Abstract
Objective
To determine whether interferon‐gamma (IFN‐γ) and interleukin‐4 (IL‐4) genes confer susceptibility for the idiopathic inflammatory myopathies (IIMs).
Methods
A large cross‐sectional study of UK caucasian adults with polymyositis (PM, n = 101), dermatomyositis (DM, n = 94) and myositis overlapping with a connective tissue disease (myositis/CTD‐overlap, n = 70) was completed. 177 ethnically matched controls were available for comparison. Single‐nucleotide polymorphisms (SNPs) within intronic regions coding for IL‐4, IFN‐γ and a microsatellite marker within intron 1 of the IFN‐γ gene were typed.
Results
Strong linkage disequilibrium was present between SNPs in each gene. In the IFN‐γ gene, a weak allelic association was observed in PM versus controls at rs1861493 (odds ratio (OR) 1.6, 95% confidence interval (CI) 1.03 to 2.4). The microsatellite IFN‐γ CA(14) allele was associated with risk for IIMs overall (OR 3.3, 95% CI 1.4 to 7.8), the strongest association being observed within the anti‐U1‐ribonucleoprotein (RNP) group (OR 6.0, 95% CI 1.5 to 23.1), and persisting after adjustment for known myositis human leucocyte antigen (HLA) class II associations.
Conclusions
Genetic markers in the IFN‐γ gene demonstrate significant allelic associations with the IIMs in a UK Caucasian population. The SNPs tested in this study within the region coding for IL‐4 fail to show significant associations with susceptibility to IIM disease.
The idiopathic inflammatory myopathies (IIMs) are a heterogeneous group of diseases which include polymyositis (PM), dermatomyositis (DM) and myositis overlapping with other connective tissue diseases (myositis/CTD‐overlap). There is increasing recognition that genetic factors are involved in the development of IIMs.1 Most myositis studies to date have only examined the human leucocyte antigen (HLA) region where, in Caucasians, HLA‐DRB1*0301 and HLA‐DQA1*0501 are significant risk factors.2,3 Few IIM genetic studies have been conducted outside of the HLA region.1
In multifactorial diseases mediated by inflammatory processes, variations in susceptibility to/expression of disease may be influenced by functional cytokine polymorphisms, or up/downregulation of genes encoding cytokine production.4 A T‐helper type 1 (Th1)/T‐helper type 2 (Th2) cytokine imbalance is postulated in various autoimmune disorders. Th1 cytokines play a role in cell‐mediated immunity, but are detrimental in organ‐specific responses; Th2 cytokines induce antibody production and mediate allergic responses. Furthermore, Th1 and Th2 cytokine responses are antagonistic and downregulate each other.
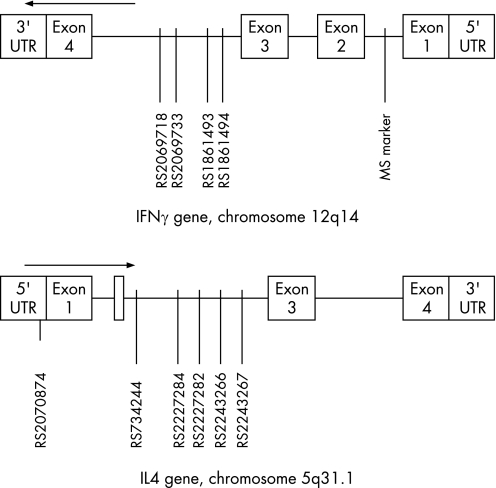
Interferon‐gamma (IFN‐γ) is a T cell‐derived Th1 cytokine and a strong inducer of major histocompatibility complex class I expression in IIM inflammatory muscle tissue.5 The IFN‐γ gene on chromosome 12q14 comprises four exons (fig 1). A variable dinucleotide repeat, (CA)n, has been shown in intron 1, acting as a non‐specific tissue enhancer element. (CA) repeat variations are associated with various autoimmune diseases.4 In contrast, interleukin‐4 (IL‐4) is a Th2 cytokine produced by activated T cells and functions as a B‐cell growth factor. The IL‐4 gene is on chromosome 5q31.1, comprising 4 exons (fig 1). IL‐4 single‐nucleotide polymorphisms (SNPs) have been associated with patients having severe asthma and multiple sclerosis.4 Variable expression of IL‐4 and IFN‐γ has been shown in IIM muscle biopsy specimens.6,7,8 Furthermore, an imbalance of Th1/Th2 cytokines in favour of Th1 is postulated in the IIMs.9
Figure 1 Schematic representations of genetic markers examined as they appear in the genome. Data obtained from the National Center for Biotechnology Information, Bethesda, Maryland, USA (www.ncbi.nlm.nih.gov). UTR, untranslated region; MS, microsatellite; arrows indicate direction of transcription.
