Abstract
Background
A genomewide scan in a DA×ACI F2 intercross studied for collagen‐induced arthritis (CIA) identified the severity quantitative trait locus Cia25 on rat chromosome 12. Cia25 co‐localises with loci regulating several forms of autoimmune diseases in rats, mice and humans, suggesting a common gene.
Objective
To characterise the effects of Cia25 on severity of arthritis in congenic rats.
Methods
DA.ACI(Cia25) congenic rats were constructed according to a genotype‐guided strategy, and tested for pristane‐induced arthritis (PIA) and CIA, induced with rat type II collagen (CII). A well‐established scoring system previously shown to correlate with histological damage, including cartilage and bone erosions, synovial hyperplasia and synovial inflammation, was used.
Results
The introgression of ACI alleles at Cia25 into DA background, as in DA.ACI(Cia25) rats, was enough to significantly reduce arthritis severity by 60% in PIA and by 40% in CIA, both in males and females compared with DA rats of the same sex. Levels of IgG anti‐CII in male DA.ACI(Cia25) rats were 83% lower than in male DA. Levels of anti‐CII in females were not affected by the congenic interval.
Conclusions
Cia25 contains a gene that regulates disease severity in two distinct models of autoimmune arthritis. Although both genders were protected in arthritis studies, only male congenic rats had a dramatic reduction in levels of anti‐CII, suggesting the possibility of a second arthritis gene in this interval that operates via the regulation of autoantibodies in a sex‐specific manner. The identification of the gene(s) accounting for Cia25 is expected to generate novel prognostic biomarkers and targets for therapy.
Rheumatoid arthritis (RA) is a chronic autoimmune disease that affects 0.5–1% of the population.1,2 Treatments targeting tumour necrosis factor α,3 T cells4 and B cells5 have substantially improved disease control, yet sustained remission is rarely achieved. The strong genetic contribution to disease suggested by the heritability of 60%2 makes the identification of genes regulating susceptibility to, and severity of, RA a promising strategy to generate novel targets for the development of more effective therapies.
Accordingly, several major histocompatibility complex (MHC) and non‐MHC RA susceptibility loci have already been mapped,6,7,8,9,10,11,12 but only a few of their respective genes have been identified13,14,15 and most are yet to be replicated. Additionally, these studies were designed to identify susceptibility genes, some of which are likely to regulate early cognate immune interactions during preclinical stages, not necessarily influencing established and chronic stages of disease, or joint damage. Increased severity predicts and correlates with disability,16 joint damage17 and premature mortality,18 and, therefore, identifying severity genes is more likely to generate useful targets for the development of new therapies. However, very little is known about the genetic regulation of RA severity.
The study of rat models of autoimmune arthritis in inbred strains has identified several arthritis severity quantitative trait loci (QTLs),19,20,21,22,23,24,25 and a disease severity gene.26 Genes identified in QTL regions will generate compelling candidates for focused RA case–control studies in well‐characterised cohorts that include severity data, and for the development of new prognostic biomarkers and therapeutic strategies. We here report the construction of rats congenic for Cia25, a QTL that we have previously identified on rat chromosome 12,20 and show evidence suggesting that this locus contains two different genes regulating the severity of arthritis induced with pristane and with collagen in both male and female rats.
Methods
Rats
Specific pathogen‐free ACI (ACI/Hsd) and DA (DA/Hsd) inbred rat strains were purchased from Harlan Sprague–Dawley (Indianapolis, Indiana, USA) and used in the breeding of the congenic strains. All the experiments involving animals were reviewed and approved by the Feinstein Institute for Medical Research Institutional Animal Care and Use Committee.
Genotype‐guided construction of Cia25‐congenic rats
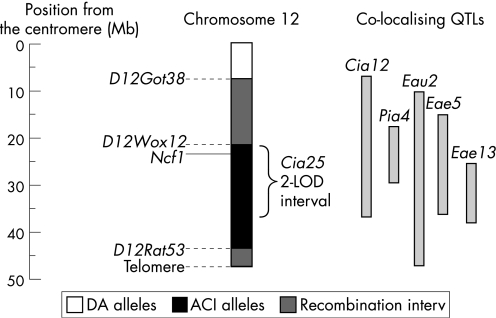
A 22.9 Mb interval containing the 15.8 Mb two‐logarithm of odds support interval comprising Cia2520 was introgressed from the arthritis‐resistant ACI strain into the arthritis‐susceptible DA background using a genotype‐guided breeding strategy to generate homozygotic DA.ACI(Cia25) rats (fig 1). There was no evidence of genomic imprinting in the DA×ACI F2 intercross genomewide scan.19,20 Therefore, two DA×ACI and two ACI×DA mating pairs (using the convention of female × male) were used to produce F1. The F1 offspring were backcrossed with DA rats, and the progeny (and the following backcross generations) were genotyped with simple sequence length polymorphic markers on chromosomes 2 (Cia7: D2Arb1, D2Mgh19, Cpb1, D2Rat38; Cia10: D2Mit12, D2Wox20, D2Rat55, D2Mgh29), 10 (Cia27: D10Rat19, D10Mit12, D10Wox7, D10Rat2) and 12 (Cia25: D12Wox12, D12Arb15, D12Mgh3, D12Arb3, D12Rat53), which are known to contain QTLs regulating arthritis severity in the DA×ACI F2 intercross.19,20 Offspring that maintained the Cia25 interval in the heterozygotic state and had homozygotic DA alleles at the other three arthritis QTLs were backcrossed again to the DA strain. After eight successive backcrosses, (BC8) rats were intercrossed to generate homozygosity at the Cia25 interval. These rats were brother–sister mated to establish and expand the DA.ACI(Cia25) congenic line. Offspring from the fifth and sixth intercrosses were used in the arthritis experiments. DA.ACI(Cia25) congenic rats were further backcrossed with DA to generate male DA.ACI(Cia25) rats heterozygotic at the Cia25 locus.
