Abstract
A universally conserved event in cell division is the formation of a cytokinetic ring at the future site of division. In the bacterium Escherichia coli, this ring is formed by the essential cell division protein FtsZ. We have used immunofluorescence microscopy to show that FtsZ assembles early in the division cycle, suggesting that constriction of the FtsZ ring is regulated and supporting the view that FtsZ serves as a bacterial cytoskeleton. Assembly of FtsZ rings was heterogeneously affected in an ftsI temperature-sensitive mutant grown at the nonpermissive temperature, some filaments displaying a striking defect in FtsZ assembly and others displaying little or no defect. By using low concentrations of the β-lactams cephalexin and piperacillin to specifically inhibit FtsI (PBP3), an enzyme that synthesizes peptidoglycan at the division septum, we show that FtsZ ring constriction requires the transpeptidase activity of FtsI. Unconstricted FtsZ rings are stably trapped at the midpoint of the cell for several generations after inactivation of FtsI, whereas partially constricted FtsZ rings are less effectively trapped. In addition, FtsZ rings are able to assemble in newborn cells in the presence of cephalexin, suggesting that newborn cells contain a site at which FtsZ can assemble (the nascent division site) and that the transpeptidase activity of FtsI is not required for assembly of FtsZ at these sites. However, aside from this first round of FtsZ ring assembly, very few additional FtsZ rings assemble in the presence of cephalexin, even after several generations of growth. One interpretation of these results is that the transpeptidase activity of FtsI is required, directly or indirectly, for the assembly of nascent division sites and thereby for future assembly of FtsZ rings.
During the division cycle of Escherichia coli, a cytokinetic ring is formed at the future site of division by the essential cell division protein FtsZ (1). FtsZ is highly conserved among the Eubacteria (1) and Archaea (2, 3) and has also been found in plant chloroplasts (4). Like tubulin, a distant homologue of FtsZ, FtsZ self-assembles into protofilaments in vitro and has an essential GTPase activity (5–11). FtsZ forms a ring structure at the future division site that remains at the leading edge of the invaginating septum (12). The FtsZ ring may serve as a contractile scaffold for assembly of other Fts proteins or may play a more active role in providing the force necessary for contraction, perhaps using energy from GTP hydrolysis. FtsZ ring assembly is likely to be highly regulated: assembly of the FtsZ ring is inhibited by SulA under conditions of DNA damage (13, 14), and the site of division, and thus the site of FtsZ assembly, is at least partly controlled by proteins encoded by the minCDE locus (15–17). However, very little is known about the structure of the FtsZ ring, the signals that initiate septum formation, or the relationship between FtsZ assembly and the other known cell division proteins (FtsA, FtsI, FtsQ, FtsL, FtsN, FtsK, and FtsW) (15, 18–23).
At the time of septation, cells must activate peptidoglycan synthesis at the division septum coordinately with constriction of the FtsZ ring and the rest of the cell envelope. Constriction of the cell envelope requires FtsI, also known as PBP3, a transpeptidase that catalyzes formation of peptide crosslinks between peptidoglycan strands in the division septum (18, 24, 25). The penicillins cephalexin and piperacillin irreversibly and specifically inactivate the transpeptidase active site of FtsI (24, 26, 27). FtsI can also be inactivated by temperature-sensitive (Ts) mutations in the ftsI gene (28). Cells without functional FtsI continue to elongate and replicate their chromosomes but fail to divide, forming multinucleate filaments. FtsI is a membrane protein with a small cytoplasmic domain, a single membrane-spanning segment, and a periplasmic domain that encodes the transpeptidase activity. FtsI is widely believed to function after FtsZ (18). For example, electron microscopy studies have shown that filaments formed by ftsI mutants have blunt constrictions, indicating that constriction is initiated but not completed. Filaments from ftsZ mutants, on the other hand, are “smooth” and lack partially constricted septa (18, 29). These results have been interpreted to mean that FtsZ is needed to initiate constriction, whereas FtsI is only needed after initiation. Furthermore, results using a radioactive peptidoglycan precursor, meso-[3H]diaminopimelic acid, to examine the topography of peptidoglycan synthesis when FtsZ or FtsI are inactivated support these conclusions (30). Finally, genetic studies using cell shape mutants have also suggested that FtsI acts after FtsZ (31).
FtsZ has been localized by immunofluorescence microscopy (IFM) in Bacillus subtilis where, during sporulation, its localization changes from the midpoint to the cell poles, coincident with a switch from medial to polar septation (32). This altered localization requires the SpoOA transcription factor and, presumably, proteins whose synthesis requires SpoOA. FtsZ has also been localized by IFM in fts mutants of E. coli, in which it was concluded that the ftsA, ftsQ, and ftsI gene products play no role in the assembly of the FtsZ ring (33). In this study, we have characterized the localization of FtsZ by using IFM in wild-type E. coli cells and in cells in which FtsI has been inactivated by either a Ts mutation or by the FtsI-specific β-lactams cephalexin and piperacillin. We show that FtsZ rings form soon after the birth of the cell and that the ftsI mutation had a highly variable effect on FtsZ ring formation in different filaments from the same culture. FtsZ ring formation in newborn cells was not inhibited by cephalexin, but filaments formed by cephalexin treatment had fewer than the expected number of FtsZ rings. These results indicate that while FtsI is not required for assembly of FtsZ in newborn cells, it may be required for the subsequent assembly of additional FtsZ rings.
