Abstract
Fusion of macrophages is an essential step in the differentiation of osteoclasts, which play a central role in the development and remodeling of bone. Osteoclasts are important mediators of bone loss, which leads, for example, to osteoporosis. Macrophage fusion receptor/signal regulatory protein α (MFR/SIRPα) and its ligand CD47, which are members of the Ig superfamily (IgSF), have been implicated in the fusion of macrophages. We show that CD200, which is not expressed in cells that belong to the myeloid lineage, is strongly expressed in macrophages at the onset of fusion. By contrast, the CD200 receptor (CD200R), which, like CD200, belongs to the IgSF, is expressed only in cells that belong to the myeloid lineage, including osteoclasts, and in CD4+ T cells. Osteoclasts from CD200−/− mice differentiated at a reduced rate. Activation of the NF-κB and MAP kinase signaling pathways downstream of RANK, a receptor that plays a central role in the differentiation of osteoclasts, was depressed in these cells. A soluble recombinant protein that included the extracellular domain of CD200 rescued the fusion of CD200−/− macrophages and their activation downstream of RANK. Conversely, addition of a soluble recombinant protein that included the extracellular domain of CD200R or short-hairpin RNA-mediated silencing of the expression of CD200R prevented fusion. Thus CD200 engagement of the CD200R at the initiation of macrophage fusion regulated further differentiation to osteoclasts. Consistent with in vitro observations, CD200−/− mice contained fewer osteoclasts and accumulated more bone than CD200+/+ mice. The CD200-CD200R axis is therefore a putative regulator of bone mass, via the formation of osteoclasts.
Keywords: fusion, macrophage, RANK, MAPK
Multinucleate osteoclasts originate from the fusion of macrophages and play a major role in the resorption of bone (1–4). Osteoclasts are essential for both the development and remodeling of bone, and increases in the number and/or activity of osteoclasts lead to diseases associated with generalized bone loss, such as osteoporosis, and others associated with localized bone loss, such as rheumatoid arthritis and periodontal disease. Because fusion is a key step in the differentiation of osteoclasts, a detailed understanding of the molecular mechanism of macrophage fusion should help develop strategies to prevent bone loss.
The adhesion of cells to one another that precedes fusion appears to involve a set of proteins similar to those exploited by viruses for fusion with host cells (5). It has been postulated, moreover, that viruses usurped the fusion-protein machinery from their target cells (1). It is now generally accepted that virus-cell fusion requires both an attachment mechanism and a fusion peptide. An example of such fusion involves gp120 of the HIV, which binds to CD4 on T lymphocytes and macrophages (6, 7), whereas the fusion molecule gp40, which is derived from the same precursor (gp160) as gp120, is thought to trigger the actual fusion event. We postulated (8) that the fusion machinery used by macrophages is similar to that used by viruses to infect cells. In 1998, we reported that the expression of macrophage fusion receptor/signal regulatory protein α (MFR/SIRPα) is induced transiently in macrophages at the onset of fusion (9). MFR/SIRPα and its receptor, CD47, belong to the superfamily of immunoglobulins (IgSF), as does CD4, and their interaction plays a role in the recognition of self and in the fusion of macrophages (10). To gain further insight into the mechanism of macrophage fusion, we subjected fusing alveolar macrophages from rats to genome-wide oligonucleotide microarray analysis, and we discovered the expression of CD200 de novo at the onset of fusion.
CD200 also belongs to the IgSF and has a short cytoplasmic tail. It is expressed on various types of mouse and human cells (see ref. 11 for a review) and on mouse osteoblasts (12), but not on macrophages. By contrast, the receptor for CD200 (CD200R), which, resembling CD200, contains two IgSF domains, is expressed predominantly in myeloid cells and includes an intracellular domain that mediates downstream signaling, typically delivering an inhibitory signal. Although five mouse CD200R-related genes, termed mCD200RLa-e, were identified, only one human homolog, called hCD200RLa, is known, yet apparently not expressed (see ref. 11 for a review). Both mCD200RLa and -Lb isoforms were characterized further and show close homology to CD200R in the extracellular region, with short cytoplasmic regions that contain a positively charged lysine residue in the transmembrane domain, possibly interacting with DAP12 to deliver an activating signal. It appears, however, that only CD200R and possibly the strain-specific CD200RLe bind CD200 (11, 13).
CD200-CD200R has a pattern of expression similar to that of MFR/SIRPα-CD47 in that CD200, like CD47, is widely expressed, whereas CD200R, like MFR/SIRPα, is expressed predominantly in cells that belong to the myeloid lineage. Therefore, we postulated that the CD200-CD200R axis might play a role in the fusion of macrophages based on the clearly defined role for the CD200-CD200R interaction to regulate macrophage function (14), and that mice that lack CD200 would have a defect in macrophage fusion and, as a result, in both osteoclast differentiation and bone remodeling.