In view of these findings, we hypothesise that Th1/Th2 cytokine responses are aetiologically involved in genetic susceptibility in myositis, and thus investigate the role of IFN‐γ and IL‐4 genes in a large cohort of subtypes of patients with IIM.
Materials and methods
Study design
A case–control, cross‐sectional, genetic association study, comparing patients with IIM with normal subjects.
Cases
Between 1999 and 2004, a UK‐wide collaboration of 56 rheumatologists and 4 neurologists comprising the Adult Onset Myositis Immunogenetic Collaboration (AOMIC) (for details, see appendix in Chinoy et al3) recruited 272 Caucasian patients, ⩾18 years at the onset of disease. The inclusion criteria for PM/DM was probable or definite disease, according to Bohan and Peter.10 For myositis/CTD‐overlap, myositis may be less rigorously diagnosed as part of another connective tissue disease (CTD). Thus, 17 of the 70 (24%) patients with myositis/CTD‐overlap were included if they: (1) met published criteria for their primary CTD; (2) possessed at least two of four Bohan–Peter criteria; and (3) possessed at least one myositis‐specific/associated antibody (MSA/MAA). The remaining 53 patients with myositis/CTD‐overlap fulfilled criteria for their primary disease and satisfied probable/definite myositis criteria. Collaborating physicians confirmed/excluded interstitial lung disease and cancer‐associated myositis (CAM) by relevant investigations. Written consent of the patients was obtained according to the Declaration of Helsinki.
Controls
In all, 177 UK caucasian control subjects were recruited from primary population registers in the UK, as part of the epidemiological studies described previously.3 Control marker frequencies are comparable with those found in the National Center for Biotechnology Information, Bethesda, Maryland, USA SNP database (http://ncbi.nlm.nih.gov/SNP) and those previously published.11 Collection of clinical data and blood was under regulation of local research ethics committees.
Serological typing
Sera were obtained from patients for determination of MSAs (anti‐transfer RNA (tRNA) synthetases: ‐Jo‐1, ‐PL‐7, ‐PL‐12, ‐EJ, ‐OJ, ‐KS; ‐Mi‐2, ‐SRP) and MAAs (anti‐PM‐Scl, ‐Ku, ‐U1‐ribonucleoprotein (RNP), ‐U3‐RNP), as described.3
Genotyping
Genetic markers were selected from the National Center for Biotechnology Information SNP database (fig 1). Assays were based on TaqMan or SNaPshot (PE Applied Biosystems (ABI), Oxford UK). The IFN‐γ microsatellite marker and insertion–deletion were genotyped using an ABI 3100 DNA analyser.12 Primers and conditions are available on a supplementary file on‐line. HLA genotyping was performed as described.3
Statistical analyses
Genotyping frequencies for each SNP were tested for Hardy–Weinberg equilibrium in controls. Allelic and genotype frequencies were compared between myositis cases and controls. Probabilities were calculated using Fisher's exact test and, unless otherwise stated, corrected for multiple comparisons using Bonferroni for the number of SNPs tested. Data were expressed as ORs with exact 95% CI. Linkage disequilibrium (LD) was calculated with r2 values, using HelixTree (V 3.1.2), (Golden Helix, Bozeman, Montana, USA). Haplotypes were estimated and constructed using the expectation/maximisation algorithm. The statistical package Stata (release 8), was used to perform statistical analysis. The study had 80% power to detect the effect of an SNP, with a range of 17–40% minor allele frequency in the tested SNPs, conferring an OR of 2.0 at the 5% significance level, assuming a dominant mode of inheritance.