MATERIALS AND METHODS
Immunolocalization of FtsZ.
Bacterial cultures were grown in Luria–Bertani (LB) broth at 37°C for the wild-type strains MG1655 and MC4100 (Fig. 1 A–D and K–R), and at 30°C for the Ts strains MM62 (ftsZ84) and LMG64 (ftsI23). The Ts strains were shifted to the nonpermissive temperature (42°C) for three generations prior to fixation. The transpeptidase activity of FtsI was inhibited by the addition of cephalexin (10 μg/ml, final concentration) or piperacillin (2 μg/ml, final concentration) to their minimal inhibitory concentrations. Both drugs had identical effects on cell division and FtsZ localization. The drugs were added to cultures at an OD600 of about 0.1, and the cultures were allowed to undergo one, two, or three doublings. All cultures were fixed directly in growth medium in a final concentration of 2.6% paraformaldehyde, 0.006% glutaraldehyde, and 40 mM sodium phosphate (pH 7.5). Samples were processed for immunofluorescence microscopy as described (34), except that due to the increased sensitivity of E. coli cells to lysozyme as compared with B. subtilis, a much lower concentration of lysozyme was used (1:1000 dilution) and to optimize the results it was necessary to titrate the lysozyme concentration in each experiment. The fixed cells were treated for 5 min at room temperature with lysozyme at 0.5, 1.0, and 2.0 μg/ml (final concentrations). The samples were incubated at 4°C overnight with a 1:5000 dilution of FtsZ-specific antisera (a gift of Joe Lutkenhaus, University of Kansas Medical Center). Fluorescein-conjugated anti-rabbit secondary antibodies (Jackson ImmunoResearch) were used at a 1:100 dilution, and the nucleoids were stained with propidium iodide (Molecular Probes) at 0.1 mg/ml. The images were recorded on Ektachrome 400 film, and the resulting slides were scanned using a Nikon CoolScan and processed using Adobe photoshop.
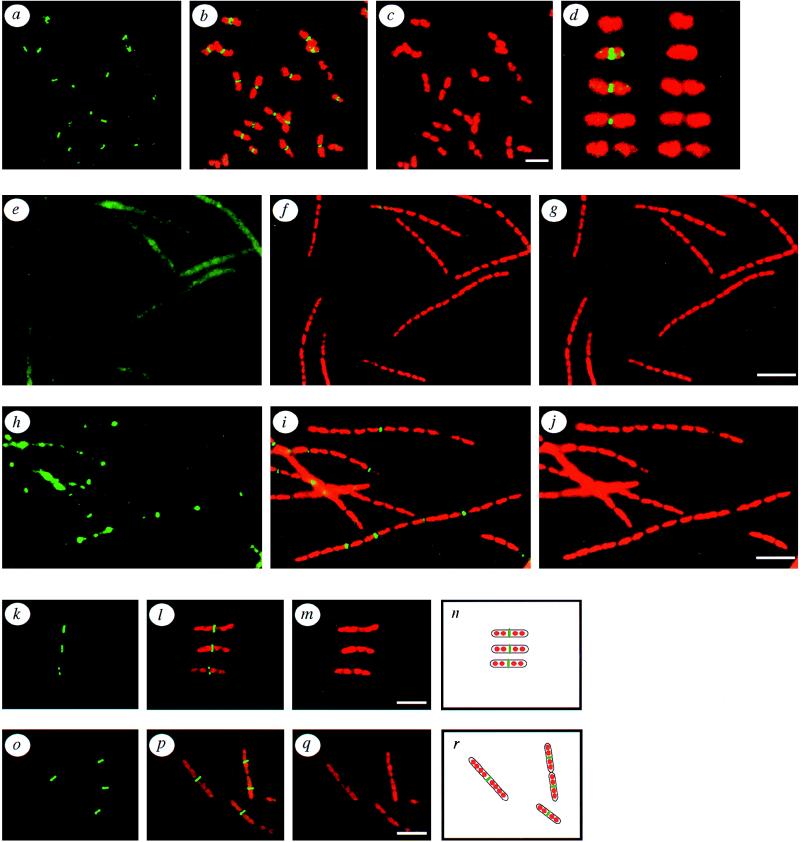
Figure 1.
