed to thymidine (T) were coloured red and underlined. (B) Unmethylated MSP (U‐MSP) and M‐MSP showing the expected results, with all 19 normal controls (N1–N19) amplifying in U‐MSP but not in M‐MSP, and the methylated positive control (P) amplifying only in M‐MSP but not in U‐MSP. MW, molecular weight marker; B, reagent blank; P, positive control of methylated DNA; N1–N19, normal marrow DNA. (C) For the MM samples, samples 2 and 3 (S2, S3) showed DAP kinase hypermethylation. B, reagent blank; P, positive control of methylated DNA; S1–4, MM marrow DNA. (D) M‐MSP of the serially diluted methylated positive control DNA, showing a sensitivity of 1 in 103. BM, bone marrow; MM, multiple myeloma.
Figure 2 (A) Methylation‐specific polymerase chain reaction (MSP) of p14 showing the sequence of methylated positive control (upper panel: wild‐type (WT) cytosine (C) residues that remained unchanged in methylated CpG are coloured blue and underlined, while those that were changed to thymidine (T) are coloured red and underlined), absence of p14 hypermethylation in normal control DNA (middle panel: presence of unmethylated MSP (U‐MSP) but absence of methylated MSP (M‐MSP) amplification in normal control DNA) and primary myeloma marrow samples (lower panel: presence of U‐MSP but absence of M‐MSP amplification in normal control DNA). (B) MSP of Apaf‐1 showing the sequence of methylated positive control (upper panel: WT cytosine (C) residues that remained unchanged in methylated CpG are coloured blue and underlined, while those that were changed to thymidine (T) are coloured red and underlined), absence of p14 hypermethylation in normal control DNA (middle panel: presence of U‐MSP but absence of M‐MSP amplification in normal control DNA) and primary myeloma marrow samples (lower panel: presence of U‐MSP but absence of M‐MSP amplification in normal control DNA). B, reagent blank; M, M‐MSP; MM, multiple myeloma; MW, molecular weight; N1–N8, normal marrow DNA; S1–S4, myeloma marrow samples; U, U‐MSP
Sensitivity of DAP kinase M‐MSP
For the positive control, the sensitivity of M‐MSP was 1×10−3 for DAP kinase (fig 1D).
5‐AC treatment of WL2 myeloma cell line
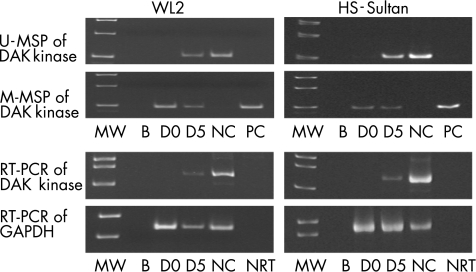
DAP kinase was completely methylated and consequently not expressed in WL2 and HS‐Sultan also. 5‐AC treatment of both cell lines resulted in partial demethylation of DAP kinase, as evidenced by positive U‐MSP amplification and gene expression on day 5 (fig 3).
Figure 3 5‐Azacytidine (5‐AC) treatment of WL2 and HS‐Sultan. Death‐associated protein (DAP) kinase was unmethylated in the normal marrow control (NC), which was associated with expression of DAP kinase transcript. On day 0 (D 0), DAP kinase was completely methylated in WL2 and HS‐Sultan, with a corresponding absence of DAP kinase transcript. At 5 days after treatment (D 5), partial de‐methylation of DAP kinase occurred, leading to positive amplification in unmethylated methyl‐specific polymerase chain reaction (U‐MSP), and expression of the DAP kinase transcript. B, blank; MW, molecular weight marker; NC, normal marrow control; NRT, non‐reagent control; PC, positive control of methylated DNA.