Figure 1 The Cia25 interval and colocalising autoimmunity loci. Numbers represent interval length and simple sequence length polymorphic marker positions relative to the centromere in megabases (Mb). The black segment identifies homozygotic ACI alleles. The positions of Ncf1 and the two‐logarithm of odds (2‐LOD) score support interval for Cia25 are shown, and colocalising rat quantitative trait loci (QTLs, Cia12,25Pia4,23Eau2,27Eae528 and Eae1329) are indicated in light grey.
Genotyping
PCR conditions have been reported previously.20,30 The tail tips were excised at age 3–4 weeks and DNA extracted with the DNeasy kit (Qiagen, Valencia, California, USA). Fluorescent‐labelled primers were used for PCR and products run along with TAMRA‐labelled size standards in an ABI 310 capillary genotyper/sequencer (ABI, Foster City, California, USA). GENESCAN V.3.1 software (ABI) was used for data extraction and allele assignment. Two readers manually checked all genotypes, and questionable readings were re‐checked or repeated. Marker details are available at the Rat Genetic database (http://niams.nih.gov/rtbc/ratgbase) and the Rat Genome database (http://rgd.mcw.edu).
Pristane‐induced arthritis
The 8–12‐week‐old rats were anaesthetised and injected intradermally with 150 μl of pristane (2,6,10,14‐tetramethylpentadecane) divided between two injection sites at the base of the tail (day 0).31,32 Pristane was initially purchased from ICN (Aurora, Ohio, USA), and then from Sigma‐Aldrich (St Louis, Missouri, USA) owing to a discontinuation in supply. Pristane from these two brands induced arthritis of similar severity (data not shown).
Collagen‐induced arthritis
Collagen‐induced arthritis (CIA) was induced according to a previously described protocol.19 Purified homologous rat type II collagen (CII, Chondrex, Redmond, Washington, USA) was dissolved overnight in 0.1 N acetic acid (2 mg/ml) at 4°C and emulsified with incomplete Freund's adjuvant (IFA, Difco, Detroit, Michigan, USA) to a final concentration of 1 mg/ml. Anaesthetised 8–12‐week‐old rats were injected intradermally with 2 mg/kg of CII at the base of the tail, divided into six injection sites (day 0). A booster injection of 100 μg CII/IFA was administered on day 7.19,33
Arthritis scoring
We used a previously described arthritis scoring system19 that evaluates individual joints and measures arthritis severity according to joint size as follows: (a) interphalangeal, metacarpophalangeal and metatarsophalangeal joints in the four lateral digits are scored: 0, no arthritis; 1, arthritis present and (b) wrist, mid‐forepaw, ankle and midfoot joints are scored: 0, normal; 1, minimal swelling; 2, moderate swelling; 3, severe swelling; 4, severe swelling and non‐weight bearing. The scores from all involved joints were added (maximum score per rat per day = 80). The same observer obtained the arthritis scores on days 0, 14, 18, 21, 24, 28 and 31 after induction. The arthritis severity index (ASI), a comprehensive measure of disease severity over time (area under the curve), was determined for each animal by adding the individual arthritis scores obtained over the course of the experiment. The ASI correlates with the degree of histological damage, including synovial inflammation, synovial hyperplasia, and cartilage and bone erosive changes.32,34,35 To dissociate the severity and susceptibility components, we analysed only those animals that developed arthritis, defined as a daily score of ⩾1 in at least two consecutive time points.
Measurement of anti‐collagen (CII) autoantibodies
Serum samples collected on day 18 after CIA induction were diluted 1:40 000 and assayed in duplicates for anti‐CII IgG, using a commercially available ELISA kit coated with highly purified CII (Chondrex), according to the manufacturer's instructions, and were reported in U/ml.
Statistical analyses
Based on previous observations of gender differences in the genetic regulation of CIA,20 male and female rats were studied for arthritis severity both separately and together. Medians were compared using the Mann–Whitney test and correlations were calculated with the Pearson's correlation coefficient using SigmaStat V.3.0. A p value of ⩽0.05 was considered significant.