Subcellular localization of FtsZ in wild-type, ftsZ84, ftsI23, and cephalexin-treated E. coli. (a–c) Localization of FtsZ (green) in exponentially growing wild-type E. coli. (a) FtsZ immunostaining (green) alone. (b) Doubly exposed micrograph of the same field showing both FtsZ (green) and the propidium iodide-stained nucleoids (red). (c) The propidium iodide-stained nucleoids alone. (d) A series of cells depicting FtsZ assembly states during the E. coli division cycle, starting from a newborn cell lacking an FtsZ ring (top) and ending with a fully constricted but as yet unseparated pair of newborn cells. The left column of cells show both FtsZ protein (green) and the nucleoids (red), while the right column of cells show the nucleoids only (red). (e–g) Localization of FtsZ in an ftsZ84 mutant at the nonpermissive temperature. (e) FtsZ staining (green) alone. (f) A double exposure of the same field of cells showing FtsZ (green) and the nucleoids (red). (g) Propidium iodine-stained nucleoids. (h–j) Localization of FtsZ in the ftsI23 mutant at the nonpermissive temperature. (h) FtsZ alone (green). (i) FtsZ (green) and the nucleoids (red). (j) Nucleoids alone (red). (k–m) Localization of FtsZ in cells treated with cephalexin for two generatations. (k) FtsZ staining (green) alone. (l) FtsZ (green) and the nucleoids (red). (m) Nucleoids alone (red). (n) Diagram of the cells in k–m, showing the inferred outlines of the cells (black), and the positions of the nucleoids (red), and the FtsZ rings (green). (o–q) A different field of cells from the same experiment as in k–m, showing additional patterns of FtsZ localization in filaments after two generations of growth in the presence of cephalexin. (o) FtsZ (green) alone. (p) A double exposure showing both FtsZ (green) and the nucleoids (red). (q) Nucleoids alone (red). (r) A diagram of the cells in o–q, showing the inferred outlines of the cells (black), and the positions of the nucleoids (red) and the FtsZ rings (green). (Bar = 5 μm.)
Calculation of Age of Appearance of FtsZ Rings.
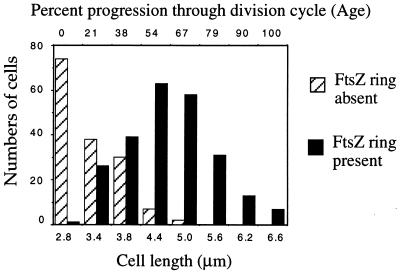
Individual cells (389 cells) from several fields were measured and scored for the presence or absence of an FtsZ ring. The numbers of cells with or without FtsZ rings is plotted versus cell length. Cell length increases exponentially during the division cycle and was used to estimate cell age, which is described as percent progression through the division cycle (35). The shortest cells (newborns) consistently (99%) lacked a visible FtsZ ring. By 20% of the way through the division cycle, a significant fraction (40%) of the cells contained a detectable FtsZ ring; presumably, FtsZ ring formation begins even earlier. Approximately 90% of the cells estimated to be midway through the division cycle contained an FtsZ ring. Rarely (<1%), cells appeared with a small dot of fluorescence at their midpoints. While these fluorescent spots could represent FtsZ assembly intermediates, such cells were scored as lacking an FtsZ ring. The relationship between length and age is not perfect; there is a 20% coefficient of variation in both the size at which division occurs and the size of newborn cells (35). Therefore, we also estimated the age at which FtsZ rings form by a method that does not rely on cell length measurements but instead assumes that the culture was in balanced growth and that all cell interdivision times were equal. The age structure of such an idealized population can be described by the equation F(x) = (ln 2)(2)(1−x) (36). F(x) is the relative frequency of cells of any given age x, ranging from 0 for newborns to 1.0 for nearly divided cells. Since FtsZ rings appeared at a certain age, x, and persisted until age 1.0, the integral ∫F(x)dx evaluated between the limits x and 1.0 is equal to the fraction of cells containing an FtsZ ring. Integrating by substitution gives ∫F(x)dx = −2(1−x)|x1.0. We observed that 238 out of 389 cells, or 61%, had an FtsZ ring (Fig. 2). The value (age) x that satisfies 0.61 = −2(1−x)|x1.0 is 0.3, indicating that FtsZ rings formed about one-third of the way through the division cycle. Early assembly was not observed by immunoelectron microscopy, probably because immunoelectron microscopy is much less sensitive than IFM (34). Our analysis is based on strain MG1655 growing in LB broth at 37°C with a doubling time of 22 min. Similar results were obtained with another commonly used wild-type strain, MC4100, although it is possible that the timing of FtsZ ring formation will vary between strains or growth conditions.
Figure 2.
Cell size distribution of FtsZ rings. Individual wild-type (MG1655) cells (389) from several fields were measured and scored for the presence or absence of an FtsZ ring. The numbers of cells with (solid bars) or without (hatched bars) FtsZ rings is plotted versus cell length. Cell length was used to estimate cell age, which is described as percent progression through the division cycle (35). The fraction of cells in each length class containing an FtsZ ring ranged from 1% for the shortest cells (newborns) to 100% for the longer cells.
Immunoprecipitation of FtsZ.
