Abstract
Recently we have demonstrated that replacing His6 by constrained amino acids in the well known antagonist SHU9119 resulted in potent and selective antagonist ligands especially at the hMC3R and hMC4 receptors. With the aim to further explore position 6 in the sequence of SHU9119 and MT-II, we have designed, synthesized, and pharmacologically characterized a series of peptide analogues of MT-II and SHU9119 at the human melanocortin receptors subtypes MC3R, MC4R and MC5R. All these peptides were modified at position 6 with constrained amino acids which are commercially available. In this study we have identified new selective ligands for the hMC4R, and an antagonist for the hMC3/hMC4 receptors. Additionally, we have discovered an interesting new selective antagonist at the hMC3R, Ac-Nle-c[Asp-βAla-DNal(2′)-Arg-Trp-Lys]-NH2 (2, PG-106) which represents an important tool in further biological investigations of the hMC3R. PG-106 will be useful in further efforts to differentiate the substructural features responsible for selectivity at the hMC3R, hMC4R, and hMC5R.
Keywords: Melanocortin receptors, Melanotropins, Structure-activity relationships, Cyclic melanotropins, Receptor selective melanotropin antagonists
Introduction
Melanotropins (α-MSH, β-MSH, γ-MSH) are peptide hormones and neurotransmitters derived through a series of proteolytic cleavages from the precursor pro-opiomelanocortin (POMC). Over the past decade, molecular cloning of the five subtype receptors (MC1R to MC5R) for these peptides and ACTH has provided tools for systematic studies of their physiological effects [7,8,9,10,22,27,30]. The natural ligands for these receptors include α-MSH, β-MSH, γ-MSH as well as ACTH. α-MSH is a linear peptide with 13 residues (Ac-Ser-Tyr-Ser-Met-Glu-His-Phe-Arg-Trp-Gly-Lys-Pro-Val-NH2). Recent discoveries have demonstrated that these peptides and their derivatives also display several other effects besides pigmentation and adrenal function including the regulation of feeding and sexual behaviour, and the modulation of the immune system [e.g. 1,5-8]. Substitution of DPhe-7 Phe-7 and Nle for Met-4 respectively yields NDP-α-MSH which was found to be a superagonist at melanocortin receptors [1,19,26].
Intensive efforts are currently being made to develop selective ligands for the melanocortin receptors to understand the physiological roles played by these receptors [6,12,17,20]. In particular, clarification of the role of melanocortin receptor subtypes, i.e., MC3R, MC4R and MC5R [4,5,23,29] is particularly important specially after the discovery that MC3R and MC4R are involved in the regulation of feeding [11,21,28]. In fact, agonists of the MC4R cause a reduction of food intake by inducing satiety, while antagonists promote feeding [11,28]. Ligands at this receptor represent an attractive pharmaceutical target in development of drugs to the treatment of obesity and other eating disorders. Previous extensive structure-activity studies on melanotropins, in particular on α-MSH, led to the small cyclic peptide MT-II, Ac-Nle-c[Asp-His-DPhe-Arg-Trp-Lys]-NH2 (1), a potent and non-selective agonist at the human melanocortin receptors (18). Similarly, the lactam analogue Ac-Nle-c[Asp-His-DNal(2′)-Arg-Trp-Lys]-NH2, SHU-9119, is a potent antagonist at human melanocortin 3 and 4 receptors (19) and a partial agonist at the hMC5R (19,120). Recently, extensive structure-activity studies performed in our laboratory have shown that the replacement of His residue in position 6 in the sequence of MT-II and SHU9119 (Ac-Nle-[Asp-His-DPhe/DNal(2)-Arg-Trp-Lys]-NH2), with some dihedral constrained amino acids led to potent and highly hMC3R and hMC4R selective ligands [2,13,14,15]. Also, the introduction of β-modified proline in position 6 of MT-II sequence resulted in analogues with enhanced hMC5R selectivity [3]. In our effort to obtain new melanotropin peptides with improved potency and selectivity, we have designed and synthesized a series of novel cyclic MT-II and SHU9119 analogues in which we investigate the impact of additional constrained amino acids in position 6 on receptor selectivity.