Results
Development of a significantly milder form of pristane‐induced arthritis in DA.ACI(Cia25) rats
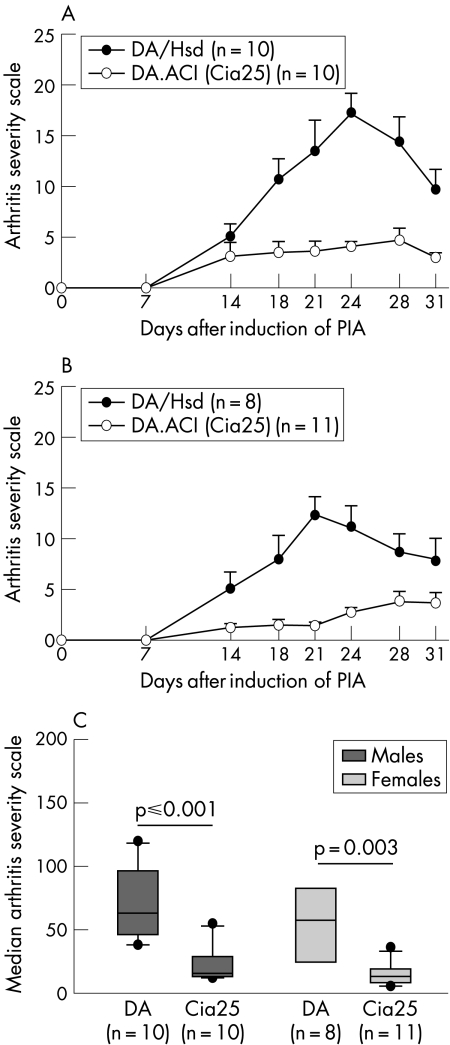
Male and female DA.ACI(Cia25) rats developed significantly milder pristane‐induced arthritis (PIA) compared with DA rats of the same gender (median ASI (25th–75th centile): DA males 63 (49–95), DA.ACI(Cia25) males 15 (13–26); p⩽0.001; DA females 57 (27–76); DA.ACI(Cia25) females 13 (8–18); p = 0.003; fig 2). ACI‐derived alleles at the Cia25 interval were associated with a 70% reduction in the median ASI of both male and female congenic rats with PIA.
Figure 2 Cia25 regulates the severity of pristane‐induced arthritis (PIA). (A) Male and (B) female DA.ACI(Cia25) rats (white circles) had significantly milder PIA than DA (black circles). Scores are plotted as mean (SEM). (C) The cumulative arthritis severity scores (ASI), which are known to correlate with histology damage, were significantly higher in DA rats compared with homozygotic DA.ACI.(Cia25) congenic rats (median ASI (25th–75th centile): DA males 63 (49–95), DA.ACI(Cia25) males 15 (13–26); p⩽0.001; DA females 57 (27–76); DA.ACI(Cia25) females 13 (8–18); p = 0.003, Mann–Whitney test). Boxes represent the 25th–75th centile, and error bars represent the 5th–95th centile.
Development of a significantly milder form of CIA in DA.ACI(Cia25) rats
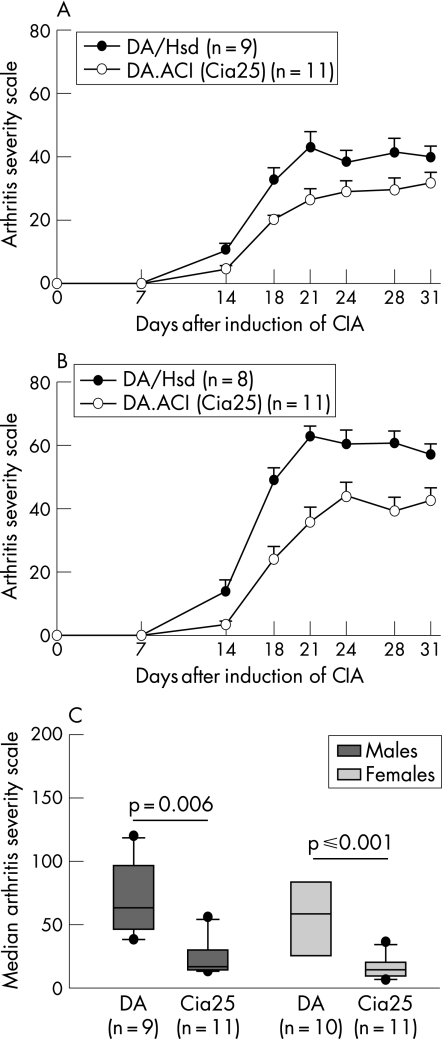
Male and female congenic rats developed significantly milder CIA compared with DA rats of the same gender (median ASI (25th–75th centile): DA males 226 (209–259), DA.ACI(Cia25) males 126 (116–166); p = 0.006; DA females 292.5 (234–330); DA.ACI(Cia25) females 208 (173–231);p⩽0.001; fig 3). The presence of ACI‐derived alleles at the Cia25 interval was associated with a 40% reduction in the median ASI of both male and female congenic rats with CIA.