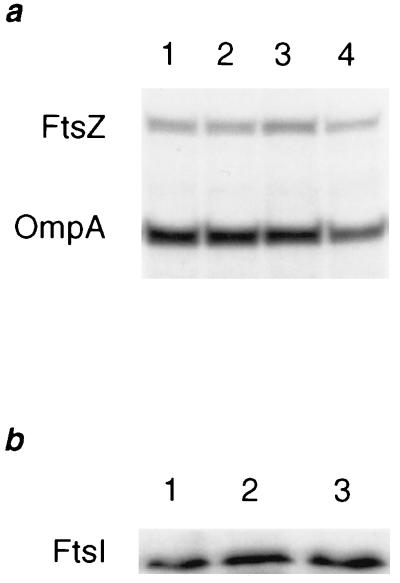
Cephalexin (10 μg/ml) was added to one of two parallel cultures of MC4100 growing exponentially in minimal medium supplemented with 18 amino acids at 37°C at an OD600 of ≈0.05. After two generations of growth with (see Fig. 5a, lanes 3 and 4) or without (see Fig. 5a, lanes 1 and 2) cephalexin (10 μg/ml), cultures were pulse-labeled with [35S]methionine for 20 sec and then chased with excess unlabeled methionine for 1 min (lanes 1 and 3) or 15 min (lanes 2 and 4). Samples were taken and incubated in ice-cold 10% trichloroacetic acid for 30 min, centifuged for 20 min at 4°C, and resuspended in 110 mM Tris (pH 8.8). FtsZ and OmpA were then immunoprecipitated and separated by SDS/PAGE in 10% gels as described (37). The counts in the FtsZ and OmpA bands were quantitated using a Molecular Dynamics PhosphorImager, and FtsZ counts were normalized to OmpA counts. In all experiments using cephalexin or piperacillin, cultures were examined by light microscopy to confirm the effects on cell division. Cells began to lyse after three generations of growth in the presence of these drugs, interefering with the examination of the effect of these drugs on FtsZ ring assembly after prolonged treatment.
Figure 5.
Cephalexin does not affect FtsZ or FtsI protein levels. (a) Pulse–chase immunoprecipitation analysis of FtsZ in cultures grown in the absence of β-lactam antibiotics (lanes 1 and 2) or grown for two generations in the presence of cephalexin (lanes 3 and 4). Radiolabeled FtsZ and OmpA, an internal control protein, were immunoprecipitated after a 1-min chase (lanes 1 and 3) and a 15-min chase (lanes 2 and 4). After quantitation of the counts in each band, the normalized value for each FtsZ band was 1.4, 1.3, 1.8, and 1.7 (arbitrary units) for lanes 1–4, respectively, indicating that cephalexin does not affect the rates of synthesis of FtsZ (lanes 1 and 3) or its stability (lanes 2 and 4). (b) Immunoblot analysis of FtsI after three generations of growth in the presence of cephalexin (lane 2) or piperacillin (lane 3) or without drugs (lane 1). The normalized values for the FtsI band were 0.69, 0.84, and 0.78 (arbitrary units) for lanes 1–3, respectively, indicating that cephalexin and piperacillin did not affect FtsI protein levels in cells grown in their presence for two (data not shown) or three (shown) generations.
Western Blot Analysis of FtsI.
A culture of MC4100 growing exponentially in LB medium at 37°C was split into three flasks containing either no drugs, cephalexin (10 μg/ml), or piperacillin (2 μg/ml). After two (data not shown) or three generations of growth, samples were taken, pelleted by centrifugation, and resuspended in SDS sample buffer, and boiled for 5 min. Equivalent cell masses (0.05 OD600 unit) were loaded onto a 10% polyacrylamide/SDS gel and proteins were separated by electrophoresis prior to transfer to nitrocellulose. Immunodetection of FtsI was performed with the ECL Western blotting system (Amersham) and a rabbit polyclonal serum (a gift of Luz-Maria Guzman, Harvard Medical School) raised against a synthetic FtsI peptide. For quantitation, film was scanned with a computing densitometer (Molecular Dynamics) and counts in the FtsI band were normalized to an internal control band (data not shown) to correct for any sampling variation.
RESULTS AND DISCUSSION
Division Cycle Assembly of FtsZ in Wild-Type Cells.
Exponentially growing cells of E. coli were rapidly fixed (34) and incubated with primary antibodies specific for FtsZ followed by fluorescein-conjugated secondary antibodies (green) and with propidium iodide to stain nucleoids (red). We could readily detect FtsZ rings, which appeared as green bands, at the midpoints of most cells (Fig. 1 a–c). In some cells, the FtsZ ring was constricting (green dot), while other cells had only a diffuse nonlocalized pattern of FtsZ immunostaining. Occasionally cells were oriented such that the ring structure of FtsZ could be seen (data not shown). To confirm that the pattern of localized staining was due to the FtsZ protein, we localized FtsZ in an ftsZ84 Ts mutant. When grown at the nonpermissive temperature, Fts mutants fail to divide and produce long multinucleate filaments. As expected, no FtsZ rings were detectable in filaments formed by the ftsZ84 mutant (Fig. 1 e–g).