Materials and Methods
Materials
Nα-Fmoc-protected amino acids and resin were purchased from Advanced ChemTech (Louisville, KY). HBTU and HOBt were purchased from Quantum Biotechnologies (Montreal, Quebec, Canada). For the Nα-Fmoc-protected amino acids, the following side chain protecting groups were used: Lys(Nε-Alloc); Arg(Ngu-Pbf); Asp(β-Allyl); His(Nim-Trt) and Trp(Nin-Boc). All protected amino acid derivatives were analyzed for purity by thin-layer chromatography before use. Peptide synthesis solvents, reagents, as well as CH3CN for HPLC were reagent grade and were acquired from commercial sources and used without further purification unless otherwise noted. TLC was done on Analtech, Inc. (Newark, DE) silica gel 60 F254 plates using the following solvent systems: (A) 1-butanol / acetic acid / pyridine / water (5:5:1:4); (B) ethyl acetate / pyridine / acetic acid / water (5:5:1:3); (C) 1-butanol / acetic acid / water (4:1:1). The peptides were detected on the TLC plates using iodine vapor. Amino acid analyses were performed at the University of Arizona Biotechnology Core Facility. The system used was an Applied Biosystems Model 420A amino acid analyzer with automatic hydrolysis (Vapor Phase at 160 °C for 1 h 40 min using 6 N HCl) and a precolumn phenylthiocarbamyl-amino acid (PTC-AA) analysis. No corrections are made for amino acid decomposition. FAB-MS analyses were performed at the University of Arizona Core Facility. The instrument was custom made in Breman, Germany, and consists of a LIQUID SIMS4 Sectors AMD mass spectrometer. The experimental conditions consisted of a glycerol matrix-scan of 200-2000 Da in the positive ion mode. The purity of the finished peptides was checked by TLC in three solvent systems and by analytical RP-HPLC at 230, 254, and 280 nm using a Hewlett-Packard 1090 Series II Liquid Chromatograph with a built-in diode array detector (Table 1). In all cases, the purity of the finished peptides was greater than 95%. The structures of the pure peptides were confirmed by FAB-MS. The analytical data for the peptide is given in Table 1.
Table 1.
Physicochemical properties of the melanocortin peptides
| Code | Sequence | Solv. Aa | Solv. Ba | Solv. Ca | M.W. | MS | HPLCb |
|---|---|---|---|---|---|---|---|
| PG105 | Ac-Nle-[Asp-βAla-DPhe-Arg-Trp-Lys]-NH2 | 0.82 | 0.04 | 0.41 | 958.09 | 959.1 | 4.47 |
| PG106 | Ac-Nle-[Asp-βAla-DNal(2′)-Arg-Trp-Lys]-NH2 | 0.78 | 0.04 | 0.38 | 1008.5 | 1009.1 | 4.15 |
| PG107 | Ac-Nle-[Asp-tBuGly-DPhe-Arg-Trp-Lys]-NH2 | 0.85 | 0.08 | 0.43 | 1000.8 | 1001.2 | 5.13 |
| PG108 | Ac-Nle-[Asp-tBuGly-DNal(2′)-Arg-Trp-Lys]-NH2 | 0.82 | 0.04 | 0.41 | 1050.6 | 1051.1 | 5.47 |
| PG103 | Ac-Nle-[Asp-Hyp(Bzl)-DPhe-Arg-Trp-Lys]-NH2 | 0.80 | 0.04 | 0.39 | 1074.2 | 1074.8 | 5.44 |
| PG104 | Ac-Nle-[Asp- Hyp(Bzl)-DNal(2′)-Arg-Trp-Lys]-NH2 | 0.92 | 0.10 | 0.55 | 1124.4 | 1124.6 | 5.23 |
| PG135 | Ac-Nle-c[Asp-Mamb-DPhe-Arg-Trp-Lys]-NH2 | 0.90 | 0.12 | 0.48 | 1020.4 | 1020.7 | 5.74 |
| PG-136 | Ac-Nle-c[Asp-Mamb-DNal(2′)-Arg-Trp-Lys]-NH2 | 0.92 | 0.13 | 0.51 | 1070.6 | 1070.9 | 5.81 |
| PG-943 | Ac-Nle-c[Asp-Tic-DPhe-Arg-Trp-Lys]-NH2 | 0.90 | 0.12 | 0.49 | 1046.3 | 1046.8 | 5.61 |
Notes:
Solvent systems:(A) 1-butanol/HOAc/pyridine/H2O (5:5:1:4); (B) EtOAc/pyridine/AcOH/H2O (5:5:1:3); (C) 1-butanol/AcOH/H2O (4:1:1)
HPLC k' = [(peptide retention time − solvent retention time)/solvent retention time] in a solvent system of 10% CH3CN in 0.1% TFA and a gradient to 90% CH3CN over 40 min. An analytical Vydac C18 (218TP104) column was used with a flow rate of 1 mL/min.