Results
Demography
The 265 cases recruited for the study included 101 patients with PM, 94 patients with DM and 70 patients with myositis/CTD‐overlap. The patients with myositis/CTD‐overlap had the following primary diagnoses: 45 with systemic sclerosis, 9 with mixed CTD, 7 with Sjögren's syndrome, 7 with systemic lupus erythematosus and 2 with rheumatoid arthritis. A detailed summary of further demographics is provided in Chinoy et al.3
Genetic associations
All markers, except rs734244, showed no deviation from the Hardy–Weinberg equilibrium. Strong LD (r2>0.64) was observed across all markers, except rs1861494 (r2<0.5). The LD detected between markers in the IFN‐γ, IL‐4 and HLA class II genes was weak (r2<0.1).
Table 1 summarises the IFN‐γ and IL‐4 genotype and allele frequencies. In the IFN‐γ gene, a weak allele association for the A allele in rs1861493 was noted in PM. The frequency of the AA genotype was increased under a recessive model. This allele was also increased in patients who were anti‐tRNA synthetase positive (78% cases vs 67% controls, OR 1.7, 95% CI 1.0 to 3.0, uncorrected probability (puncorr 0.04)). No further significant associations were observed in the other subgroups, groups combined, or in the IL‐4 markers.
Table 1 Genotype and allele frequencies of interferon‐gamma ‐γ and interleukin‐4 polymorphisms in patients with myositis, and controls.
| IL‐4 markers | Controls | PM | DM | Overlap |
|---|---|---|---|---|
| rs2070874 | n = 156 | n = 88 | n = 84 | n = 65 |
| CC, CT, TT | 75.6, 23.1, 1.3 | 79.5, 20.5, 0 | 77.4, 21.4, 1.2 | 69.2, 29.2, 1.6 |
| C, T | 87.2, 12.8 | 89.8, 10.2 | 88.1, 11.9 | 83.8, 16.2 |
| rs734244 | n = 161 | n = 97 | n = 90 | n = 70 |
| CC, CT, TT | 77.6, 12.4, 9.9 | 75.3. 10.3, 14.4 | 77.6, 10.3, 12.1 | 71.4, 18.6, 10.0 |
| C, T | 83.8, 16.2 | 80.4, 19.6 | 80.7, 19.3 | 81.3, 18.7 |
| rs2227284 | n = 156 | n = 87 | n = 83 | n = 64 |
| GG, GT, TT | 59.0, 36.5, 4.5 | 59.8, 35.6, 4.6 | 68.7, 6.0, 25.3 | 56.3, 35.9, 7.8 |
| G, T | 77.2, 22.8 | 77.6, 22.4 | 81.3, 18.7 | 78.0, 22.0 |
| rs2227282 | n = 151 | n = 88 | n = 84 | n = 67 |
| GG, GC, CC | 58.3, 37.1, 4.6 | 55.7, 38.6, 5.7 | 70.2, 23.8, 6.0 | 58.2, 34.3, 7.5 |
| G, C | 76.8, 23.2 | 75.0, 25.0 | 82.1, 17.9 | 75.4, 24.6 |
| rs2243266 | n = 156 | n = 87 | n = 85 | n = 61 |
| CC, CT, TT | 76.9, 21.8, 1.3 | 78.2, 21.8, 0 | 77.6, 21.2, 1.2 | 70.5, 27.9, 1.6 |
| C, T | 87.8, 12.2 | 89.1, 10.9 | 88.2, 11.8 | 84.4, 15.6 |
| rs2243267 | n = 156 | n = 90 | n = 85 | n = 65 |
| GG, GC, CC | 76.3, 22.4, 1.3 | 79.0, 21.2, 0.8 | 77.6, 21.2, 1.2 | 69.2, 29.2, 1.6 |
| G, C | 87.5, 12.5 | 89.4, 10.6 | 88.2, 11.8 | 83.8, 16.2 |
| IFN‐γ markers | ||||
| rs1861494 | n = 151 | n = 85 | n = 72 | n = 60 |
| TT, TC, CC | 47.0, 42.2, 10.6 | 56.5, 36.5, 7.0 | 50.0, 37.5, 12.5 | 56.7, 35.0, 8.3 |