A series of cells that represent a progression through the division cycle is shown in Fig. 1d. Two statistical methods were used to estimate when in the division cycle FtsZ assembles (Fig. 2 and Materials and Methods); both indicate that FtsZ ring formation occurs about one-third of the way through the division cycle. Cells inferred to be newborns, based on their length, rarely contained detectable FtsZ rings. Early in the division cycle, FtsZ rings appeared and remained assembled until the onset of septation. These results confirm that FtsZ undergoes a cycle of assembly and disassembly during the division cycle and that the FtsZ ring is assembled early in the division cycle. A similar result was obtained in E. coli and B. subtilis, in which FtsZ rings were observed in most cells (32, 33). The early assembly of the FtsZ ring in both E. coli and B. subtilis implies that its constriction is regulated subsequent to its assembly and that this regulation is conserved among the eubacteria. Further evidence for the regulation of constriction was found in B. subtilis sporulation, in which two apparently identical FtsZ rings are formed but only one constricts during normal development (32).
From our characterization of FtsZ localization in wild-type cells, we could not determine when the assembly of FtsZ rings begins. We note that assembled FtsZ rings were not detected in nascent daughter cells and were rarely detected in newborn cells. This suggests that the FtsZ ring does not fully assemble before cell separation. However, partially assembled rings containing only a few hundred molecules of FtsZ may not be detectable above the background of unassembled FtsZ protein. Therefore, the beginning of the assembly process may occur at any time prior to when fully assembled FtsZ rings appear.
FtsZ Ring Formation in ftsI Mutants.
To gain further insight into how and when FtsZ rings assemble, we investigated the effect of inactivating FtsI on FtsZ ring assembly dynamics. First, we localized FtsZ in the ftsI23 Ts mutant strain in parallel to its isogenic control after growth at either the permissive temperature or after three mass doublings at the nonpermissive temperature (Fig. 1 h–j). Unlike the ftsZ84 Ts mutant, ftsI mutants displayed a heterogeneous population of filaments. Some filaments had only one or two FtsZ rings despite having 16 nucleoids, while other filaments of similar length contained many more (four to eight) FtsZ rings (Fig. 1 h–j and data not shown). Clearly, FtsZ rings form at only a fraction of the potential division sites in many of the filaments formed by the Ts mutant at 42°C, raising the possibility that FtsI is involved in FtsZ ring assembly. These results are similar to those recently reported, in which 10% of the filaments contained no FtsZ rings, while another 10% of the filaments contained six FtsZ rings (33).
Establishing the role that FtsI plays in FtsZ ring dynamics was not possible with the ftsI mutant, due to the heterogeneity of the filaments. Furthermore, the pattern of FtsZ rings in these filaments appeared to be random, making it difficult to distinguish new FtsZ rings from old FtsZ rings. One possible interpretation of these results is that inactivation of FtsI prevents FtsZ rings from forming and that residual FtsI activity allows some FtsZ rings to assemble. An alternative interpretation is that FtsI plays no role in FtsZ ring assembly, as suggested by the filaments with multiple FtsZ rings assembled. However, this interpretation fails to explain why 10% of the filaments have no FtsZ ring and another 10% have only one FtsZ ring. Although FtsZ rings could assemble normally and then be unstable, this possibility cannot be distinguished from a defect in new ring assembly due to the random placement of FtsZ rings in the Ts mutant and the consequent inability to distinguish new FtsZ rings from old FtsZ rings.
A further difficulty is that the effect of the ftsI mutation on FtsI protein is unknown. Some Ts mutants cause the inactivity or instability of the mutant protein at the nonpermissive temperature, thereby causing the rapid inactivation of the mutant protein. Others cause a Ts defect in folding of the mutant protein and require several generations at the nonpermissive temperature before the protein is completely inactivated, as the functional protein synthesized at the permissive temperature must be replaced by nonfunctional protein. With either kind of mutation, if the protein is only partially inactivated by the mutation, if very little functional protein is required, or if the mutation affects only one of several functions of the protein, then studies to determine the precise function of the protein will be difficult to interpret. In our analysis, these effects could cause the variable numbers of FtsZ rings in ftsI Ts mutant filaments. An additional class of conditional mutations does not conditionally affect the protein whose function they impair but rather affect a protein whose activity is more essential at the nonpermissive temperature. Such mutants include the htr genes of E. coli, whose function is only required at high temperature (38, 39); the secG, secD, and secF genes (40–42), whose products are essential for viability only at low temperatures; and the dds gene of B. subtilis, whose product is required for cell division only at high temperatures (43). A functional analysis of such mutations may reveal the nature of the conditional inhibition of the pathway in which they function but may not reveal the precise biochemical functions of the mutant proteins (40).
Inactivation of the Transpeptidase Activity of FtsI Inhibits FtsZ Ring Constriction and FtsZ Ring Formation in Filaments.
