Abstract
Polycomb-mediated repression and DNA methylation are important epigenetic mechanisms of gene silencing. Recent evidence suggests a functional link between the polycomb repressive complex (PRC) and Dnmts in cancer cells. Here we provide evidence that Lsh, a regulator of DNA methylation, is also involved in normal control of PRC-mediated silencing during embryogenesis. We demonstrate that Lsh, a SNF2 homolog, can associate with some Hox genes and regulates Dnmt3b binding, DNA methylation, and silencing of Hox genes during development. Moreover, Lsh can associate with PRC1 components and influence PRC-mediated histone modifications. Thus Lsh is part of a physiological feedback loop that reinforces DNA methylation and silencing of PRC targets.
Keywords: chromatin, DNA methylation, polycomb, Lsh
Recently, several connections have been suggested between DNA methylation (1, 2) and polycomb repressive complex (PRC-mediated silencing (3) in cancer cells. First, it was demonstrated that genes that are CpG methylated in cancer cells are frequently marked by PRC binding and histone 3 K27 methylation early in development (4–6). In addition, it was reported that Ezh2 (a PRC2 component) controls Dnmt binding and DNA methylation at the Myt1 gene in cancer cell lines (7). These results suggest that PRC may premark sites for de novo DNA methylation at genes methylated in cancer, but this may be a rare and aberrant event in cells predisposed to cancer.
In this study, we tested whether PRC-mediated silencing and DNA methylation are normally linked during development, using the Lsh knockout model (8). Lsh, a member of the SNF2 chromatin remodeling family (9, 10), is involved in the control of DNA methylation patterns during embryonic development (11, 12). Lsh-mediated DNA methylation is crucial for retroviral silencing and repression of selected imprinted loci (13, 14). Lsh may be directly involved in the control of de novo methylation via association with Dnmt3a and -3b in embryonic stem cells (15). Because Lsh−/− mice die at birth, we used Lsh−/− embryos to examine a functional link between Lsh, DNA methylation, and silencing of selected PRC targets such as Hox genes.
Results
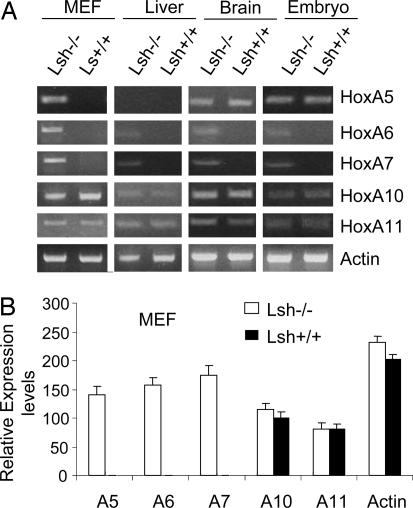
We first examined Hox gene expression in various tissues from Lsh−/− mice. RT-PCR analysis shows HoxA5, HoxA6, and HoxA7 genes are silenced in wild-type murine embryonic fibroblasts (MEFs) but reactivated in Lsh−/− MEFs (Fig. 1A). In contrast, HoxA10 and HoxA11 genes are already expressed in wild-type MEFs and not significantly changed in the absence of Lsh, as revealed by conventional RT-PCR analysis as well as real-time PCR analysis (Fig. 1). Similarly, liver, brain, and whole-embryo tissues showed derepression of HoxA6 and HoxA7 genes after Lsh depletion but unchanged HoxA10 and HoxA11 gene expression levels (Fig. 1A). In addition, HoxB4, HoxC6, HoxC8, and HoxD13 were derepressed in Lsh−/− MEFs (Table 1), indicating that Lsh does not exclusively affect silencing of the HoxA gene cluster. Thus, Lsh is an important transcriptional regulator of selected PRC targets during normal development.
Fig. 1.
Lsh deletion causes de-repression of Hox genes in various tissues. (A) RT-PCR analysis for detection of the indicated HoxA genes derived from Lsh−/− and Lsh+/+ MEFs, liver, brain, or whole embryo tissue (day 18 of gestation). (B) Real-time PCR analysis of HoxA gene expression comparing wild-type MEFs with Lsh−/− MEFs.