We found that the expression of CD200 was potently induced de novo in macrophages at the onset of fusion, and that osteoclasts deficient in CD200 had a defect in multinucleation and in signaling downstream of receptor activator of NF-κB (RANK), which are essential for osteoclastogenesis. We also found that CD200-deficient mice had a lower number of osteoclasts and a higher bone density than wild-type mice. Together, our observations indicate that the CD200-CD200R axis plays a central role in the fusion of macrophages and the formation of osteoclasts.
Results
Expression of CD200 de Novo in Macrophages at the Onset of Fusion.
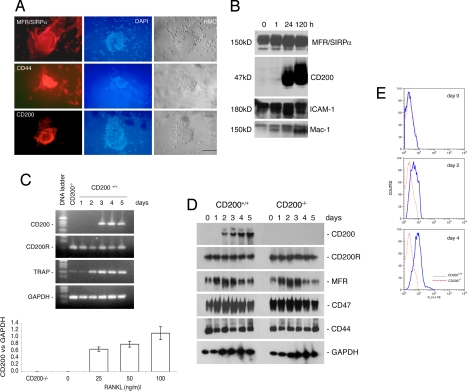
To identify previously undescribed components of the machinery of macrophage fusion, we submitted fusing alveolar macrophages from rats to genome-wide microarray analysis. Such macrophages provide an efficient and homogeneous model system for studies of macrophage fusion (refs. 8–10, 15; see ref. 2 for a review), because they are “naïve” and fuse spontaneously in vitro when plated confluently, without the addition of cytokines. Barely any transcripts encoding CD200 (GenBank accession no. X01785) were detected in freshly isolated macrophages, but the levels of transcripts in fusing macrophages were 0.6 ± 1.4, 34.9 ± 7.2, and 61.6 ± 23.4 times higher than those in freshly isolated cells 1, 24, and 120 h after plating, respectively (mean ± SD; n = 3). To confirm the cell-surface expression of CD200, we reacted multinucleated alveolar macrophages with a monoclonal antibody raised against the extracellular domain of CD200. In parallel, we subjected fusing alveolar macrophages to Western blot analysis at different times. We used antibodies directed against MFR/SIRPα as a control, because the expression of this protein is induced at the onset of macrophage fusion (8). Our results confirmed the strong and de novo expression of CD200 as early as 24 h after plating (Figs. 1 A and B). However, unlike MFR/SIRPα, CD200 was not expressed in mononucleate macrophages (Fig. 1A).
Fig. 1.
Rat alveolar macrophages and mouse bone marrow-derived macrophages express CD200 upon multinucleation. (A) Freshly isolated rat alveolar macrophages were plated at confluency over 50% of the surface of each well to promote fusion and multinucleation. After 5 days, they were subjected to immunohistochemical analysis. Note that mononucleated macrophages were positive for MFR/SIRPα and CD44 but not for CD200. (Scale bar, 1 mm.) Also note that multinucleate rat alveolar macrophages contained hundreds of nuclei that were stained with DAPI (blue). (B) Freshly isolated rat alveolar macrophages were plated as in A and subjected to Western blot analysis at the indicated times. Note that CD200 was not detected in macrophages for the first 24 h. (C) Mouse bone marrow-derived macrophages were cultured in the presence of M-CSF (30 ng/ml) and RANKL (50 ng/ml) for the indicated times to induce the differentiation of multinucleate osteoclasts. Cells were analyzed by RT-PCR. Note that mouse bone marrow-derived macrophages expressed transcripts for CD200 receptor (CD200R) but not for CD200. The abundance of CD200 mRNA relative to that of GAPDH, in response to M-CSF (30 ng/ml) and increasing doses of RANKL, was determined. (Scale bars represent standard deviations; n = 3.) (D) Mouse bone marrow-derived macrophages were cultured in the presence of M-CSF (30 ng/ml) and RANKL (50 ng/ml) for the indicated times to induce the differentiation of multinucleate osteoclasts. Cells were subjected to Western blot analysis by using antibodies directed against the indicated antigens. (E) Flow-cytometric analysis (in a FACS) of the expression of CD200. Mouse bone marrow-derived macrophages were isolated from CD200+/+ and CD200−/− mice, cultured in the presence of M-CSF (30 ng/ml) and RANKL (50 ng/ml) and subjected to flow-cytometric analysis at the indicated times with an antibody directed against CD200 and a control isotype antibody. Bone marrow-derived macrophages expressed increasing amounts of CD200 with time in the presence of M-CSF and RANKL, which promote fusion, multinucleation and osteoclastogenesis.