General Method for Peptide Synthesis and Purification
The protected peptide resins used to make the cyclic melanotropins were prepared using 0.5 g of Rink amide resin (0.7 mmol of NH2/g of resin) by first coupling Nα-Fmoc-Lys(Nε-Alloc)-OH to the resin previously deprotected by a 25% piperidine solution in DMF for 30 min. In this strategy automated solid phase synthesis was performed on the Advanced ChemTech ACT 396 instrument. The following protected amino acids were then added stepwise Nα-Fmoc-Trp(Nin-Boc)-OH, Nα-Fmoc-Arg(Nγ-Pbf)-OH, Nα-Fmoc-DNal(2′)-OH, Fmoc-DPhe-OH, Nα-Fmoc-His-OH or Nα-Fmoc-Xaa-OH Nα-Fmoc-Asp(β-Allyl)-OH, and Nα-Fmoc-Nle-OH. Each coupling reaction was accomplished using a 3-fold excess of amino acid with HBTU and HOBt in the presence of diisopropylethyl amine (DIEA, 6-fold excess). The Nα-Fmoc protecting groups were removed by treating the protected peptide resin with 25% piperidine solution in DMF twice (1 × 5 min and 1 × 25 min). The peptide resin was then washed three times with DMF and the next coupling step was then initiated in a stepwise manner. All reactions were done under an Argon atmosphere. Following the assembly of the protected peptide resin, the terminal Nα-Fmoc group was removed in the usual manner. The Nα amino group was acetylated with 25% acetic anhydride in dichloromethane for 20 min. The next step was removal of the Nγ-Alloc group of Lys and the β-allyl group of Asp under well controlled conditions [24]. For this the peptide resin was washed with CH2Cl2 under Argon atmosphere and was added a solution of PhSiH3 (24 equiv.) in 2 mL of CH2Cl2 all under Argon. Then a solution of Pd(PPh3)4 (0.25 equiv.) in 6 mL of CH2Cl2 was added and reaction allowed to proceed under Argon for 30 min. Then the peptide resin was washed with CH2Cl2 (3x), with DMF (3x) and with DCM (4x), and the deprotection process repeated. The macrocyclic lactam ring formation was the mediated by addition of HBTU (6 equiv.), HOBt (6 equiv.) and DIEA (12 equiv.) for 2 h. The process was repeated if necessary (Kaiser test was used to monitor completion). The peptide was then cleaved from the resin using trifluoracetic acid/triethylsilane/H2O (9.0:0.5:0.5) for 3 h. The resin was removed by filtration and the crude peptide recovered by precipitation using chilled anhydrous ethyl ether to give a white powder which was purified by HPLC on a C18-bonded silica column (Vydac 218TPP1010, 1.0 × 25 cm) eluting with a linear gradient of acetonitrile in aqueous 0.1% TFA. The products were obtained by lyophilization of the appropriate fractions after removal of the acetonitrile by rotary evaporation. Analysis by analytical HPLC and TLC (3 solvents) showed the peptides to be pure (>98%) (Table 1). The structures were further confirmed by high resolution mass spectroscopy and amino acid analysis.
Receptor Binding Assay
Competition binding experiments were carried out using whole HEK293 cells stably expressing human MC3, MC4, and MC5 receptors. HEK293 cells transfected with hMCRs [3] were seeded on 96-well plates 48 hours before assay (100,000 cells/well). For the assay, the cell culture medium was aspirated and cells were washed twice with a freshly prepared binding buffer containing 100% minimum essential medium with Earle's salt (MEM, GIBCO), 25 mM HEPES (pH 7.4), 0.2% bovine serum albumin, 1mM 1,10-phenanthrolone, 0.5 mg/L leupeptin, 200 mg/L bacitracin. Next, cells were incubated with different concentrations of unlabeled peptide and labeled [125I]-[Nle4,D-Phe7]-α-MSH (Perkin-Elmer Life Science, 100,000 cpm/well, 0.1386 nM) for 40 min at 37°C. The assay medium was subsequently removed and each well was washed twice with the binding buffer. The cells were then lysed by the addition of 250 μL of 0.1M NaOH and 250 μL of 1% Triton X-100. The lysed cells were transferred to 12×75 mm glass tubes and the radioactivity was measured by a Wallac 1470 WIZARD Gamma Counter.
Adenylate Cyclase Assay
HEK 293 cells transfected with human melanocortin receptors [3] were grown to confluence in MEM medium (GIBCO) containing 10% fetal bovine serum, 100 units/mL penicillin and streptomycin, and 1 mM sodium pyruvate. The cells were seeded on 96-well plates 48 hours before assay (100,000 cells/well). For the assay, the cell culture medium was removed and the cells were rinsed with 1 mL of MEM buffer (GIBCO) or with Earle's balanced salt solution (EBSS, GIBCO). An aliquot (0.4 mL) of the Earle's balanced salt solution was placed in each well along with 5 μL 0.5 mM isobutylmethylxanthine (IBMX) for 1 min at 37°C. Next, varying concentration of aliquots of melanotropin peptides (0.1 mL) were added, and the cells were incubated for 3 min at 37°C. The reaction was stopped by aspirating the assay buffer and adding 0.15 mL ice-cold Tris/EDTA buffer to each well. After dislodging the cells with the help of trypsin, the cells were transferred to polypropylene micro-centrifuge tubes and placed in a boiling water bath for 15 min. The cell lysate was then centrifuged for 2 min at 6500 rpm, and 50 μL of the supernatant was aliquoted into an Eppendorf tube. The total cAMP content was measured by competitive binding assay according to the TRK 432 assay kit instructions (Amersham Corp., Piscataway, NJ).