Table 1.
Summary of the gene expression patterns at different Hox clusters in Lsh−/− or Lsh+/+ tissues
| MEF> |
Brain |
Liver |
||||
|---|---|---|---|---|---|---|
| Lsh+/+ | Lsh−/− | Lsh+/+ | Lsh−/− | Lsh+/+ | Lsh−/− | |
| HoxA2 | + | + | − | + | + | + |
| HoxA5 | − | + | + | + | − | − |
| HoxA6 | − | + | − | + | − | + |
| HoxA7 | − | + | − | + | − | + |
| HoxA10 | + | + | + | + | + | + |
| HoxB3 | + | + | − | − | − | + |
| HoxB4 | − | + | − | + | − | + |
| HoxB6 | + | + | + | + | + | + |
| HoxC6 | − | + | − | + | + | + |
| HoxC8 | − | + | − | − | − | − |
| HoxC9 | + | + | + | + | + | + |
| HoxD10 | + | + | + | + | + | + |
| HoxD13 | − | + | − | − | − | − |
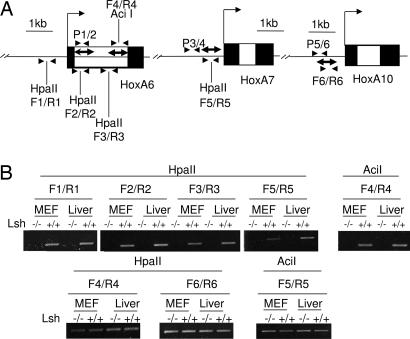
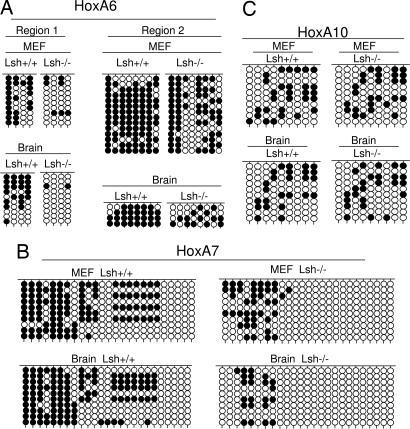
To address the molecular mechanism, we examined the DNA methylation pattern at Hox genes. Using methylation-sensitive PCR, genomic DNA derived from Lsh−/− MEFs and Lsh−/− liver samples was found sensitive to HpaII or AciI digestion in comparison with wild-type controls, indicating loss of methylation at several sites located at the promoter region of HoxA6 and HoxA7 genes. In addition, several sites in the gene body of HoxA6 that were previously shown to bind PRC components (16) were hypomethylated in the absence of Lsh (Fig. 2). To confirm and quantify the methylation results bisulphite sequencing was used, examining the same CpG regions of the HoxA6, HoxA7 genes as well as the control HoxA10 promoter region (Fig. 2A). DNA methylation levels were reduced at two HoxA6 sites, and a HoxA7 region comparing Lsh+/+ MEFs to Lsh−/− MEFs (58% vs. 18% and 81% vs. 54% at HoxA6 and 43% vs. 16% at HoxA7) (Fig. 3). Moreover, Lsh−/− brain tissue revealed an even more pronounced loss of CpG methylation compared with wild-type controls (55% vs. 5%, 88% vs. 44% at HoxA6 and 44% vs. 9% at HoxA7). In contrast, methylation levels at the HoxA10 promoter (that showed no change in gene expression after Lsh depletion) were unaltered in the absence of Lsh in either MEFs or brain tissue (32% vs. 30% and 28% vs. 28%) (Fig. 3C). Thus alterations in the DNA methylation pattern were correlated with transcriptional changes and controlled by Lsh.
Fig. 2.