| T, C | 68.2, 31.8 | 74.7, 25.3 | 68.8, 31.2 | 74.2, 25.8 |
| rs1861493* | n = 148 | n = 83 | n = 74 | n = 63 |
| AA, AG, GG | 44.6, 45.3, 10.1 | 59.0, 35.0, 6.0 | 48.6, 37.8, 13.6 | 58.7, 31.8, 9.5 |
| A, G | 67.2, 32.8 | 76.5, 23.5 | 67.6, 32.4 | 74.6, 25.4 |
| rs2069733 | n = 159 | n = 101 | n = 90 | n = 69 |
| G/G, G/–, –/– | 52.8, 39.0, 8.2 | 60.4, 35.6, 4.0 | 48.9, 43.3, 7.8 | 62.3, 30.4, 7.3 |
| G, ‐ | 72.3, 27.7 | 78.2, 21.8 | 70.6, 29.4 | 77.5, 22.5 |
| rs2069718 | n = 156 | n = 87 | n = 82 | n = 66 |
| GG, GA, AA | 35.9, 51.9, 12.2 | 47.1, 41.4, 11.5 | 37.8, 41.5, 20.7 | 33.3, 57.6, 9.1 |
| G, A | 61.9, 38.1 | 67.8, 32.2 | 58.5, 41.5 | 62.1, 37.9 |
DM, dermatomyositis; IFN‐γ, interferon‐γ; IL‐4, interleukin‐4; PM, polymyositis.
n may vary between markers due to genotyping failures.
X, Y refers to percentage allele frequencies. XX, XY, YY refers to percentage genotype frequencies.
Allele percentage frequencies are based on 2n. *rs1861493 A allele, PM versus controls, puncorr = 0.04, pcorr>0.05, OR 1.6, 95% CI 1.0 to 2.4. rs1861493 AA genotype, PM versus controls, puncorr = 0.04, pcorr>0.05, OR 1.8, 95% CI 1.0 to 2.3.2.
Sequencing analysis of the IFN‐γ microsatellite marker revealed six allelic variants, ranging from 11 to 16 (CA). Table 2 provides a summary of these frequencies in clinical subgroups. The overall microsatellite marker allelic distribution and, specifically, the (CA)14 polymorphism was significantly different in patients with myositis/CTD‐overlap versus controls (table 2). The (CA)14 polymorphism was increased in patients who were anti‐U1‐RNP positive versus controls (10% vs 2% controls, OR 6.0, 95% CI 1.5 to 23.1, puncorr = 0.005), losing significance after correction for tested MSA/MAAs. Weaker associations for the (CA)14 polymorphism were observed in patients with PM, patients with DM and patients who were anti‐tRNA synthetase positive. A multivariate logistic regression model was created, including the microsatellite marker and known myositis HLA class II alleles as predictors (DRB1*03, DQA1*05, DQB1*023). The (CA)14 polymorphism remained a significant risk factor in overall and myositis/CTD‐overlap groups after adjusting for the HLA alleles, but no additive or multiplicative effects were observed. Only one (CA)14/(CA)14 homozygote was observed, in the DM group. The frequency of the heterozygous genotype (CA)12/(CA)14 was significantly increased in patients with myositis/CTD‐overlap versus controls (6.8% overlap vs 1.3% controls, OR 13.4, 95% CI 2.7 to 128.1, pcorr = 0.002). No other genotypic associations, including homozygous states, were observed for the microsatellite marker. No significant associations were observed in patients with other MSA/MAAs, overlap subgroups, CAM or interstitial lung disease.