Statistical analysis
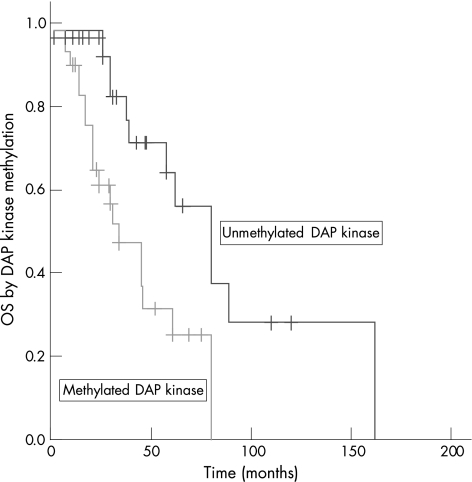
DAP kinase hypermethylation was not associated with age (p = 0.16), sex (p = 0.59), Durie–Salmon stage (p = 0.76) and paraprotein subtypes (p = 0.37; table 2). However, DAP kinase hypermethylation was significantly associated with an inferior median OS (methylated 34 months vs unmethylated 80 months, p = 0.013; fig 4).
Table 2 Correlation of death‐associated protein kinase hypermethylation and patient characteristics.
| DAP kinase | Methylated | Unmethylated | p Value |
|---|---|---|---|
| Number | 29 | 26 | |
| Median age | 60.0 | 54.5 | 0.16 |
| Sex | 0.59 | ||
| Male | 19 | 15 | |
| Female | 10 | 11 | |
| Ig subtype | 0.37 | ||
| IgG | 19 | 15 | |
| IgA | 7 | 4 | |
| IgD | 0 | 1 | |
| Light chain | 3 | 6 | |
| Durie–Salmon stage | 0.76 | ||
| ⩽II | 11 | 9 | |
| III | 18 | 17 |
DAP, death‐associated protein.
Figure 4 Overall survival (OS) of patients with and without death‐associated protein (DAP) kinase hypermethylation. DAP kinase hypermethylation was associated with an inferior OS.
Discussion
We showed that DAP kinase was frequently methylated in MM samples, whereas p14 and Apaf‐1 were not. Therefore, abrogation of the putative tumour suppressor genes involved in the DAP kinase‐related apoptotic pathway in some cases might collaborate with other antiapoptotic signals from the microenvironment to enhance the survival of neoplastic plasma cells.1,2 Moreover, as concomitant abrogation of multiple tumour suppressors of this apoptosis pathway does not confer additional survival benefit, our findings suggested that hypermethylation of tumour suppressor components of this apoptosis pathway appears to be mutually exclusive.
Despite the sensitivity of DAP kinase MSP being only 1 in 103, the methylation status of the neoplastic plasma cells should preferably be studied by CD138‐sorted plasma cells.
Take‐home messages
Death‐associated protein (DAP) kinase, a tumour suppressor gene regulating p53‐mediated apoptosis, is potentially important in myeloma pathogenesis because of its frequent methylation.
DAP kinase hypermethylation might be of prognostic significance.
In the myeloma cell lines WL2 and HS‐Sultan, DAP kinase hypermethylation was associated with the absence of transcript, which was re‐expressed after 5‐AC treatment. As 5‐AC is a DNA methyltransferase inhibitor, and thus a demethylating agent, re‐expression of the transcript after 5‐AC treatment confirmed gene silencing‐associated gene promoter hypermethylation.
In this study, DAP kinase hypermethylation was associated with an inferior OS but not with Durie–Salmon stage. The latter could be due to the retrospective nature of the study, the small number of patients and the heterogeneity of treatment. For instance, 29 (52.7%) of our patients have undergone bone marrow transplantation. Therefore, the prognostic value of DAP kinase methylation needs verification in future prospective clinical trials with a larger number of patients receiving uniform treatment.