To investigate whether CD200 was also expressed in osteoclasts, we cultured mouse bone marrow-derived macrophages in the presence of macrophage colony-stimulating factor (M-CSF) (30 ng/ml) and RANK ligand (RANKL) (50 ng/ml) for 5 days to generate osteoclasts (16). Unlike MFR/SIRPα and CD44, neither transcripts encoding CD200 nor CD200 protein were detected in macrophages, but strong expression of such transcripts and of CD200 was induced by RANKL as early as day 2. Moreover, the induction of expression of CD200 transcripts depended on the dose of RANKL (Fig. 1C). By contrast, the expression of CD200R was clearly constitutive (Fig. 1 C and D). Of note, MFR/SIRPα, CD47, and CD44 were expressed in mouse osteoclasts during their differentiation, and the levels of these proteins were unaffected by disruption of the expression of CD200, because osteoclasts from mice deficient in CD200 expressed similar levels of these proteins. This observation suggests that the expression of these fusion molecules is regulated by a mechanism that is independent of and different from CD200.
To confirm that CD200 was expressed on the surface of osteoclasts, we cultured bone macrophages as described (16), reacted them at different times with a monoclonal antibody that recognized the extracellular domain of CD200, and subjected them to flow-cytometric analysis. The results, shown in Fig. 1E, confirm the strong and de novo cell-surface expression of CD200 at the onset of osteoclast fusion/multinucleation.
Together, our results indicate that CD200 might be a previously unrecognized component of the macrophage fusion machinery. Therefore, we postulated that the deletion of CD200 could affect the differentiation of osteoclasts.
The Absence of CD200 Impaired the Differentiation of Osteoclasts.
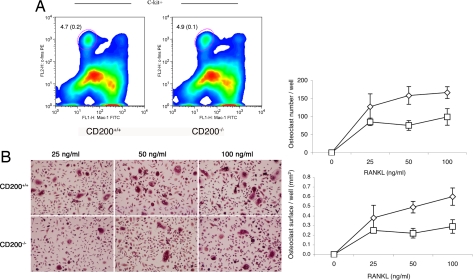
To examine first whether CD200 affects the number of osteoclast precursors, we subjected freshly isolated bone marrow cells to flow-cytometric analysis using surface markers expressed by preosteoclasts (17, 18). The percentage of precursor cells relative to the total number of bone marrow cells was similar in CD200+/+ and CD200−/− mice (Fig. 2A). We then compared the rates of osteoclastogenesis in vitro in CD200+/+ and CD200−/− mice. We cultured mouse bone marrow macrophages in the presence of M-CSF (30 ng/ml) and increasing concentrations of RANKL for 5 days to generate osteoclasts. The absence of CD200 resulted in a dose-dependent decrease in the number of osteoclasts and in the surface area covered by osteoclasts (Fig. 2B). These data strongly supported our hypothesis that CD200 plays a role in the formation of osteoclasts. Although fewer and smaller, bone-marrow-derived CD200−/− macrophages differentiated into osteoclasts that formed an actin ring and resorbed dentin; hence, they appeared morphologically and functionally mature [supporting information (SI) Fig. 6]. In addition, CD200−/− osteoclasts, like wild types, expressed mature osteoclast markers, such as calcitonin receptors, and osteoclast-associated receptor (OSCAR) and to a lesser level cathepsin K (SI Table 1). Together, our data suggest that CD200 plays a role in the multinucleation of osteoclasts but not in their ability to differentiate into active osteoclasts (SI Fig. 6).
Fig. 2.
Osteoclasts and their precursors are affected by the absence of CD200. (A) Bone marrow cells from 6-week-old CD200-deficient and wild-type mice were subjected to flow-cytometric analysis with antibodies directed against c-fms, Mac-1, and C-kit, as surface markers. Note that the absence of CD200 did not affect the number of osteoclast precursor cells (Left). (Scale bars, SD; n = 5.) (B) Bone-marrow-derived macrophages from 6-week-old CD200-deficient mice were cultured in the presence of M-CSF (30 ng/ml) and increasing concentrations of RANKL for 5 days to induce the differentiation of osteoclasts (Left). Bone marrow macrophages that lacked CD200 formed fewer osteoclasts than wild-type cells (Right). (Scale bars, SD; n = 5.)