Decreased CpG methylation after Lsh deletion at selected Hox gene sites. (A) Map of the HoxA6, HoxA7, and HoxA10 genes illustrating the location of primers (black triangles). The methylation-sensitive restriction enzyme sites HpaII and AciI are shown, as well as the primers (designated F1/R1 to F6/R6) used for methylation-sensitive PCR analysis. The ChIP primers are designated P1/2, P3/4, and P5/6. The regions analyzed for bisulphate sequencing are indicated with a double arrow. (B) Methylation-sensitive PCR analysis using genomic DNA derived from Lsh−/− and Lsh+/+ MEFs or liver after digestion with HpaII or AciI. PCR analysis amplifying a region around an AciI site after HpaII digestion (or HpaII site after AciI digestion) served as a control for equal input of DNA. The primers F6/R6 amplify a region lacking methylation sensitive restriction enzyme sites.
Fig. 3.
Lsh deletion reduces DNA methylation at HoxA6 and HoxA7 sites. (A) Genomic DNA derived from Lsh−/− and Lsh+/+ MEFs or brain was subjected to bisulfite sequencing and examined at regions for two regions of the HoxA6 gene, as indicated in the map of Fig. 2A. Methylated CpG are presented by black circles and unmethylated sites by open circles. (B) Bisulfite sequencing analysis for MEFs and brain at the HoxA7 gene. (C) Bisulfite sequencing analysis for MEFs and brain at the HoxA10 gene.
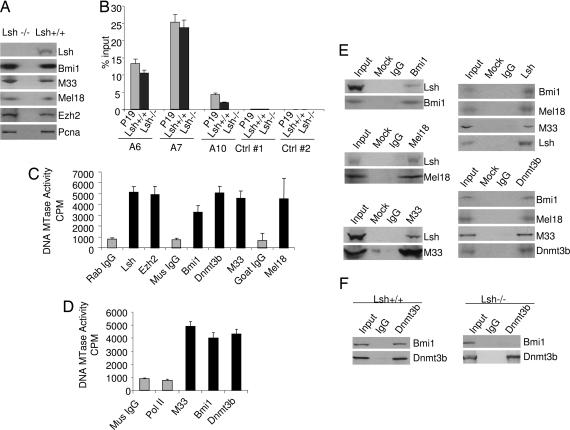
To investigate whether Lsh is directly involved in Hox gene silencing, we initially examined the expression levels of known PRC1 components to test whether Lsh deletion affects their expression level. However, there was no evidence that either PRC1 components such as Bmi1 (Pcgf4), M33 (Cbx2, mPc1), Mel18 (Pcgf2, Rnf110, and Zfp144), or PRC2 components such as Ezh2 (PRC2) were differentially expressed when comparing nuclear extracts derived from Lsh−/− and Lsh+/+ MEFs (Fig. 4A). Next we examined whether Lsh could directly associate with HoxA genes in P19 cells or MEFs (Fig. 4B). Using ChIPs with anti-Lsh antibodies followed by real-time PCR, we compared genes that were reactivated by Lsh deletion (HoxA6 and HoxA7) with those that were not affected (HoxA10). Whereas the HoxA6 and HoxA7 genes that were affected by Lsh deletion revealed binding of Lsh, the promoter regions of HoxA10 showed reduced association, and hardly any association was detected with intergenic control regions. As expected, no significant binding for Lsh was detected for either HoxA sites in Lsh−/− MEFs. This suggested that Lsh may play a direct role in the methylation at some HoxA sites. Next, we tested whether Lsh and Dnmts can associate with PRC1 components. Using nuclear extracts derived from P19 embryonal carcinoma cells (because these cells are high in de novo methyltransferase activity and Lsh protein levels), DNA methyltransferase activity was found to be associated with immunoprecipitates after using specific antibodies against Bmi1, M33, and Mel18 but was not detectable after precipitation using antibodies against Pol II as control (Fig. 4 C and D). The activity was comparable to that found after precipitation of Dnmt3b, the PRC2 subunit Ezh2 (7), or Lsh (15). In addition, immunoprecipitations of PRC1 components demonstrated a specific association between Bmi1, Mel18, or M33 with Lsh (Fig. 4E) and vice versa, specific immunoprecipitation of Lsh or Dnmt3b demonstrated an interaction with PRC1 components (Fig. 4E). The association between Dnmt3b and Bmi1 was readily detectable in wild-type MEF nuclear extracts but reduced in extracts derived from Lsh−/− MEFs (Fig. 4F). This suggested that the interaction of Dnmt3 with PRC at least in part depends on the presence of Lsh, and that it may be Lsh rather than DNA that performs a scaffolding-like function and promotes this interaction. Taken together, these data suggest a model in which Lsh plays a direct role in the control of Hox gene silencing.