Figure 3 Cia25 regulates the severity of collagen‐induced arthritis (CIA). (A) Male and (B) female DA.ACI(Cia25) rats (white circles) had significantly milder CIA compared with DA rats (black circles). Scores are plotted as mean (SEM). (C) The cumulative arthritis severity scores (ASI), which are known to correlate with histology damage, were significantly higher in DA rats, compared with homozygotic congenic rats (median ASI (25th–75th centile): DA males 211 (187–246), DA.ACI(Cia25) males 126 (116–166); p = 0.006; DA females 306 (263–340); DA.ACI(Cia25) females 208 (173–231); p⩽0.001). Boxes represent 25th–75th centile, and error bars represent the 5th–95th centile.
Regulation of disease severity throughout the course of arthritis by Cia25
The arthritis‐protective effect associated with ACI alleles at Cia25 was detected as early as 14 days after induction for both PIA (males and females combined, median score at day 14 (25th–75th centile): DA 8 (4–13), DA.ACI(Cia25) 1 (1–2); p⩽0.001) and CIA (males and females combined, median score at day 14 (25th–75th centile): DA 13 (3–18), DA.ACI(Cia25) 4 (2–6); p = 0.004), and persisted throughout the 31‐day observation period.
Autoantibodies against type II collagen
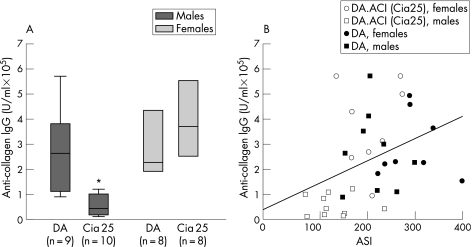
Autoantibodies against CII were detectable in DA and DA.ACI(Cia25) animals immunised with CII, indicating appropriate immunisation (fig 4A). Levels of IgG against CII in male DA.ACI(Cia25) rats were 83% lower than those in male DA (median IgG, mg/ml (25th–75th centile): DA 32.2 (13.9–44.8); DA.ACI(Cia25) 5.3 (2.2–12.2); p⩽0.001). Levels of IgG against CII in female DA.ACI(Cia25) rats were similar to those in DA (fig 4A). These observations suggest that Cia25 also contains a gene that operates in a male‐specific manner, where ACI alleles significantly reduce levels of autoantibodies against CII. Levels of anti‐collagen IgG significantly correlated with the ASI (Pearson's correlation coefficient = 0.365, p = 0.03, n = 35; fig 4B).
Figure 4 Cia25 influences the levels of anti‐collagen autoantibodies in males. Levels of anti‐collagen IgG were determined by ELISA in sera collected 18 days after immunisation with type II collagen (CII). (A) Levels of anti‐collagen autoantibodies in male DA.ACI(Cia25) (shown as Cia25) rats were 80% lower than in male DA, but no difference was observed in females. (B) Levels of anti‐collagen IgG correlated with the arthritis severity scores (ASI; Pearson's correlation coefficient = 0.365, p = 0.03, n = 35). *p⩽0.001, Mann–Whitney test.
Discussion
The study of intercrosses between rat inbred strains discordant for arthritis susceptibility and severity provides a powerful tool for the identification of loci regulating disease characteristics such as severity,20,33 chronicity20,23 and the production of rheumatoid factors.36 The phenotypic effect of loci identified in linkage studies can be definitively confirmed in congenic strains30,32,35,37,38,39 such as the one described herein, thus providing the basis for the gene identification effort.26 Findings from studies in rats can then be analysed in focused candidate gene RA case–control association studies.
We have previously mapped five QTLs regulating CIA in an intercross between DA and ACI rats,19,20 two strains that share the same arthritis‐favouring MHC RT1av1 haplotype, yet differ in susceptibility to, and severity of, disease. In the present study, the Cia25 locus was studied in DA.ACI(Cia25) congenic rats, which had significantly reduced severity of PIA and CIA compared with DA, confirming the presence of an arthritis‐regulatory gene in this locus. The cumulative arthritis severity score (ASI) used here was previously shown to correlate with the degree of histological synovial inflammation, and cartilage and bone erosive changes.32,34,35 Therefore, the protective effect of Cia25 on clinical disease severity suggests that this gene is also relevant to joint damage.
The arthritis‐protective effect associated with ACI alleles at Cia25 was significant in both models, but more pronounced in PIA. The difference in the magnitude of protection between PIA and CIA suggests that the Cia25 gene modulates pathways more directly relevant to PIA than to CIA.
Cia25 was originally identified as a female‐specific QTL in the DA×ACI F2 intercross studied for CIA.20 However, both male and female congenic rats had reduced CIA severity. Although discrepancy between studies in the F2 intercross and congenic rats remains incompletely understood, it has been described before.39 It is considered that epistatic interactions between ACI alleles at Cia25 and ACI alleles elsewhere in the genome of the F2 offspring could have been a factor. The congenic data demonstrating protection in both genders has in fact broader relevance, as it suggests that more patients could benefit from therapies targeting the Cia25 gene.