To better understand the role that FtsI plays in FtsZ ring dynamics, we used the FtsI-specific β-lactam antibiotics cephalexin and piperacillin to inhibit the transpeptidase activity of FtsI. We localized FtsZ in filaments formed by growing wild-type cultures for one, two, or three generations in the presence of cephalexin. Cephalexin treatment had several effects on FtsZ localization (Fig. 1 k–r). One striking class of filaments had a single centrally positioned FtsZ ring (Fig. 1 k–m). Such filaments were the shortest filaments in the population, suggesting that they were derived from newborn cells. Since newborn cells in the untreated culture lack an FtsZ ring, this suggests that the FtsZ ring assembled after inactivation of the transpeptidase activity of FtsI. This agrees with previous conclusions that FtsI activity is not necessary for assembly of FtsZ rings at future division sites. This centrally located FtsZ ring persisted in such filaments for up to three generations in the presence of cephalexin or piperacillin, demonstrating that old and unconstricted FtsZ rings are not inherently unstable in the absence of FtsI activity. The persistence of these rings also directly demonstrated that cephalexin inhibits FtsZ ring constriction and thereby that FtsI is required for constriction. Occasionally, cephalexin-treated filaments with partially constricted FtsZ rings were seen, although they appeared much less frequently than in untreated cultures. This suggests that the constricting FtsZ ring may be unstable in the presence of cephalexin or that at some stages of septation FtsI is either less sensitive to cephalexin or, as seems less likely, less essential for continued constriction. The results obtained with cephalexin indicate that the transpeptidase activity of FtsI is required after the assembly of the FtsZ ring in newborn cells.
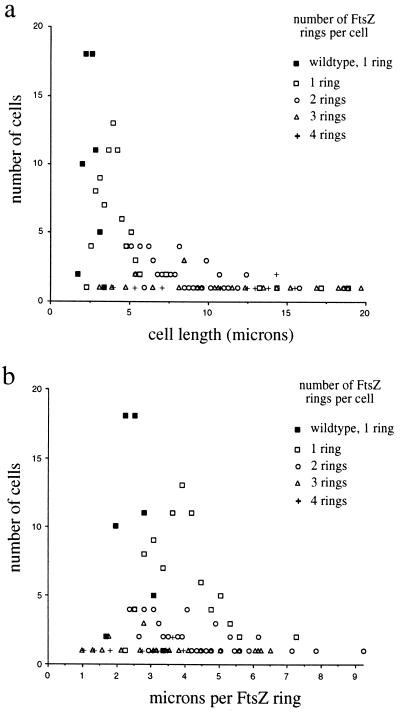
The total number of FtsZ rings present in filaments formed by inactivation of FtsI with cephalexin or piperacillin was surprisingly low. If an FtsZ ring formed between each nucleoid, then filaments growing for two or three generations would be expected to have between three and seven FtsZ rings. However, filaments with only one or two FtsZ rings made up a substantial fraction of the cephalexin-treated populations in each experiment (Fig. 3a). The deficit in FtsZ ring formation is also clearly evident when the number of FtsZ rings in each filament is normalized to cell length (Fig. 3b). More than 60% of all filaments have more micrometers per FtsZ ring than the maximum observed for untreated wild-type cells. Thus, although FtsI is not needed for assembly of FtsZ in newborn cells, FtsZ assembly at additional sites appears to be significantly delayed in filaments treated with cephalexin. The effect of cephalexin on FtsZ ring formation is summarized in a model in Fig. 4. The delay does not appear to be due to an effect of cephalexin on the FtsZ or FtsI protein levels, as cephalexin did not affect the rates of synthesis or stability of FtsZ (Fig. 5a) or the steady-state levels of FtsI (Fig. 5b).
Figure 3.
Decreased occurrence of FtsZ rings in filaments produced by cephalexin treatment. FtsZ was localized in wild-type MC4100 cells (solid squares) or in filaments after growth in the presence of cephalexin for one, two, or three generations of growth at 37°C (open symbols). The length of each filament was measured, the numbers of FtsZ rings in each filament were counted, and the data are presented in two different ways. (a) Number of cells of a given length with one, two, three, or four FtsZ rings per cell. (b) The number of FtsZ rings in each cell or filament was normalized to its length and expressed as microns per FtsZ ring. Four filaments longer than 10 μm (13, 14, 17, and 19 μm) with a single FtsZ ring are not shown. Filaments generally contained from one to four FtsZ rings, although one filament with five rings (13 μm long, 2.6 μm per ring), one filament with six rings (18 μm long, 3 μm per ring), and one filament with seven rings (17 μm long, 2.4 μm per ring) were observed (data not shown). Similar results were obtained with MG1655, except for the difference in cell lengths observed for the two wild-type strains.
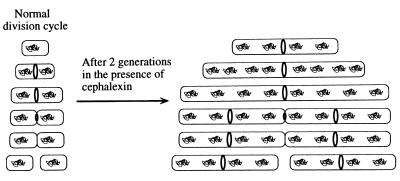
Figure 4.