Fig. 4.
Lsh is associated with PRC1 components. (A) Western blot analysis for detection of Bmi1, M33, Mel18, and Ezh2 using nuclear extracts derived from Lsh−/− and Lsh+/+ MEFs. Detection of Pcna and Lsh served as controls. (B) ChIP assays were performed on chromatin extracts derived from P19 (gray bar), Lsh+/+ (black bar), and Lsh−/− (open bar) MEFs using anti-Lsh antibodies and control IgG to detect association to specific HoxA6, HoxA7, and HoxA10 sites. Primers used for real-time PCR analysis are illustrated in Fig. 2A. In addition, two control primers were designed that are located within the HoxA cluster but >3,000 bp away from the HoxA10 or HoxA11 genes. The percentage of input was calculated for each precipitate. The values for the IgG control were <0.1% of input. (C and D) Nuclear extracts of P19 cells were immunoprecipitated with the indicated specific antibodies and assayed for DNA methyltransferase activity. (E) Western blot analysis for detection of Lsh after immunoprecipitation with anti-Bmi1, anti-Mel18, or anti-M33 antibodies using P19 nuclear extracts. Species-matched IgG or omission of antibody (Mock) served as controls. Western blot analysis for detection of Mel18, Bmi1, M33 after IP using anti-Lsh, or anti-Dnmt3b antibodies. (F) Western blot analysis for detection of Bmi1 after immunoprecipitation with anti-Dnmt3b comparing extracts derived from Lsh+/+ or Lsh−/− MEFs.
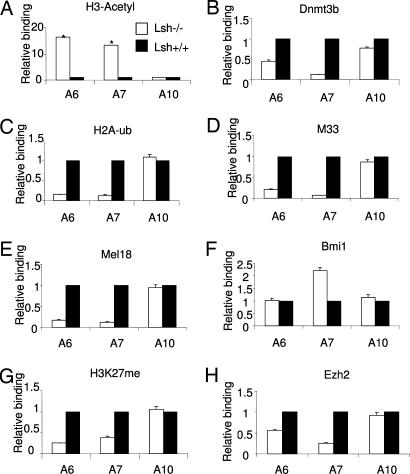
To understand whether Lsh can affect association of Dnmt3b to target sites and whether the presence of Lsh can modulate PRC-associated activities, histone modifications were examined by ChIP. Using quantitative PCR analysis, first histone acetylation, a marker for transcriptional activation, was examined comparing genes whose expression levels were affected by the absence of Lsh (such as HoxA6 and HoxA7) with HoxA10 that was unaffected by Lsh (Fig. 5A). H3 acetylation was enhanced in Lsh−/− MEFs at HoxA6 and HoxA7 loci but not at the HoxA10 gene and thus correlated well with transcriptional changes. In contrast to histone acetylation, Dnmt3b binding was reduced at HoxA6 and HoxA7 genes (2- and 9-fold, respectively) when comparing Lsh−/− MEFs to wild type (Fig. 5B). These data support the idea that Lsh at least in part promotes association of Dnmt3b to specific sites at Hox genes. To investigate whether Lsh and DNA methylation affect PRC-mediated histone modifications, we first analyzed H2A ubiquitylation mediated by PRC1 (17) (Fig. 5C). Whereas HoxA6 and HoxA7 sites revealed a reduction of H2A-K116 ubiquitylation of ≈7-fold in Lsh−/− samples compared with wild-type controls, HoxA10 sites were unchanged. Lsh deletion resulted in reduced association of M33 and Mel18 to HoxA6 and HoxA7 loci (ranging from 5- to 13-fold), in contrast to HoxA10 sites that were unaffected (Fig. 4 D and E). Bmi1 binding, though, did not show a reduction, suggesting that the recruitment of Bmi1 is independent of PRC2 activity and not sufficient to maintain silencing (Fig. 5F). However, decrease of M33 and Mel18 binding and reduced H2A ubiquitylation suggest that Lsh/DNA methylation was important for complete assembly and activity of the PRC1 complex.