Data Analysis
IC50 and EC50 values represent the mean of two experiments performed in triplicate. IC50 and EC50 estimates and their associated standard errors were determined by fitting the data using a nonlinear least squares analysis, with the help of GraphPad Prism 4 (GraphPad Software, San Diego, CA).
Results
The melanotropin analogues listed in Table 1, were prepared by solid-phase peptide synthesis and evaluated for their binding affinities to the human melanocortin receptors 3–5 in competitive binding assays using the radiolabeled ligand [125I]-NDP-α-MSH and for their agonist or antagonist potency in cAMP assays employing the HEK293 cells expressing these receptors (Table 2).. Replacement of His6 with a βAla residue in the MT-II sequence, yielded analogue 1 (Ac-Nle-c[Asp-βAla-DPhe-Arg-Trp-Lys]-NH2, PG-105), with partial agonist activity at the hMC3R, but with 140-fold lower in binding affinity and 170-fold lower cAMP activity (EC50 = 320 and 1.85 nM, respectively) compared to MT-II. Analogue 1 was found to have no binding affinity for the hMC4 and hMC5 receptors at 10−5M. Analogue 2 (Ac-Nle-c[Asp-βAla-DNal(2′)-Arg-Trp-Lys]-NH2, PG-106), which differs from 1 by having a D-Nal(2′) in position 7, was a selective antagonist at the hMC3R and totally inactive as a ligand at the hMC4R and hMC5R. These results confirm that simple, unconstrained amino acid residues in the 6 position of MT-II could generate a selective ligand at hMC3R.
Table 2.
Binding and intracellular cAMP accumulation of the α-melanotropin analogues at human melanocortin receptors
| No | Peptide Code |
Sequence | hMC3R | hMC4R | hMC5R | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IC50 (nM) |
EC50 (nM) |
% Activity |
IC50 (nM) |
EC50 (nM) |
% Activity |
IC50 (nM) |
EC50 (nM)b |
% Activity |
|||
| 1 | PG105 | Ac-Nle-[Asp-βAla-DPhe-Arg-Trp-Lys]-NH2 | 170±20 | 320±50 | 30 | n.b. | 1500±200 | / | n.b. | n.a. | / |
| 2 | PG106 | Ac-Nle-[Asp-βAla-DNal(2′)-Arg-Trp-Lys]-NH2 | 210±30 | n.a. | / | n.b. | 9900 | / | n.b. | n.a. | / |
| 3 | PG107 | Ac-Nle-[Asp-tBuGly-DPhe-Arg-Trp-Lys]-NH2 | 41±5 | 160±22 | 100 | 550±60 | 740±80 | / | 200.±30 | 1100±150 | 100 |
| 4 | PG108 | Ac-Nle-[Asp-tBuGly-DNal(2′)-Arg-Trp-Lys]-NH2 | 18±2 | 66±7 | 5 | 26±3 | 2400±300 | / | 6.3±1 | 1000±100 | 100 |
| 5 | PG103 | Ac-Nle-[Asp-Hyp(Bzl)-DPhe-Arg-Trp-Lys]-NH2 | 2.8±3 | 19±2 | 34 | 4.6±0.5 | 470±50 | 17 | 13±1 | 110±12 | 105 |
| 6 | PG104 | Ac-Nle-[Asp- Hyp(Bzl)-DNal(2′)-Arg-Trp-Lys]-NH2 | 15±2 | n.a. | / | 17±2 | >5000 | / | 15±5 | 70±10 | 23 |
| 7 | PG-135 | Ac-Nle-c[Asp-Mamb-DPhe-Arg-Trp-Lys]-NH2 | 670±250 | 800±69 | 83 | 930±69 | n.a. | 4 | 1800±100 | 710±100 | 50 |
| 8 | PG-136 | Ac-Nle-c[Asp-Mamb-DNal(2)-Arg-Trp-Lys]-NH2 | 67±13 | n.a. | 1.7 | >3000 | n.a. | / | 2500±200 | n.a. | / |
| 9 | PG-943 | Ac-Nle-c[Asp-Tic-DPhe-Arg-Trp-Lys]-NH2 | 2.6±0.4 | 20±2 | 102 | 15±2 | 6±1 | 99 | 10±2 | 1000 | 100 |
| MTII | Ac-Nle-c[Asp-His-D-Phe-Arg-Trp-Lys]- NH2 | 1.25±0.20 | 1.85±0.20 | 100 | 1.07±0.3 | 2.87±0.52 | 100 | 7.47±0.2 | 3.3±0.7 | 100 | |
| SHU9119 | Ac-Nle-c[Asp-His-DNal(2)-Arg-Trp-Lys]- NH2 | 0.23±0.02 | - | 0 | 0.06±0.01 | - | 0 | 0.09±0.02 | 0.12±0.01 | 97 | |
IC50 = concentration of peptide at 50% specific binding (N = 4); EC50 = effective concentration of peptide that was able to generate 50% maximal intracellular cAMP accumulation (N = 4). The peptides were tested at a range of concentration from 10−10 to 10−5 M; n.b.= no binding affinity at the receptor indicated at 10−5M conc. of ligand; n.a. = no cAMP activity at the receptor indicated at 10−5M conc. of ligand.