The oncogenic role of DAP kinase/p14/Apaf‐1 has been demonstrated in previous studies. DAP kinase expression was shown to suppress the metastatic potential of tumour cells in animal models.21 Modest frequency of DAPkinase hypermethylation (10–20%) has been identified in lung, colon, and head and neck cancers.22 In contrast, frequent (66–72%) DAP kinase hypermethylation was demonstrated in B‐cell lymphoma and MM.23,24 Moreover, DAP kinase has been shown to be methylated in patients with monoclonal gammopathy of undetermined significance at a frequency comparable with that with MM.25p14 hypermethylation was found in 10–30% of renal, gastric and colorectal cancers.22 In haematological malignancies, p14 hypermethylation was present in 40% of chronic myeloid leukaemia cases in acceleration,26 but in only 8% of acute lymphoblastic leukaemia cases,27 and in none of the therapy‐related myelodysplastic syndrome/acute myeloid leukaemia cases.28 In MM, one previous study showed p14 hypermethylation in 9% of the patients.29 The infrequent p14 hypermethylation was consistent with results from another study, which showed p14 expression in MM.30 Furthermore, as the p14/p16 locus was not found to be deleted in MM,31p14 does not appear to be targeted in MM. Finally, while hypermethylation of Apaf‐1 has been detected in melanoma,32 and recently in leukaemia,26 this is the first report demonstrating the absence of Apaf‐1 hypermethylation in MM.
In conclusion, of the putative tumour suppressor genes in the DAP kinase/p14/HDM2/p53/Apaf‐1 apoptosis pathway, only DAP kinase is frequently methylated in MM, which is associated with gene silencing. p14 and Apaf‐1 were not methylated in MM. The high sensitivity of DAP kinase MSP might be a useful marker for monitoring of minimal residual disease.
Acknowledgements
We thank Dr R Robert Orlowski (Department of Medicine, Division of Hematology/Oncology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA) for his critical review of the manuscript.
Abbreviations
5‐AC - 5‐azacytidine
DAP - death‐associated protein
HSCT - haematopoietic stem cell transplantation
MM - multiple myeloma
MSP - methylation‐specific polymerase chain reaction
OS - overall survival
Footnotes
Competing interests: None declared.
References
- 1.Kuehl W M, Bergsagel P L. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer 20022175–187. [DOI] [PubMed] [Google Scholar]
- 2.Hideshima T, Bergsagel P L, Kuehl W M.et al Advances in biology of multiple myeloma: clinical applications. Blood 2004104607–618. [DOI] [PubMed] [Google Scholar]
- 3.Raveh T, Droguett G, Horwitz M S.et al DAP kinase activates a p19ARF/p53‐mediated apoptotic checkpoint to suppress oncogenic transformation. Nat Cell Biol 200131–7. [DOI] [PubMed] [Google Scholar]
- 4.Sherr C J. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol 20012731–737. [DOI] [PubMed] [Google Scholar]
- 5.Lowe S W, Cepero E, Evan G. Intrinsic tumour suppression. Nature 2004432307–315. [DOI] [PubMed] [Google Scholar]
- 6.Hengartner M O. The biochemistry of apoptosis. Nature 2000407770–776. [DOI] [PubMed] [Google Scholar]
- 7.Peller S, Rotter V. TP53 in hematological cancer: low incidence of mutations with significant clinical relevance. Hum Mutat 200321277–284. [DOI] [PubMed] [Google Scholar]
- 8.Avet‐Loiseau H, Li J Y, Godon C.et al P53 deletion is not a frequent event in multiple myeloma. Br J Haematol 1999106717–719. [DOI] [PubMed] [Google Scholar]
- 9.Herman J G, Baylin S B. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 20033492042–2054. [DOI] [PubMed] [Google Scholar]
- 10.Chim C S, Liang R, Kwong Y L. Gene promoter hypermethylation in hematologic malignancies. Hematol Oncol 200220167–176. [DOI] [PubMed] [Google Scholar]
- 11.Chim C S, Fung T K, Liang R. Disruption of INK4/CDK/Rb cell cycle pathway by gene hypermethylation in multiple myeloma and MGUS. Leukemia 2003172533–2535. [DOI] [PubMed] [Google Scholar]