Both DA.ACI(Cia25) congenic rats and DA rats immunised with CII and IFA produced autoantibodies against CII, indicating adequate immunisation. Male DA.ACI(Cia25) congenic rats had a significant reduction of 83% in the levels of anti‐CII autoantibodies, showing that the presence of ACI alleles at Cia25 significantly suppresses levels of autoantibodies. This observation suggests that a gene within Cia25 affects pathways leading to the activation of autoreactive B cells and/or the production of antibodies. Levels of anti‐CII antibodies, particularly in males, correlated with disease severity, although in female DA.ACI(Cia25) congenic rats anti‐CII levels were similar to those in DA rats, and did not correlate with disease severity. Anti‐CII autoantibodies participate in the pathogenesis of CIA,40,41 and their levels are genetically regulated and affect arthritis severity, as we have observed in DA.F344(Cia5a) rats congenic for a chromosome 10 QTL.35 Therefore, these results raise the possibility that the Cia25 interval contains two arthritis severity genes, one of which regulates arthritis and autoantibodies in males, and the other operates via antibody‐independent mechanisms to regulate disease in females, and perhaps in males as well. Studies in recombinant subcongenic rats should determine whether two independent gender‐specific arthritis genes are in fact located within this interval.
Preliminary experiments suggest that Cia25 operates in a dominant manner (data not shown). This knowledge will be important during the process of reducing the critical interval and positionally cloning the Cia25 gene(s), since the arthritic phenotype will need to be screened in homozygotic recombinant subcongenic rats, and not in heterozygotic offspring.
The Cia25 22.3 Mb interval on rat chromosome 12 contains a number of potentially interesting candidate genes, such as Ccl24, Nos1, Ptpn11 and Ncf1. Ncf1 was previously found to account for Pia4.26 The same disease‐promoting and resistance Ncf1 polymorphisms described by Olofsson et al26 also segregate in the DA and ACI strains,20 and these variants are capable of modulating the arthritic phenotype in DA rats.42 Although the Ncf1 polymorphisms that we have previously reported20 probably account for part of the Cia25 effect detected in the congenic rats, we consider that there is yet a second arthritis severity gene within Cia25. Support for this hypothesis comes from two pieces of information. First, the difference in levels of anti‐CII autoantibodies between male DA and DA.ACI(Cia25) rats, which was not detected in females, suggests two different pathogenic processes. Second, several loci regulating different autoimmune diseases colocalise with Cia25, and there is a strong possibility that there will be a common gene accounting for all these loci, and Ncf1 does not seem to be the gene. Specifically, we have genotyped Ncf1 in the strains used in the identification of the Cia25 colocalising locus Eau2, which regulates experimental autoimmune uveitis (EAU).27 Both F344 (EAU‐resistant) and LEW (EAU‐susceptible) have identical alleles at Ncf1 codons 103 and 153 (Gulko et al, unpublished observations), and thus these arthritis regulatory single nucleotide polymorphisms do not explain the Eau2 locus. Therefore, it can be conceived that the same as yet unknown gene accounting for Eau2 will be the gene accounting for part of the Cia25 effect. Studies of recombinant subcongenic rats that exclude Ncf1 will be critical to confirm the existence of a second arthritis and autoimmunity gene within Cia25. We are in the process of generating these subcongenic rats.
Cia25 is of great interest, as it is in a region known to harbour several autoimmune disease‐regulatory loci in rats,27,28,29 in mouse chromosome 5,43,44,45 and in the human syntenic regions on chromosomes 7q, 12q and 22q46,47,48,49 genomes, including the arthritis loci Pia423 and Cia12.25 Taken together, these data further support the concept that the same gene may regulate arthritis and other autoimmune diseases in rats, mice and humans. Although this clustering of non‐MHC autoimmunity regulatory loci does not prove that these loci are accounted for by the same gene, it certainly raises that possibility.50 In fact, recent human studies have identified CTLA451,52 and PTPN2215,53,54 as two genes involved in the regulation of multiple autoimmune diseases, and Cia25 could be the third one.
In conclusion, we have demonstrated that Cia25 contains at least one gene that significantly influences arthritis severity. The identification of the Cia25 gene(s) has the potential to generate an important novel prognostic biomarker, and candidate gene, or candidate pathway, for focused case–control analyses in RA cohorts designed to study disease severity. Ultimately, the identification of the Cia25 gene(s) is expected to generate new targets for arthritis therapy.
Acknowledgements
We thank Bill Rall and Carl Hansen (National Institutes of Health; NIH) for their assistance with embryo re‐derivation of this strain.