On the other hand , the analogue 3 (Ac-Nle-[Asp-tBuGly-DPhe-Arg-Trp-Lys]-NH2, PG-107) with a tButGly residue in position 6, that is, a residue more bulky and constrained in the side chain moiety, was found to have affinity for all melanocortin receptors tested in this study. Analogue 3 was a full and selective agonist at the hMC3R, almost 13-fold more selective for the hMC3R than the hMC4R, and 5-fold selective with respect to the hMC5R (IC50 = 41 nM, 550 nM, and 200 nM, respectively). Surprisingly, the analogue 4 (Ac-Nle-[Asp-tBuGly-DNal(2′)-Arg-Trp-Lys]-NH2, PG-108) which differs from 3 by having a DNal(2′) in position 7, was found to have slight agonist activity at all melanocortin receptors tested. In fact, analogue 4 has a similar high affinity for both the hMC3R and hMC4R, and is slightly selective for the hMC5R (IC50s = 18.4 nM, 25.7 nM, and 6.3 nM, respectively). However, analogue 4 was 37-fold more potent at the hMC3R than the hMC4R, and 15-fold more with respect to hMC5R (EC50 = 66 nM, 2400 nM, and 1000 nM, respectively) in functional cAMP assays. Analogue 5 (Ac-Nle-c[Asp-Hyp(Bzl)-DPhe-Arg-Trp-Lys]-NH2, PG-103) which substitutes in position 6, a more hydrophilic Pro derivative, was found to be highly potent but slightly less potent than MT-II, exhibiting similar binding affinity at the hMC3R and the hMC4R (IC50 = 2.8 nM and 4.6 nM, respectively) as for the hMC5R. These results suggest that the Bzl residue does not significantly affect binding affinity at melanocortin receptors. Interestingly, analogue 6 (Ac-Nle-c[Asp-Hyp(Bzl)-DNal(2′)-Arg-Trp-Lys]-NH2, PG-104) was found to be a potent antagonist at the hMC4R (IC50 = 17 nM) and the hMC3R (IC50 = 15 nM), but a partial agonist at the hMC5R (IC50 = 15 nM, EC50 = 70 nM). Analogue 7 (Ac-Nle-c[Asp-Mamb-DPhe-Arg-Trp-Lys]-NH2, PG-135) with a 3-aminomethyl-benzoic acid (Mamb) residue in position 6 instead of His, had very weak binding at the hMC3R, the hMC4R and the hMC5R. Interestingly, analogue 8 (Ac-Nle-c[Asp-Mamb-DNal(2′)-Arg-Trp-Lys]-NH2, PG-135) which differs from 7 by having DNal(2′) in position 7, was a potent and highly selective antagonist for the hMC3R. These data suggest that the Mamb residue has a considerable impact in the formation of ligand-receptor complexes but only for antagonists at the hMC3R. It is possible that the same residue could destabilize the ligand-receptor interactions for other melanocortin receptors when DPhe7 is present in position 7. Analogue 9 (Ac-Nle-c[Asp-Tic-DPhe-Arg-Trp-Lys]-NH2, PG-943) containing the Tic residue in position 6, resulted in an agonist at hMC3R, hMC4R and hMC5R with a very high binding affinity for all the receptor subtypes (IC50 = 2.6 nM, 15 nM and 10 nM respectively). This was in contrast with our earlier findings [15] where substitution of DNal(2')7 was done in place of DPhe7. Interestingly, this analogue was a very potent antagonist at hMC3R and hMC4R and a full agonist at hMC5R.