Assembly cycle of FtsZ in wild-type and cephalexin-treated cells. (Left) Model of the assembly cycle of FtsZ during one generation of growth. (Right) Effect on FtsZ assembly of inactivating FtsI with cephalexin for two generations. The newborn cell lacks a detectable FtsZ ring, which assembles soon after birth. The FtsZ ring disassembles after constriction but before cell separation. While wild-type newborn cells lack FtsZ rings (see Figs. 1 a–d and 2), even the shortest filaments (which have four nucleoids) in the cephalexin-treated population contain FtsZ rings (see Fig. 1 k–n). This suggests that newborn cells, although they generally lack detectable FtsZ assembly intermediates, are capable of nucleating FtsZ assembly in the presence of cephalexin. Assembly of additional FtsZ rings in these filaments appears to be inhibited. For example, after cephalexin treatment, many filaments contain only a single FtsZ ring, despite having eight well-separated nucleoids, while cells that are nearly constricted produce filaments with eight nucleoids and with FtsZ rings at the midpoints of the nascent daughter cells (see Fig. 1 o–r). These filaments suggest that the FtsI transpeptidase activity may be required prior to the completion of septation for assembly of FtsZ at the future sites of cell division within each daughter cell.
Several different models, each of which is consistent with some of our observations, can be proposed to account for our results. One model suggests that the concentration of FtsZ is too low to support formation of more FtsZ rings in filaments. At issue in this model is the pool of free FtsZ. The total FtsZ in the filaments is essentially normal (Fig. 5a), but some of this is assembled into rings and, therefore, not available for future ring assembly. Consistent with this model, ftsA and ftsQ mutants behave similarly to ftsI mutants (33), as might be expected if the effect on FtsZ is a secondary consequence of filamentation. However, it is unlikely that the drop in FtsZ concentration would be very large (more than 50%), and there is conflict in the literature as to whether a 50% decrease in FtsZ concentration severely compromises division in E. coli (44, 45). Interestingly, during sporulation in B. subtilis, two FtsZ rings assemble in one cell in wild-type cells and also in spoOH mutant cells, which probably have only about 25% of the normal levels of FtsZ (32, 46).
A second model suggests that additional FtsZ rings do not assemble unless the current one constricts. For example, a checkpoint could exist in which regulatory proteins monitor the constriction of FtsZ rings and prevent assembly of additional rings until septation is complete. Alternatively, a factor that stimulates assembly of FtsZ at the next division site could be sequestered in the inactive FtsZ complex and may be released when constriction commences to allow assembly of the future division sites. These two models would be consistent with the fts mutants forming multiple rings if one assumes that some of the apparently unconstricted rings contain enough residual Fts activity to progress past the checkpoint step. These models are also consistent with the apparently identical effects of different fts mutants on FtsZ ring formation, since all of the Fts proteins are likely to be required for constriction and progression past a possible checkpoint. Precedent for the regulation of FtsZ ring assembly by SulA or MinCD is well established, as are the checkpoints that monitor the eukaryotic division cycle. More recently, two component regulatory proteins have been implicated in regulating cell division in Caulobacter crescentus (47–49).
A third possibility is that synthesis of the future site of FtsZ assembly requires FtsI activity at this site (rather than at the cell midpoint as in the preceding model). For example, FtsI could form a complex with other Fts proteins that assembles either before or simultaneously with a few molecules of FtsZ. The fact that ftsA, ftsQ, and ftsI mutations all have a similar effect on FtsZ ring formation may indicate that all of these Fts proteins are necessary for FtsZ to assemble. Initiation of assembly of this complex would have to begin well before birth of the cell, possibly before or coincident with the onset of constriction of the existing FtsZ ring. Heterogeneity in FtsZ ring formation in the ftsI mutants would reflect the leakiness of ftsI inactivation. This model suggests that at sometime prior to the separation of daughter cells, the future site of septation is formed in nascent daughter cells in a reaction requiring FtsI. This raises the possibility that in a cell near the completion of septation, two additional cell division complexes are at work to create the new sites in the nascent daughter cells. In the undivided cell, these putative nascent sites would be localized to the future midpoints of the nascent daughter cells and may serve as sites of assembly for FtsZ in minicell mutants.
Acknowledgments
We gratefully acknowledge Joe Lutkenhaus and Luz-Maria Guzman for antibodies and stimulating discussions. This work was supported by a MERIT Award from the National Institutes of General Medical Sciences to J.B. and by National Institutes of Health Grant GM18568 to R.L. K.P. is supported by the Cancer Research Fund of the Daymon Runyon Walter Winchell Foundation Fellowship and D.S.W. is a Department of Energy Energy Biosciences Fellow of the Life Sciences Research Foundation. J.B. is supported by an American Cancer Society Research Professorship.
Footnotes
Abbreviations: IFM, immunofluorescence microscopy; Ts, temperature-sensitive.