Abbreviations
ASI - arthritis severity index
CIA - collagen‐induced arthritis
CII - type II collagen
EAU - experimental autoimmune uveitis
IFA - incomplete Freund's adjuvant
MHC - major histocompatibility complex
PIA - pristane‐induced arthritis
QTL - quantitative trait locus
RA - rheumatoid arthritis
Footnotes
Funding: This work was funded by NIH grants number R01‐AR46213, R01‐AR052439 (NIAMS) and R01‐AI54348 (NIAID) to PSG.
Competing interests: None.
References
- 1.Seldin M F, Amos C I, Ward R, Gregersen P K. The genetics revolution and the assault on rheumatoid arthritis. Arthritis Rheum 1999421071–1079. [DOI] [PubMed] [Google Scholar]
- 2.MacGregor A J, Snieder H, Rigby A S, Koskenvuo M, Kaprio J, Aho K.et al Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum 20004330–37. [DOI] [PubMed] [Google Scholar]
- 3.Moreland L W, Baumgartner S W, Schiff M H, Tindall E A, Fleischmann R M, Weaver A L.et al Treatment of rheumatoid arthritis with a recombinant human tumor necrosis factor receptor (p75)‐Fc fusion protein. N Engl J Med 1997337141–147. [DOI] [PubMed] [Google Scholar]
- 4.Genovese M C, Becker J C, Schiff M, Luggen M, Sherrer Y, Kremer J.et al Abatacept for rheumatoid arthritis refractory to tumor necrosis factor alpha inhibition. N Engl J Med 20053531114–1123. [DOI] [PubMed] [Google Scholar]
- 5.Edwards J C, Szczepanski L, Szechinski J, Filipowicz‐Sosnowska A, Emery P, Close D R.et al Efficacy of B‐cell‐targeted therapy with rituximab in patients with rheumatoid arthritis. N Engl J Med 20043502572–2581. [DOI] [PubMed] [Google Scholar]
- 6.Jawaheer D, Seldin M F, Amos C I, Chen W V, Shigeta R, Etzel C.et al Screening the genome for rheumatoid arthritis susceptibility genes: a replication study and combined analysis of 512 multicase families. Arthritis Rheum 200348906–916. [DOI] [PubMed] [Google Scholar]
- 7.Jawaheer D, Seldin M F, Amos C I, Chen W V, Shigeta R, Monteiro J.et al A genomewide screen in multiplex rheumatoid arthritis families suggests genetic overlap with other autoimmune diseases. Am J Hum Genet 200168927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Etzel C J, Chen W V, Shepard N, Jawaheer D, Cornelis F, Seldin M F.et al Genomewide meta‐analysis for rheumatoid arthritis. Hum Genet 2006119634–641. [DOI] [PubMed] [Google Scholar]
- 9.Shiozawa S, Hayashi S, Tsukamoto Y, Goko H, Kawasaki H, Wada T.et al Identification of the gene loci that predispose to rheumatoid arthritis. Int Immunol 1998101891–1895. [DOI] [PubMed] [Google Scholar]
- 10.Osorio y Fortea J, Bukulmez H, Petit‐Teixeira E, Michou L, Pierlot C, Cailleau‐Moindrault S.et al Dense genomewide linkage analysis of rheumatoid arthritis, including covariates. Arthritis Rheum 2004502757–2765. [DOI] [PubMed] [Google Scholar]
- 11.Eyre S, Barton A, Shephard N, Hinks A, Brintnell W, MacKay K.et al Investigation of susceptibility loci identified in the UK rheumatoid arthritis whole‐genome scan in a further series of 217 UK affected sibling pairs. Arthritis Rheum 200450729–735. [DOI] [PubMed] [Google Scholar]
- 12.John S, Shephard N, Liu G, Zeggini E, Cao M, Chen W.et al Whole‐genome scan, in a complex disease, using 11,245 single‐nucleotide polymorphisms: comparison with microsatellites. Am J Hum Genet 20047554–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M.et al Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet 200334395–402. [DOI] [PubMed] [Google Scholar]
- 14.Okamoto K, Makino S, Yoshikawa Y, Takaki A, Nagatsuka Y, Ota M.et al Identification of I kappa BL as the second major histocompatibility complex‐linked susceptibility locus for rheumatoid arthritis. Am J Hum Genet 200372303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Begovich A B, Carlton V E, Honigberg L A, Schrodi S J, Chokkalingam A P, Alexander H C.et al A missense single‐nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet 200475330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mau W, Bornmann M, Weber H, Weidemann H F, Hecker H, Raspe H H. Prediction of permanent work disability in a follow‐up study of early rheumatoid arthritis: results of a tree structured analysis using RECPAM. Br J Rheumatol 199635652–659. [DOI] [PubMed] [Google Scholar]
- 17.Mottonen T, Paimela L, Leirisalo‐Repo M, Kautiainen H, Ilonen J, Hannonen P. Only high disease activity and positive rheumatoid factor indicate poor prognosis in patients with early rheumatoid arthritis treated with “sawtooth” strategy. Ann Rheum Dis 199857533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pincus T, Brooks R H, Callahan L F. Prediction of long‐term mortality in patients with rheumatoid arthritis according to simple questionnaire and joint count measures. Ann Intern Med 199412026–34. [DOI] [PubMed] [Google Scholar]