Discussion
Structure-Activity Relationships
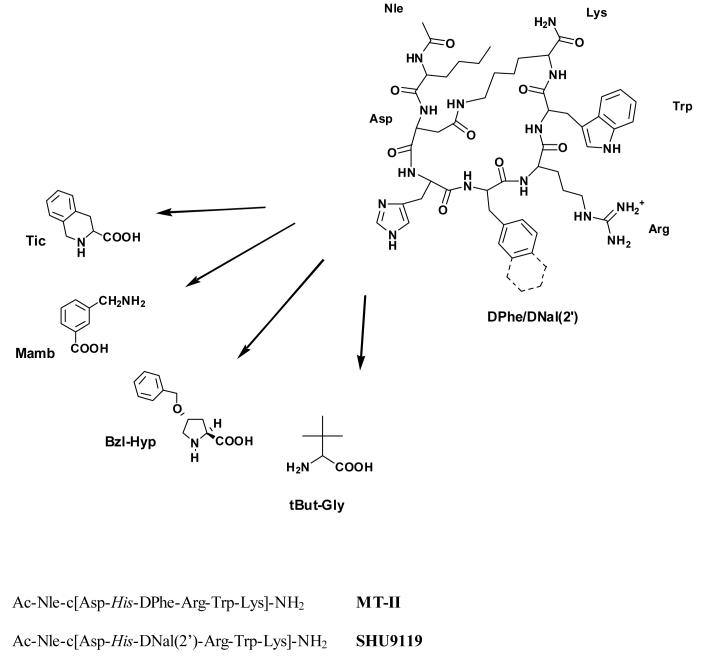
Our previous results have demonstrated that incorporation of the conformationally constrained proline residue and its analogues into the lactam bridge might stabilize bioactive conformations, and can improve selectivity of melanotropin peptides at the MCRs [13,15]. In fact, replacing His6 by Pro6 in the well known antagonist SHU9119 we have obtained a potent agonist at the hMC5R (EC50 = 0.072 nM) with full antagonist activity at hMC3 and hMC4 receptors. We also have demonstrated that the same substitution in MT-II led to a compound with agonist activity at the hMCRs substantially similar to that of MT-II. Based on these and other results [2,18] we have come to the conclusion that the imidazole group of histidine is not essential for the binding of MT-II and SHU9119 at the hMC4R, and that the restriction of conformational freedom at histidine position by proline and related analogues did not affect interactions of MT-II and SHU-9119 at the hMC4R. To expand on these important observations, additional analogues of these melanotropins substituted in position 6 with other unconventional amino acid residues were synthesized and tested at the hMC3R, hMC4R and hMC5R (Table 2).
These results demonstrate that the presence of an appropriate residue in position 6 can change the biological profile of melanocortin peptides at the hMC3–5 receptors. The structure–activity relationships information provided by this set of synthetic melanocortin analogues supports further the hypothesis that the position 6 could be a factor for selectivity and potency at central melanocortin receptors.
In conclusion, structure-activity studies on synthetic melanocortin analogues at the human MC3, MC4 and MC5 receptors have identified new ligands as antagonists at the hMC3R/hMC4R (analogues 2 and 6) but most importantly we have discovered a new selective antagonist for the hMC3R (analogue 2) which is potentially useful in further biological investigations of the hMC3R. Since it is selective for the hMC3R over the hMC4R and hMC5R, it can help us to differentiate the substructural features responsible for selectivity at melanocortin receptors.
Fig. 1.
Structure of the potent monocyclic peptides MT-II and SHU9119, and the conformationally constrained amino acids considered for the modification at position 6.
Acknowledgements
This research was supported primarily by the U.S. Public Health Service, National Institute of Health, DK17420 and DA06284.
The following additional abbreviations are used:
Abbreviations used for amino acids and designation of peptides follow the rules of the IUPAC-IUB Commission of Biochemical Nomenclature in J. Biol. Chem. 1972, 247, 977-983.
- AAA
amino acid analysis
- Boc
tert-butyloxycarbonyl
- BSA
bovine serum albumin
- Bzl
benzyl
- tBu
tert-butyl
- cAMP
adenosine 3',5'-cyclic monophosphate
- DCM
dichloromethane
- DIPEA
N,N-diisopropylethylamine
- DMF
N,N-dimethylformamide
- Et3SiH
triethylsilane
- FAB-MS
fast-atom bombardment mass spectrometry
- Fmoc
9-fluorenylmethoxycarbonyl
- HOBt
N-hydroxybenzotriazole
- HBTU
2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- HEPES
N-(2-hydroxyethyl)-piperazine-N'-(2-ethanesulfonic acid)
- Pbf
2,2,4,6,7-pentamethyldihydrobenzo-furan-5-sulfonyl
- RP-HPLC
reversed-phase high performance liquid chromatography
- TFA
trifluoroacetic acid; Tris, 2-amino-2-(hydroxymethyl)-1,3-propanediol
- Trt
triphenylmethyl (trityl).