Fig. 5.
Lsh controls Dnmt3b recruitment and PRC-mediated histone modifications at Hox sites. ChIP assays were performed from chromatin extracts of Lsh−/− (open bar) and Lsh+/+ (black bar) MEFs using the indicated antibodies to detect specific histone modifications or association of specific proteins at HoxA genes. Primers used for real-time PCR analysis are illustrated in Fig. 2A. The percentage of input was calculated for each precipitate and the values expressed as ratio of Lsh−/− samples over wild type. The following antibodies were used: (A) Antiacetylation of H3. The asterix indicate the minimum ratio above wild-type controls, because the actual values for Lsh−/− samples exceeded the range of the standard curve. (B) Anti-Dnmt3b. (C) Anti-H2A-K116 ubiquitylation. (D) Anti-M33. (E) Anti-Mel18. (F) Anti-Bmi1. (G) Anti-H3-K27 trimethylation. (H) Anti-Ezh2.
Because PRC1 recruitment via M33 (mPc1) is thought to depend on PRC2-mediated histone methylation (18), we examined Ezh2 binding and H3-K27 trimethylation levels. Both marks were decreased in Lsh−/− MEFs at HoxA6 and HoxA7 sites (2- to 4-fold) and unperturbed at HoxA10 sites compared with wild-type samples (Fig. 5 G and H). These data suggest that PRC2 cannot fully assemble in the absence of Lsh and DNA methylation.
DNA methylation has long been known to participate in genomic imprinting, X inactivation, repression of repeats, and silencing of tumor suppressor genes (1, 2). Here we provide evidence that Lsh can associate with some Hox genes, controls DNA methylation levels at Hox genes, and is also crucial for normal developmental regulation of Hox gene expression pattern. We further demonstrate that PRC1 and PRC2 activities are tightly correlated to DNA methylation, and that there may be a feedback loop between DNA methylation and PRC-mediated histone modifications. This supports the idea of a complex network of diverse epigenetic modifications rather than a simple sequential activation cascade during mammalian embryogenesis.
Based on Lsh homology with SNF2 family members, part of its activity may depend on presumed nucleosomal remodeling activity that may allow for better access of DNA-binding proteins to their nucleosomal target sites (9, 10). In addition, Lsh may also have chromatin remodeling-independent or scaffold-like functions, for example in promoting Dnmt3 activity or stabilizing the interactions of proteins. The observation that the association of Dnmt3b with Bmi1 is influenced by Lsh (Fig. 4F) would be consistent with a role of Lsh in scaffolding function. Possibly, via its association with PRC components, Lsh may bind to Hox loci and promote targeting of Dnmt. Alternatively, other not-yet-defined factors may recognize PRC-mediated histone modifications and lead to Lsh and subsequent Dnmt3b recruitment. Increased DNA methylation may result in decreased histone acetylation levels and a decline in transcription. Histone 3-K4 methyltransferases coupled to Pol II may alter H3-K4 methylation levels and may ultimately prevent spreading of the repressive H3-K27me mark (19). As another possibility, noncoding RNA transcripts that are prevalent in mammalian Hox clusters (20) may demarcate regions of gene silencing by regulating PRC2 occupancy and H3-K27me levels (21). A rise in H3-K27me may enhance PRC1 targeting to Hox genes (18). Alternatively, DNA methylation itself may affect PRC1 binding, as has been shown for reduced Bmi1 localization to PcG bodies after Dnmt1 depletion (22). More PRC1 binding may further promote DNA methylation and subsequent H3-K27methylation reinforcing the silencing marks in several feedback loops.