- 19.Gulko P S, Kawahito Y, Remmers E F, Reese V R, Wang J, Dracheva S V.et al Identification of a new non‐major histocompatibility complex genetic locus on chromosome 2 that controls disease severity in collagen‐induced arthritis in rats. Arthritis Rheum 1998412122–2131. [DOI] [PubMed] [Google Scholar]
- 20.Meng H C, Griffiths M M, Remmers E F, Kawahito Y, Li W, Neisa R.et al Identification of two novel female‐specific non‐major histocompatibility complex loci regulating collagen‐induced arthritis severity and chronicity, and evidence of epistasis. Arthritis Rheum 2004502695–2705. [DOI] [PubMed] [Google Scholar]
- 21.Kawahito Y, Cannon G, Gulko P, Remmers E, Longman R, Reese V.et al Localization of quantitative trait loci regulating adjuvant induced arthritis in rats: evidence for genetic factors common to multiple autoimmune diseases. J Immunol 19981614411–4419. [PubMed] [Google Scholar]
- 22.Lorentzen J C, Glaser A, Jacobsson L, Galli J, Fakhrai‐rad H, Klareskog L.et al Identification of rat susceptibility loci for adjuvant‐oil‐induced arthritis. Proc Natl Acad Sci USA 1998956383–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vingsbo‐Lundberg C, Nordquist N, Olofsson P, Sundvall M, Saxne T, Pettersson U.et al Genetic control of arthritis onset, severity and chronicity in a model for rheumatoid arthritis in rats. Nat Genet 199820401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furuya T, Salstrom J L, McCall‐Vining S, Cannon G W, Joe B, Remmers E F.et al Genetic dissection of a rat model for rheumatoid arthritis: significant gender influences on autosomal modifier loci. Hum Mol Genet 200092241–2250. [DOI] [PubMed] [Google Scholar]
- 25.Griffiths M M, Wang J, Joe B, Dracheva S, Kawahito Y, Shepard J S.et al Identification of four new quantitative trait loci regulating arthritis severity and one new quantitative trait locus regulating autoantibody production in rats with collagen‐induced arthritis. Arthritis Rheum 2000431278–1289. [DOI] [PubMed] [Google Scholar]
- 26.Olofsson P, Holmberg J, Tordsson J, Lu S, Akerstrom B, Holmdahl R. Positional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat Genet 20033325–32. [DOI] [PubMed] [Google Scholar]
- 27.Sun S H, Silver P B, Caspi R R, Du Y, Chan C C, Wilder R L.et al Identification of genomic regions controlling experimental autoimmune uveoretinitis in rats. Int Immunol 199911529–534. [DOI] [PubMed] [Google Scholar]
- 28.Bergsteinsdottir K, Yang H T, Pettersson U, Holmdahl R. Evidence for common autoimmune disease genes controlling onset, severity, and chronicity based on experimental models for multiple sclerosis and rheumatoid arthritis. J Immunol 20001641564–1568. [DOI] [PubMed] [Google Scholar]
- 29.Dahlman I, Jacobsson L, Glaser A, Lorentzen J C, Andersson M, Luthman H.et al Genomewide linkage analysis of chronic relapsing experimental autoimmune encephalomyelitis in the rat identifies a major susceptibility locus on chromosome 9. J Immunol 19991622581–2588. [PubMed] [Google Scholar]
- 30.Joe B, Remmers E F, Dobbins D E, Salstrom J L, Furuya T, Dracheva S.et al Genetic dissection of collagen‐induced arthritis in chromosome 10 quantitative trait locus speed congenic rats: evidence for more than one regulatory locus and sex influences. Immunogenetics 200051930–944. [DOI] [PubMed] [Google Scholar]
- 31.Vingsbo C, Sahlstrand P, Brun J G, Jonsson R, Saxne T, Holmdahl R. Pristane‐induced arthritis in rats: a new model for rheumatoid arthritis with a chronic disease course influenced by both major histocompatibility complex and non‐major histocompatibility complex genes. Am J Pathol 19961491675–1683. [PMC free article] [PubMed] [Google Scholar]
- 32.Brenner M, Meng H C, Yarlett N C, Griffiths M M, Remmers E F, Wilder R L.et al The non‐major histocompatibility complex quantitative trait locus Cia10 contains a major arthritis gene and regulates disease severity, pannus formation, and joint damage. Arthritis Rheum 200552322–332. [DOI] [PubMed] [Google Scholar]
- 33.Remmers E F, Longman R E, Du Y, O'Hare A, Cannon G W, Griffiths M M.et al A genome scan localizes five non‐MHC loci controlling collagen‐induced arthritis in rats. Nat Genet 19961482–85. [DOI] [PubMed] [Google Scholar]
- 34.Brenner M, Braun C, Oster M, Gulko P S. Thermal signature analysis as a novel method for evaluating inflammatory arthritis activity. Ann Rheum Dis 200665306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner M, Meng H C, Yarlett N C, Joe B, Griffiths M M, Remmers E F.et al The non‐MHC quantitative trait locus Cia5 contains three major arthritis genes that differentially regulate disease severity, pannus formation, and joint damage in collagen‐ and pristane‐induced arthritis. J Immunol 20051747894–7903. [DOI] [PubMed] [Google Scholar]