Amino acid symbols denote L-configuration unless indicated otherwise. Mamb, 3-aminomethyl-benzoic acid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Al-Obeidi F, Hadley ME, Pettitt BM, Hruby VJ. Design of a new class of superpotent cyclic α-Melanotropins based on quenched dynamic stimulations. J Am Chem Soc. 1989;111:3413–6. [Google Scholar]
- 2.Bednarek MA, MacNeil T, Tang R, Kalyani RN, Van der Ploeg LHT, Weinberg DH. Potent and selective peptide agonists of α-melanotropin action at human melanocortin receptor 4: Their synthesis and biological evaluation in vitro. Biochem Biophy Res Comm. 2001;286:641–5. doi: 10.1006/bbrc.2001.5444. [DOI] [PubMed] [Google Scholar]
- 3.Cai M, Cai C, Mayorov AV, Xiong C, Cabello CM, Soloshonok VA, Swift JR, Trivedi D, Hruby VJ. Biological and conformational study of β-substituted prolines in MT-II template: Steric effects leading to human MC5 receptor selectivity. J Peptide Res. 2004;63:116–31. doi: 10.1111/j.1399-3011.2003.00105.x. [DOI] [PubMed] [Google Scholar]
- 4.Cone RD, Mountjoy KG, Robbins LS, Nadeau JH, Johnson KR, Roselli-Rehfuss L, Mortrud MT. (1993) Cloning and functional characterization of a family of receptors for the melanotropic peptides. Ann NY Acad Sci. 1993;680:342–63. doi: 10.1111/j.1749-6632.1993.tb19694.x. [DOI] [PubMed] [Google Scholar]
- 5.Cone RD, editor. The melanocortin receptors. Humana Press; Totowa, New Jersey: 2000. [Google Scholar]
- 6.Cowley MA, Cone RD, Enriori P, Louiselle I, Williams SM, Evans AE. Electrophysiological actions of peripheral hormones on melanocortin neurons. Ann NY Acad Sci. 2003;994:175–86. doi: 10.1111/j.1749-6632.2003.tb03178.x. [DOI] [PubMed] [Google Scholar]
- 7.De Wied D, Jolles J. Neuropeptides derived from proopiomelanocortin: behavioral, physiological, and neurochemical effects. Physiol. Rev. 1982;62:976–1059. doi: 10.1152/physrev.1982.62.3.976. [DOI] [PubMed] [Google Scholar]
- 8.De Wied D. Behavioral pharmacology of neuropeptides related to melanocortins and the neurohypophyseal hormones. Eur J Pharmacol. 1999;375:1–11. doi: 10.1016/s0014-2999(99)00339-8. [DOI] [PubMed] [Google Scholar]
- 9.De Wied D, Croiset G. Stress modulation of learning and memory processes. Methods Achieve Exp Phatol. 1991;15:167–199. [PubMed] [Google Scholar]
- 10.Eberle AN, editor. The Melanotropins: Chemistry, Physiology, and Mechanisms of Action. Karger; Basel, Switzerland: 1988. pp. 210–319. [Google Scholar]
- 11.Fan W, Boston BA, Kesterson RA, Hruby VJ, Cone RD. Role of melanocortigenic neurons in feeding and the agouti obesity syndrome. Nature. 1997;385:165–8. doi: 10.1038/385165a0. [DOI] [PubMed] [Google Scholar]
- 12.Fung S, Hruby VJ. Design of cyclic and other templates for potent and selective peptide α-MSH analogues. Curr Opinion Chem Biol; Next-Generation Therapeutics. 2005;9:352–8. doi: 10.1016/j.cbpa.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grieco P, Han G, Weinberg D, MacNeil T, Van der Ploeg LHT, Hruby VJ. Design and synthesis of highly potent and selective melanotropin analogues of SHU9119 modified at position 6. Biochem Biophys Research Commun. 2002;292:1075–80. doi: 10.1006/bbrc.2002.6739. [DOI] [PubMed] [Google Scholar]
- 14.Grieco P, Han G, Hruby VJ. New dimensions in the design of potent and receptor selective melanotropin analogues. In: Fields GB, Tam JP, Barany G, editors. Peptides for the new millenium. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2000. pp. 541–2. [Google Scholar]
- 15.Grieco P, Lavecchia A, Cai M, Trivedi D, Weinberg D, MacNail T, Van der Ploeg LHT, Hruby VJ. Structure-activity studies of the melanocortin peptides: Discovery of potent and selective antagonists at hMC3 and hMC4 receptors. J Med Chem. 2002;45:5287–94. doi: 10.1021/jm0202526. [DOI] [PubMed] [Google Scholar]