Although PRC silencing is highly conserved in different species, Drosophila and Caenorhabditis elegans do not show significant levels of genomic DNA methylation and lack an Lsh homolog. Thus the involvement of Lsh-mediated DNA methylation in PRC silencing shows a complexity that is unique to higher organisms. We hypothesize that Lsh associates only with a subset of polycomb complexes, because PRC components can assemble into various functionally distinct complexes and, as we report here, Lsh affects only some but not all examined Hox genes. More than 1,000 potential target sites have been reported for PRC components, and Lsh may also affect some of them (16, 23, 24). Thus the biologic activities of Lsh may be partially overlapping with PRC activities and may include effects on stem cell properties of breast epithelium, hematopoietic, and neuronal precursor potential and effects on skeletal development (3, 25, 26).
Cancer cells are long known to show aberrant DNA methylation patterns (1, 2) and recent evidence suggests that hypermethylation at promoter regions is linked to PRC binding (3–7). This study suggests that PRC-mediated silencing and DNA methylation are not aberrantly connected in cancer cells but part of an ordinary regulatory pathway involving Lsh. Whether this link is unique for embryonic cells or is also present in adult differentiated cells and why these pathway are targeted in cancer to loci that are usually unmethylated remains unknown, but Lsh is one possible candidate that could play a role in aberrant recruitment of Dnmts. On the other hand, cancer is also associated with global DNA hypomethylation, which in turn may derepress some Hox genes. Deregulation of Hox genes has been implied in hematopoietic malignancies and ectopic Hox gene expression determines the phenotype in ovarian epithelial cell cancer (3, 27, 28). The suggested connection between the two epigenetic pathways may shed new light on the molecular mechanisms involved in tumorigenesis and may prove helpful to improving strategies for cancer treatment or prevention in future.
Methods
Western Blot Analysis and Immunoprecipitations.
Samples were separated on 4–12% Tris-glycine SDS/PAGE gels and blotted onto Immobilon P membrane (Millipore, Bedford, MA). Western blotting was performed according to standard procedures by using ECL detection reagents, according to the manufacturer's instructions (Amersham, Piscataway, NJ). Nuclear extracts were prepared as described (29). For immunoprecipitations, the nuclear extract buffer was adjusted to a final concentration of 50 mM Tris (pH 7.5)/150 mM NaCl/1 mM EDTA/0.5% Nonidet P-40. Nuclear extracts (200 μg) were precleared for 30 min with protein G agarose (Invitrogen, Carlsbad, CA) and then incubated with 20 μl of antibodies for 2 h or overnight at 4°C. Washing was performed three times in 500 μl of buffer [50 mM Tris (pH 7.5)/150 mM NaCl/1 mM EDTA/0.5% Nonidet P-40] at 4°C, 5 min each cycle on a rotator. Antibodies used for immunoprecipitation or Western analysis were species-matched normal IgG (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-Lsh recombinant protein affinity-purified antibody, anti-Dnmt3b (Alexis, San Diego, CA), anti-Bmi1 (Upstate Biotechnology, Lake Placid, NY), anti-Ezh2 (Upstate Biotechnology), anti-Mel18 (Abcam, Cambridge, MA), anti-M33 antibody (BD Transduction Laboratories, Franklin Lanes, NJ), anti-Pcna (Santa Cruz Biotechnology), and anti-Pol II antibody (Upstate Biotechnology). The following secondary antibodies were used: goat anti-rabbit HRP-conjugated IgG, goat anti-mouse HRP-conjugated IgG, and rabbit anti-goat HRP-conjugated IgG (Santa Cruz Biotechnology).
In Vitro DNA Methyltransferase Activity Assay.