- 36.Wernhoff P, Olofsson P, Holmdahl R. The genetic control of rheumatoid factor production in a rat model of rheumatoid arthritis. Arthritis Rheum 2003483584–3596. [DOI] [PubMed] [Google Scholar]
- 37.Wester L, Olofsson P, Ibrahim S M, Holmdahl R. Chronicity of pristane‐induced arthritis in rats is controlled by genes on chromosome 14. J Autoimmun 200321305–313. [DOI] [PubMed] [Google Scholar]
- 38.Remmers E F, Joe B, Griffiths M M, Dobbins D E, Dracheva S V, Hashiramoto A.et al Modulation of multiple experimental arthritis models by collagen‐induced arthritis quantitative trait loci isolated in congenic rat lines: different effects of non‐major histocompatibility complex quantitative trait loci in males and females. Arthritis Rheum 2002462225–2234. [DOI] [PubMed] [Google Scholar]
- 39.Holm B C, Wei Xu H, Jacobsson L, Larsson A, Luthman H, Lorentzen J C. Rats made congenic for Oia3 on chromosome 10 become susceptible to squalene‐induced arthritis. Hum Mol Genet 200110565–572. [DOI] [PubMed] [Google Scholar]
- 40.Takagishi K, Kaibara N, Hotokebuchi T, Arita C, Morinaga M, Arai K. Serum transfer of collagen arthritis in congenitally athymic nude rats. J Immunol 19851343864–3867. [PubMed] [Google Scholar]
- 41.Nandakumar K S, Svensson L, Holmdahl R. Collagen type II‐specific monoclonal antibody‐induced arthritis in mice: description of the disease and the influence of age, sex, and genes. Am J Pathol 20031631827–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gelderman K A, Hultqvist M, Holmberg J, Olofsson P, Holmdahl R. T cell surface redox levels determine T cell reactivity and arthritis susceptibility. Proc Natl Acad Sci USA 200610312831–12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weis J J, McCracken B A, Ma Y, Fairbairn D, Roper R J, Morrison T B.et al Identification of quantitative trait loci governing arthritis severity and humoral responses in the murine model of Lyme disease. J Immunol 1999162948–956. [PubMed] [Google Scholar]
- 44.Kono D H, Burlingame R W, Owens D G, Kuramochi A, Balderas R S, Balomenos D.et al Lupus susceptibility loci in New Zealand mice. Proc Natl Acad Sci USA 19949110168–10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahler M, Bristol I J, Sundberg J P, Churchill G A, Birkenmeier E H, Elson C O.et al Genetic analysis of susceptibility to dextran sulfate sodium‐induced colitis in mice. Genomics 199955147–156. [DOI] [PubMed] [Google Scholar]
- 46.Spritz R A, Gowan K, Bennett D C, Fain P R. Novel vitiligo susceptibility loci on chromosomes 7 (AIS2) and 8 (AIS3), confirmation of SLEV1 on chromosome 17, and their roles in an autoimmune diathesis. Am J Hum Genet 200474188–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Satsangi J, Parkes M, Louis E, Hashimoto L, Kato N, Welsh K.et al Two stage genomewide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet 199614199–202. [DOI] [PubMed] [Google Scholar]
- 48.Xu C, Dai Y, Fredrikson S, Hillert J. Association and linkage analysis of candidate chromosomal regions in multiple sclerosis: indication of disease genes in 12q23 and 7ptr‐15. Eur J Hum Genet 19997110–116. [DOI] [PubMed] [Google Scholar]
- 49.Gaffney P M, Ortmann W A, Selby S A, Shark K B, Ockenden T C, Rohlf K E.et al Genome screening in human systemic lupus erythematosus: results from a second Minnesota cohort and combined analyses of 187 sib‐pair families. Am J Hum Genet 200066547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Becker K G, Simon R M, Bailey‐Wilson J E, Freidlin B, Biddison W E, McFarland H F.et al Clustering of non‐major histocompatibility complex susceptibility candidate loci in human autoimmune diseases. Proc Natl Acad Sci USA 1998959979–9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueda H, Howson J M, Esposito L, Heward J, Snook H, Chamberlain G.et al Association of the T‐cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003423506–511. [DOI] [PubMed] [Google Scholar]
- 52.Plenge R M, Padyukov L, Remmers E F, Purcell S, Lee A T, Karlson E W.et al Replication of putative candidate‐gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet 2005771044–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M.et al A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 200436337–338. [DOI] [PubMed] [Google Scholar]
- 54.Kyogoku C, Langefeld C D, Ortmann W A, Lee A, Selby S, Carlton V E.et al Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet 200475504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]