- 16.Grieco P, Gitu PM, Hruby VJ. Preparation of “side-chain-to-side-chain” cyclic peptides by allyl and alloc strategy: Potential for library synthesis. J Peptide Res. 2001;57:250–6. doi: 10.1111/j.1399-3011.2001.00816.x. [DOI] [PubMed] [Google Scholar]
- 17.Haskell-Luevano C, Todorovic A. A review of melanocortin receptor small molecule ligands. Peptides. 2005;26:2026–36. doi: 10.1016/j.peptides.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 18.Haskell-Luevano C, Lim S, Yuan W, Cone RD, Hruby VJ. Structure activity studies of the melanocortin antagonist SHU9119 modified at the 6, 7, 8, and 9 positions. Peptides. 2000;21:49–57. doi: 10.1016/s0196-9781(99)00167-9. [DOI] [PubMed] [Google Scholar]
- 19.Hruby VJ, Lu D, Sharma SD, Castrucci AM, Kesterson RA, Al-Obeidi F, Hadley ME, Cone RD. Cyclic lactam α-melanotropin analogues of Ac-Nle4-c[Asp5,DPhe7,Lys10]-α-MSH(4-10)-NH2 with bulky aromatic amino acids at position 7 show high antagonist potency, and selectivity at specific melanocortin receptor. J Med Chem. 1995;38:3454–61. doi: 10.1021/jm00018a005. [DOI] [PubMed] [Google Scholar]
- 20.Hruby VJ, Cai M, Grieco P, Han G, Kavarana M, Trivedi D. Exploring the stereostructural requirements of peptide ligands for the melanocortin receptors. Ann NY Acad Sci. 2003;994:12–20. doi: 10.1111/j.1749-6632.2003.tb03157.x. [DOI] [PubMed] [Google Scholar]
- 21.Huznar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeyer LR, Gu W, Kesterson RA, Boston BA, Cone RD, Smith FJ, Campfield LA, Burn P, Lee F. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–41. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 22.Li SJ, Varga K, Archer P, Hruby VJ, Sharma SD, Kesterson R, Cone RD, Kunos G. Melanocortin antagonists define two distinct pathways of cardiovascular control by α- and γ-melanocyte stimulating hormones. J Neurosci. 1996;16:5182–8. doi: 10.1523/JNEUROSCI.16-16-05182.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mountjoy KG, Robbins LS, Mortrud MT, Cone RD. The cloning of a family of genes that encode the melanocortin receptors. Science. 1992;257:1248–51. doi: 10.1126/science.1325670. [DOI] [PubMed] [Google Scholar]
- 24.Pearson DA. Trialkylsilanes as scavengers for the trifluoroacetic acid deblocking of protecting groups in peptide-synthesis. Tetrahedron Lett. 1989;30:2739–42. [Google Scholar]
- 25.Prusis P, Muceniece R, Mutule I, Mutulis F, Wikberg JES. Design of new small cyclic melanocortin receptor-binding peptides using molecular modelling: role of the His residue in the melanocortin peptide core. Eur J Med Chem. 2001;36:137–46. doi: 10.1016/s0223-5234(00)01200-9. [DOI] [PubMed] [Google Scholar]
- 26.Sawyer TK, Sanfilippo PJ, Hruby VJ, Engle MH, Heward CR, Burnett J, Hadley M. [Nle4,DPhe7]α-Melanocyte stimulating hormone: A highly potent α-melanotropin with ultralong biological activity. Proc Natl Acad Sci USA. 1980;77:5754–8. doi: 10.1073/pnas.77.10.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith EM, Hughes TK, Hashemi F, Stefano GB. Immunosuppressive effects of corticotropin and melanotropin and their possible significance in human immunodeficiency virus infection. Proc Natl Acad Sci USA. 1992;89:782–6. doi: 10.1073/pnas.89.2.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vergoni AV, Bertolini A. Role of melanocortins in the central control of feeding. Eur J Pharmacol. 2000;405:25–32. doi: 10.1016/s0014-2999(00)00538-0. [DOI] [PubMed] [Google Scholar]
- 29.Wikberg JES, Muceniece R, Mandrika I, Prusis P, Linblom J, Post C, Skottner A. New aspects on the Melanocortin and their receptors. Pharmacological Res. 2000;42:393–420. doi: 10.1006/phrs.2000.0725. [DOI] [PubMed] [Google Scholar]
- 30.Wikberg JES. Melanocortin receptors: Perspectives for novel drugs. Eur J Pharmacol. 1999;375:295–310. doi: 10.1016/s0014-2999(99)00298-8. [DOI] [PubMed] [Google Scholar]