Table 2 Allele frequencies in the interferon‐gamma microsatellite marker.
| Amplicon size (bp) | IFN‐γ (CA)n* | Controls | PM | DM | Overlap | Overall |
|---|---|---|---|---|---|---|
| 2n = 316 | 2n = 186 | 2n = 178 | 2n = 136 | 2n = 500 | ||
| 122 | 11 | 0 | 0.5 | 0 | 0 | 0.2 |
| 124 | 12 | 49.7 | 49.5 | 47.2 | 43.4 | 47.0 |
| 126 | 13 | 45.2 | 40.3 | 44.4 | 44.8 | 43.0 |
| 128 | 14† | 1.9 | 5.4 | 5.0 | 8.1 | 6.0 |
| 130 | 15 | 2.9 | 3.8 | 3.4 | 3.7 | 3.6 |
| 132 | 16 | 0.3 | 0.5 | 0 | 0 | 0.2 |
| p‡ | 0.1 | 0.3 | 0.02 | 0.05 |
bp, base pairs; DM, dermatomyositis; IFN‐γ, interferon‐gamma; PM, polymyositis.
*n represents the (CA) dinucleotide repeat number.
†Overlap versus controls, OR 4.5 (1.7–12.1), puncorr = 0.005, pcorr = 0.03.
‡Comparison of distribution of alleles between cases and controls using Fisher's exact test.
Haplotypes
Haplotype frequencies were calculated for the IFN‐γ markers, capturing >93% of the variation. After correction for multiple comparisons, no significant differences between cases and controls were noted.
Discussion
We report evidence for disease association of the IFN‐γ gene in the largest SNP IIM study to date. The IFN‐γ associations for disease are found primarily in the microsatellite marker, where the CA(14) repeat represents a risk factor, specifically in patients with myositis‐CTD/overlap and patients who are anti‐U1‐RNP positive. This association is independent of HLA class II members of the 8.1 ancestral haplotype and shows no interaction with these alleles.3 However, the strength of the CA(14) repeat association is less than that of HLA class II alleles.3 These findings are unlikely to be causal, as the tested associations are weak or not present for individual SNPs, and generally no longer present after correction for multiple comparisons. Certainly, testing multiple genetic markers in several subgroups may lead to potential problems in analyses, especially when associations are weak. Myositis/CTD‐overlap is a somewhat heterogeneous group, hence the described genetic associations may refer to general autoimmunity rather than specifically for myositis. IFN‐γ microsatellite polymorphism associations have been described in other autoimmune disorders. In American patients with systemic lupus erythematosus,13 the CA(16) polymorphism is associated with anti‐U1‐RNP and severe disease markers. In giant cell arteritis, IFN‐γ associations correlate with specific clinical severity features.14 The CA(12) and CA(13) repeats are also associated with increased and reduced serum IFN‐γ production, respectively.11,15 We have not examined whether the described genetic polymorphisms correlate with aberrant serum cytokine production.
The major genetic associations so far detected in myositis lie within the HLA region.1 New technologies,—for example, whole genome association studies, may locate new disease susceptibility genes. We conclude that the tested markers within the IFN‐γ, but not the IL‐4 gene shows significant genetic associations in UK Caucasians with IIM. The IFN‐γ gene requires further investigation, including on a functional basis, to elucidate the genetic role of the Th1/Th2 cytokine responses in the aetiopathogenesis of IIMs.
Supplementary file is available at http://ard.bmj.com/supplemental
Supplementary Material
Acknowledgements
Dr Hector Chinoy is an Arthritis Research Campaign Clinical Research Fellow (grant number 16082). We thank the Myositis Support Group UK, who funded the myositis autoantibody testing. We also thank the 60 participating physicians in the AOMIC (see appendix in Chinoy et al3), without whom this research would not have been possible.
Abbreviations
anti‐U1‐RNP - anti‐U1‐ribonucleoprotein
CAM - cancer‐associated myositis
CTD - connective tissue disease
DM - dermatomyositis
HLA - human leucocyte antigen
IFN‐γ - interferon‐gamma
IIM - idiopathic inflammatory myopathy
IL‐4 - interleukin‐4
LD - linkage disequilibrium
MSA/MAA - myositis‐specific/associated antibody
myositis/CTD‐overlap - myositis overlapping with other connective tissue diseases
PM - polymyositis
SNP - single‐nucleotide polymorphism
Th - T‐helper
Footnotes
Competing interests: None declared.