- 12.Chim C S, Kwong Y L, Fung T K.et al Methylation profiling in multiple myeloma. Leukemia Res 200428379–385. [DOI] [PubMed] [Google Scholar]
- 13.Chim C S, Fung T K, Cheung J.et alSOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway. Blood 20041034630–4635. [DOI] [PubMed] [Google Scholar]
- 14.Chim C S, Liang R, Fung T K.et al Infrequent epigenetic dysregulation of CIP/KIP family of cyclin‐dependent kinase inhibitors in multiple myeloma. Leukemia 2005192352–2355. [DOI] [PubMed] [Google Scholar]
- 15.Salmon S, Cassady J R. Plasma cell neoplasms. In: DeVita VT, Hellman S, Rosenberg S, eds. Cancer, principles and practice of oncology. Philadelphia: JB Lippincott, 19972344
- 16.Chim C S, Liang R, Tam C.et alP15 and P16 promoter methylation in acute promyelocytic leukemia. J Clin Oncol 2001192033–2040. [DOI] [PubMed] [Google Scholar]
- 17.Chim C S, Tam C, Liang R, Kwong Y L.P15 and P16 gene methylation in adult acute leukemia: lack of prognostic significance. Cancer 2001912222–2229. [PubMed] [Google Scholar]
- 18.Esteller M, Tortola S, Toyota M.et al Hypermethylation‐associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res 200060129–133. [PubMed] [Google Scholar]
- 19.Chan E C, Lam S Y, Tsang K W.et al Aberrant promoter methylation in Chinese patients with non‐small cell lung cancer: patterns in primary tumors and potential diagnostic application in bronchoalveolar lavage. Clin Cancer Res 200283741–3746. [PubMed] [Google Scholar]
- 20.Inbal B, Cohen O, Polak‐Charcon S.et al DAP kinase links the control of apoptosis to metastasis. Nature 1997390180–184. [DOI] [PubMed] [Google Scholar]
- 21.Esteller M, Corn P G, Baylin S B.et al A gene hypermethylation profile of human cancer. Cancer Res 2001613225–3229. [PubMed] [Google Scholar]
- 22.Katzenellenbogen R A, Baylin S B, Herman J G. Hypermethylation of the DAP‐kinase CpG island is a common alteration in B‐cell malignancies. Blood 1999934347–4353. [PubMed] [Google Scholar]
- 23.Ng M H, To K W, Lo K W.et al Frequent death‐associated protein kinase promoter hypermethylation in multiple myeloma. Clin Cancer Res 200171724–1729. [PubMed] [Google Scholar]
- 24.Chim C S, Liang R, Leung M H.et al Aberrant gene methylation implicated in the progression of monoclonal gammopathy of undetermined significance to multiple myeloma. J Clin Pathol 200760104–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy E, Beck Z, Kiss A.et al Frequent methylation of p16INK4A and p14ARF genes implicated in the evolution of chronic myeloid leukaemia from its chronic to accelerated phase. Eur J Cancer 2003392298–2305. [DOI] [PubMed] [Google Scholar]
- 26.Roman‐Gomez J, Jimenez‐Velasco A, Castillejo J A.et al Promoter hypermethylation of cancer‐related genes: a strong independent prognostic factor in acute lymphoblastic leukemia. Blood 20041042492–2498. [DOI] [PubMed] [Google Scholar]
- 27.Christiansen D H, Andersen M K, Pedersen‐Bjergaard J. Methylation of p15INK4B is common, is associated with deletion of genes on chromosome arm 7q and predicts a poor prognosis in therapy‐related myelodysplasia and acute myeloid leukemia. Leukemia 2003171813–1819. [DOI] [PubMed] [Google Scholar]
- 28.Seidl S, Ackermann J, Kaufmann H.et al DNA‐methylation analysis identifies the E‐cadherin gene as a potential marker of disease progression in patients with monoclonal gammopathies. Cancer 20041002598–2606. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi T, Chikatsu N, Takahashi S.et al Expression of p16INK4A and p14ARF in hematological malignancies. Leukemia 1999131760–1769. [DOI] [PubMed] [Google Scholar]
- 30.Drexler H G. Review of alterations of the cyclin‐dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemia‐lymphoma cells. Leukemia 199812845–859. [DOI] [PubMed] [Google Scholar]
- 31.Soengas M S, Capodieci P, Polsky D.et al Inactivation of the apoptosis effector Apaf‐1 in malignant melanoma. Nature 2001409207–211. [DOI] [PubMed] [Google Scholar]