References
- 1.Lutkenhaus J. Mol Microbiol. 1993;9:403–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- 2.Margolin W, Wang R, Kumar M. J Bacteriol. 1996;178:1320–1327. doi: 10.1128/jb.178.5.1320-1327.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Lutkenhaus J. Mol Microbiol. 1996;21:313–319. doi: 10.1046/j.1365-2958.1996.6421360.x. [DOI] [PubMed] [Google Scholar]
- 4.Osteryoung K W, Vierling E. Nature (London) 1995;376:473–474. doi: 10.1038/376473b0. [DOI] [PubMed] [Google Scholar]
- 5.Bramhill D, Thompson C M. Proc Natl Acad Sci USA. 1994;91:5813–5817. doi: 10.1073/pnas.91.13.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer P, Crossley R, Rothfield L. Nature (London) 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 7.Erickson H P, Taylor D W, Taylor K A, Bramhill D. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson H P. Cell. 1995;80:367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee A, Lutkenhaus J. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.RayChaudhuri D, Park J T. Nature (London) 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 11.RayChaudhuri D, Park J T. J Biol Chem. 1994;269:22941–22944. [PubMed] [Google Scholar]
- 12.Bi E, Lutkenhaus J. Nature (London) 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 13.Bi E, Lutkenhaus J. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai K, Mukherjee A, Xu Y, Lutkenhaus J. J Bacteriol. 1994;175:130–136. doi: 10.1128/jb.176.1.130-136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Boer P, Crossley R E, Rothfield L I. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 16.Rothfield L I, Zhao C. Cell. 1996;84:183–186. doi: 10.1016/s0092-8674(00)80971-x. [DOI] [PubMed] [Google Scholar]
- 17.Akerlund T, Bernander R, Nordstrom K. Mol Microbiol. 1992;6:2073–2083. doi: 10.1111/j.1365-2958.1992.tb01380.x. [DOI] [PubMed] [Google Scholar]
- 18.Nanninga N. Mol Microbiol. 1991;5:791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 19.Taschner P E, Huls P G, Pas E, Woldringh C L. J Bacteriol. 1988;170:1533–1540. doi: 10.1128/jb.170.4.1533-1540.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khattar M M, Begg K J, Donachie W D. J Bacteriol. 1994;176:7140–7147. doi: 10.1128/jb.176.23.7140-7147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guzman L M, Barondess J J, Beckwith J. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 22.Carson M J, Barondess J, Beckwith J. J Bacteriol. 1991;173:2187–2195. doi: 10.1128/jb.173.7.2187-2195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Begg K J, Dewar S J, Donachie W D. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Botta G A, Park J T. J Bacteriol. 1981;145:333–340. doi: 10.1128/jb.145.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spratt B G, Pardee A B. Nature (London) 1975;254:516–517. doi: 10.1038/254516a0. [DOI] [PubMed] [Google Scholar]
- 26.Spratt B G. Eur J Biochem. 1977;72:341–342. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- 27.Hedge P J, Spratt B G. Nature (London) 1985;318:478–480. doi: 10.1038/318478a0. [DOI] [PubMed] [Google Scholar]
- 28.Spratt B G. J Bacteriol. 1977;131:293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutkenhaus J. Curr Opin Genet Dev. 1993;3:783–788. doi: 10.1016/s0959-437x(05)80099-1. [DOI] [PubMed] [Google Scholar]
- 30.Wientjes F B, Nanninga N. J Bacteriol. 1989;171:3412–3419. doi: 10.1128/jb.171.6.3412-3419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Begg K J, Donachie W D. J Bacteriol. 1985;163:615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin P, Losick R. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 33.Addinall S G, Bi E, Lutkenhaus J. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pogliano K, Harry E, Losick R. Mol Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- 35.Woldringh C L. J Bacteriol. 1976;125:248–257. doi: 10.1128/jb.125.1.248-257.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper S. Bacterial Growth and Division: Biochemistry and Regulation of Prokaryotic and Eukaryotic Division Cycles. San Diego: Academic; 1991. [Google Scholar]
- 37.Froshauer S, Green N, Boyd D, McGovern K, Beckwith J. J Mol Biol. 1988;200:501–511. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- 38.Raina S, Georgopoulos C. J Bacteriol. 1990;172:3417–3426. doi: 10.1128/jb.172.6.3417-3426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raina S, Missiakas D, Baird L, Kumar S, Georgopoulos C. J Bacteriol. 1993;175:5009–5021. doi: 10.1128/jb.175.16.5009-5021.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pogliano K J, Beckwith J. Genetics. 1993;133:763–773. doi: 10.1093/genetics/133.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishiyama K, Hanada M, Tokuda H. EMBO J. 1994;13:3272–3277. doi: 10.1002/j.1460-2075.1994.tb06628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pogliano J A, Beckwith J. EMBO J. 1994;13:554–561. doi: 10.1002/j.1460-2075.1994.tb06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beall B, Lutkenhaus J. J Bacteriol. 1989;171:6821–6834. doi: 10.1128/jb.171.12.6821-6834.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai K, Lutkenhaus J. J Bacteriol. 1991;173:3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tetart F, Bouche J. Mol Microbiol. 1992;6:615–620. doi: 10.1111/j.1365-2958.1992.tb01508.x. [DOI] [PubMed] [Google Scholar]
- 46.Gonzy-Treboul G, Karmazyn-Campelli C, Stragier P. J Mol Biol. 1992;224:967–979. doi: 10.1016/0022-2836(92)90463-t. [DOI] [PubMed] [Google Scholar]
- 47.Wingrove J A, Gober J W. Dev Biol. 1995;6:325–333. [Google Scholar]
- 48.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 49.Ohta N, Newton A. Trends Microbiol. 1996;4:326–332. doi: 10.1016/0966-842x(96)10050-0. [DOI] [PubMed] [Google Scholar]