Supplementary file is available at http://ard.bmj.com/supplemental
References
- 1.Chinoy H, Ollier W E, Cooper R G. Have recent immunogenetic investigations increased our understanding of disease mechanisms in the idiopathic inflammatory myopathies? Curr Opin Rheumatol 200416707–713. [DOI] [PubMed] [Google Scholar]
- 2.O'Hanlon T P, Carrick D M, Arnett F C, Reveille J D, Carrington M, Gao X.et al Immunogenetic risk and protective factors for the idiopathic inflammatory myopathies: distinct HLA‐A, ‐B, ‐Cw, ‐DRB1 and ‐DQA1 allelic profiles and motifs define clinicopathologic groups in caucasians. Medicine (Baltimore) 200584338–349. [DOI] [PubMed] [Google Scholar]
- 3.Chinoy H, Salway F, Fertig N, Shephard N, Tait B D, Thomson W.et al In adult onset myositis, the presence of interstitial lung disease and myositis specific/associated antibodies are governed by HLA class II haplotype, rather than by myositis subtype. Arthritis Res Ther 20068R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vandenbroeck K, Goris A. Cytokine gene polymorphisms in multifactorial diseases: gateways to novel targets for immunotherapy? Trends Pharmacol Sci 200324284–289. [DOI] [PubMed] [Google Scholar]
- 5.Figarella‐Branger D, Civatte M, Bartoli C, Pellissier J F. Cytokines, chemokines, and cell adhesion molecules in inflammatory myopathies. Muscle Nerve 200328659–682. [DOI] [PubMed] [Google Scholar]
- 6.Lundberg I, Ulfgren A K, Nyberg P, Andersson U, Klareskog L. Cytokine production in muscle tissue of patients with idiopathic inflammatory myopathies. Arthritis Rheum 199740865–874. [DOI] [PubMed] [Google Scholar]
- 7.Adams E M, Kirkley J, Eidelman G, Dohlman J, Plotz P H. The predominance of beta (CC) chemokine transcripts in idiopathic inflammatory muscle diseases. Proc Assoc Am Physicians 1997109275–285. [PubMed] [Google Scholar]
- 8.Lepidi H, Frances V, Figarella‐Branger D, Bartoli C, Machado‐Baeta A, Pellissier J F. Local expression of cytokines in idiopathic inflammatory myopathies. Neuropathol Appl Neurobiol 19982473–79. [DOI] [PubMed] [Google Scholar]
- 9.Megens‐de Letter M A, Visser L H, van Doorn P A, Savelkoul H F. Cytokines in the muscle tissue of idiopathic inflammatory myopathies: implications for immunopathogenesis and therapy. Eur Cytokine Netw 199910471–478. [PubMed] [Google Scholar]
- 10.Bohan A, Peter J B. Polymyositis and dermatomyositis (first of two parts). N Engl J Med 1975292344–347. [DOI] [PubMed] [Google Scholar]
- 11.Pravica V, Asderakis A, Perrey C, Hajeer A, Sinnott P J, Hutchinson I V. In vitro production of IFN‐gamma correlates with CA repeat polymorphism in the human IFN‐gamma gene. Eur J Immunogenet 1999261–3. [DOI] [PubMed] [Google Scholar]
- 12.Chinoy H, Salway F, Fertig N, Tait B D, Oddis C V, Ollier W E.et al Monocyte chemotactic protein‐1 single nucleotide polymorphisms do not confer susceptibility for the development of adult onset polymyositis/dermatomyositis in UK Caucasians. Rheumatology (Oxford) 200746604–607. [DOI] [PubMed] [Google Scholar]
- 13.Lee J Y, Goldman D, Piliero L M, Petri M, Sullivan K E. Interferon‐gamma polymorphisms in systemic lupus erythematosus. Genes Immun 20012254–257. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez‐Gay M A, Hajeer A H, Dababneh A, Garcia‐Porrua C, Amoli M M, Llorca J.et al Interferon‐gamma gene microsatellite polymorphisms in patients with biopsy‐proven giant cell arteritis and isolated polymyalgia rheumatica. Clin Exp Rheumatol 200422S18–S20. [PubMed] [Google Scholar]
- 15.Miyake K, Nakashima H, Akahoshi M, Inoue Y, Nagano S, Tanaka Y.et al Genetically determined interferon‐{gamma} production influences the histological phenotype of lupus nephritis. Rheumatology 200241518–524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.