After 150 μl of nuclear extracts (29) derived from P19 cells was incubated with antibodies and protein G agarose for 2 h or overnight at 4°C with rotation, agarose beads were washed three times with nuclear extraction buffer and then again incubated with 150 μl of fresh nuclear extracts and antibodies to improve the yield of DNA methyltransferase activity. After a second round of incubation and washes, the beads were rinsed with DNA methyltransferase assay buffer (50 mM Tris, pH 7.8/1 mM EDTA/1 mM DTT/10% glycerol/1% Tween) and frozen at −80°C until future analysis. Assays were performed with immunoprecipitated material still attached to agarose beads. DNA methyltransferase activity was analyzed by the standard glass fiber method using S-adenosyl-l-(3H-methyl)-methionine (Amersham–Amersham Pharmacia) as the methyl donor and poly(d[I-C]) as the DNA substrate with an incubation time of 1.5 h. After washing, filters containing 3H were placed in scintillation fluid, and the level of radioactivity was counted. Two immunoprecipitations were performed on independent nuclear extracts for each antibody and normal species IgG controls. The average of two immunoprecipitations using independent nuclear extracts was graphed with error bars representing the standard deviation.
Methylation-Sensitive PCR.
DNA was extracted by using the DNeasy kit (Qiagen, Valencia, CA). DNA was completely digested with HpaII or AciI. To analyze the methylation status of the genomic DNA, the following PCR primer pairs A6 (F1/R1), A6(F2/R2), A6(F3/R3), A6(F4/R4), A7(F5/R5), and A10 (F6/R6) were used, as listed in supporting information (SI) Text.
PCRs were carried out as follows: 5 min at 94°C, 35 cycles of 60 s at 94°C, 30 s at 60°C, and 60 s at 72°C, and finally 5 min at 72°C. The PCR products were electrophoresed on 1% agarose gels, stained with ethidium bromide, and photographed.
RT-PCR.
Total RNA was prepared from MEF cells, smashed embryos (day 18 of gestation), liver, and brain tissue (day 18 of gestation) using TRIzol reagent (Invitrogen), according to the manufacturer's instructions. Any genomic DNA present was eliminated with TURBO DNA-free Kit (Ambion, Austin, TX). Approximately 1 μg of total RNA was reverse-transcribed by using iScript reverse transcriptase (Bio-Rad, Hercules, CA). Omission of reverse transcriptase served as a negative control. cDNA was amplified by using Platinum PCR SuperMix (Invitrogen). PCR followed by agarose gel electrophoresis using Hox primers (17) was performed as follows: 5 min at 94°C, 35 cycles of 60 s at 94°C, 60 s at 56–59°C, and 60 s at 72°C, followed by one cycle of 5 min at 72°C. For real-time PCR, the following primers were used as listed in SI Text.
ChIP.
For ChIP, cells were cross-linked with 1% formaldehyde, lysed, and sonicated on ice to generate DNA fragments with an average length of 200–800 bp. After preclearing, 1% of each sample was saved as input fraction. Immunoprecipitation was performed by using specific antibodies against the indicated proteins or IgG of different species used as control. After reversal of cross-linking, nucleic acids were prepared from the eluted complex, and PCR analysis was performed. Amplification conditions were as follows: 94°C for 4 min; 94°C for 1 min; 55°C for 1 min; 72°C for 1 min (35 cycles) and 72°C for 7 min. The following antibodies were used for ChIPs: H3K27 triM, triM Acetyl-H3 (Lys-9/14), ubiquityl-histone H2A, Bmi1, Ezh2 antibodies (Upstate Biotechnology), anti-Lsh recombinant protein affinity-purified antibody, Dnmt3b antibody (Alexis), Mel18 antibody (Abcam), and M33 antibody (BD Transduction Laboratories).
Real-time PCR primer pairs for ChIPs analysis are listed in SI Text.
Real-Time PCR Analysis.
For real-time PCR analysis, the MyiQ Single-Color Real-Time PCR machine (Bio-Rad) and Platinum SYBR Green qPCR SuperMix UDG (Invitrogen) were used. The PCR for ChIPs was initiated with one cycle of 95°C for 3 min, followed by 45 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. PCR for the RT-PCR analysis was initiated with one cycle of 95°C for 3 min, followed by 45 cycles of 95°C for 30 s, 59°C for 30 s, and 72°C for 30 s. The negative control without template was carried out for each PCR analysis. To quantify the amount of the template using real-time PCR data, standard titration experiments for each template and each primer set were performed, and linear regression equation and the calculation for DNA amounts were established by using Prism 3.0 software (GraphPad, San Diego, CA) and Microsoft (Redmond, WA) Excel. Every ChIP experiment includes species-specific IgG controls. The results have been calculated as percentage of Input (which lay usually between 5% and 20%) and then expressed as ratio of Lsh−/− over wild type. For better comparison, the wild-type samples were set to one, and the values expressed as ratio of Lsh−/− samples over wild type.
Bisulphite Sequencing.
Genomic DNA from MEF cells and brain tissue (day 18 of gestation) was subjected to bisulfite treatment by using CpGenome DNA modification kit (Chemicon International, Temecula, CA) according to the manufacturer's instructions. The PCR products were separated in agarose gels and purified by using the QIAEX II gel extraction kit (Qiagen). Amplified fragments were subcloned into the pCR2.1-TOPO vector with the TOPO TA Cloning Kit (Invitrogen). Independent clones for each fragment were sequenced by using the M13 F or M13 R and only sequences with individual fingerprint selected from analysis. The primers used are listed in SI Text.
Supplementary Material
Acknowledgments
We thank Rodney Wiles and Terry Stull for excellent technical assistance. We thank Nancy Colburn and Peter Johnson for helpful discussion of the manuscript. This project has been funded in whole or part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. N01-C0-12400. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Abbreviations
- MEF
murine embryonic fibroblasts
- PRC
polycomb repressive complex.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703669104/DC1.
References
- 1.Jones PA. Semin Hematol. 2005;42:S3–8. doi: 10.1053/j.seminhematol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Goll MG, Bestor TH. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 3.Sparmann A, van Lohuizen M. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 4.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 5.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 6.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohannad HP, Chen W, Daniel VC, Berman DM, et al. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 8.Geiman TM, Muegge K. Proc Natl Acad Sci USA. 2000;97:4772–4777. doi: 10.1073/pnas.97.9.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narlikar GJ, Fan HY, Kingston RE. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 10.Muegge K. Biochem Cell Biol. 2005;83:548–554. doi: 10.1139/o05-119. [DOI] [PubMed] [Google Scholar]
- 11.Dennis K, Fan T, Geiman TM, Yan QS, Muegge K. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun LQ, Lee DW, Zhang Q, Xiao W, Raabe EH, Meeker A, Miao D, Huso DL, Arceci RJ. Genes Dev. 2004;18:1035–1046. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. Nat Cell Biol. 2006;8:1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- 14.Fan T, Hagan JP, Kozlov SV, Stewart CL, Muegge K. Development (Cambridge, UK) 2005;132:635–644. doi: 10.1242/dev.01612. [DOI] [PubMed] [Google Scholar]
- 15.Zhu H, Geiman TM, Xi S, Schmidtmann A, Jiang Q, Chen T, Li E, Muegge K. EMBO J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 17.Cao R, Tsukada Y, Zhang Y. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papp B, Muller J. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemons D, McGinnis W. Science. 2006;313:1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- 21.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez-Munoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. Mol Cell Biol. 2005;25:11047–11058. doi: 10.1128/MCB.25.24.11047-11058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng W, Liu J, Yoshida H, Rosen D, Naora H. Nat Med. 2005;11:531–537. doi: 10.1038/nm1230. [DOI] [PubMed] [Google Scholar]
- 28.Owens BM, Hawley RG. Stem Cells. 2002;20:364–379. doi: 10.1634/stemcells.20-5-364. [DOI] [PubMed] [Google Scholar]
- 29.Sadowski HB, Shuai K, Darnell JE, Jr, Gilman MZ. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.