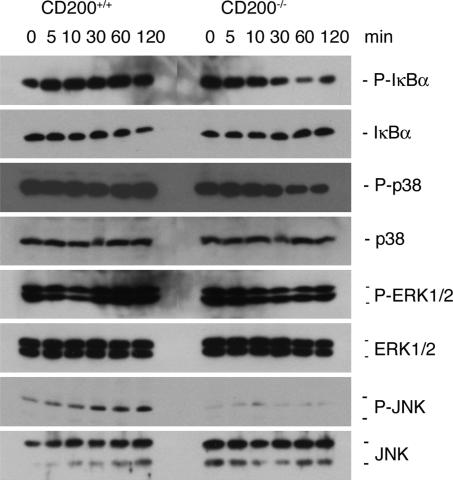
Because RANKL, which activates the NF-κB and MAP kinase signaling pathways that operate downstream of RANK, is essential for osteoclastogenesis, we next asked whether a deficiency in CD200 might affect signaling downstream of RANK. We cultured bone marrow cells from CD200-deficient and wild-type mice in the presence of M-CSF (30 ng/ml) for 2 days, then, after starving them for 2 h, we treated them with RANKL (50 ng/ml) up to 2 h and, finally, we subjected them to Western blot analysis with phosphorylated form-specific and control antibodies directed against IkBα, p38, ERK1/2, and JNK. Although activation of IkBα was slightly decreased, activation of JNK was almost completely abolished in cells that lacked CD200 (Fig. 3). These results revealed that the absence of CD200 attenuated the transduction of signals downstream of RANK and suggested that the CD200-CD200R interaction might play a role in this signaling pathway and in the formation of osteoclasts.
Fig. 3.
In osteoclasts deficient in CD200, the activation of signaling molecules downstream of RANK is suppressed. Bone marrow macrophages isolated from CD200-deficient, and wild-type mice were cultured in the presence of M-CSF (5 ng/ml) for 12–18 h. Nonadherent cells were further cultured for 2 days in 24-well dishes, starved for 2 h, and then stimulated with 50 ng/ml RANKL for the indicated times. Cells were subjected to Western blot analysis with antibodies directed against the indicated antigens. The activation, by phosphorylation, of IκB and JNK was less extensive in cells that lacked CD200 than in wild-type cells. This experiment was repeated three times with similar results.
The CD200-CD200R Axis Is a Component of the Fusion Machinery.
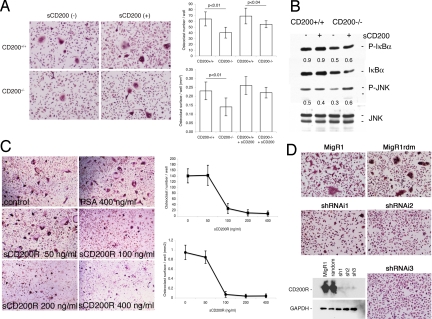
To address the putative role of the CD200-CD200R axis in the fusion of macrophages, we used several complementary strategies. First, we asked whether exogenous CD200 could rescue the differentiation of osteoclasts in vitro in cells that lack CD200. We generated a histidine-tagged soluble recombinant protein that included the extracellular domain of mouse CD200 (sCD200). We cultured bone marrow cells isolated from CD200-deficient and wild-type mice in the presence of M-CSF (30 ng/ml), RANKL (50 ng/ml), and sCD200 (0.5 μg/ml). The addition of sCD200 rescued multinucleation of CD200-deficient macrophages (Fig. 4A), whereas soluble histidine-tagged recombinant prostate-specific antigen, used as a control, did not (data not shown). We next asked whether sCD200-induced fusion resulted from the activation of JNK and IκB activation, which is suppressed in the absence of CD200. We cultured bone marrow cells from CD200-deficient and wild-type mice in the presence of M-CSF (5 ng/ml) for 2 days. After starving them for 2 h, we treated them for 30 min with RANKL (50 ng/ml), in the presence and absence of sCD200 (0.5 μg/ml) and, finally, we subjected them to Western blot, as described above. The addition of sCD200 restored the activation of JNK, but not of IκB, supporting a role for CD200 in the differentiation of osteoclasts via CD200R-mediated downstream signaling (Fig. 4B).
Fig. 4.
The CD200-CD200R axis is required for osteoclast fusion/multinucleation. (A) Bone-marrow-derived macrophages from 6-week-old wild-type mice were cultured in the presence of M-CSF (30 ng/ml) and RANKL (50 ng/ml) with or without the recombinant extracellular domain of CD200 (sCD200; 0.5 μg/ml) or recombinant prostate-specific antigen as a control. sCD200 allowed the differentiation of osteoclasts in macrophages that lacked CD200 (SD; n = 3). (B) Bone marrow macrophages isolated from CD200-deficient and wild-type mice were cultured in the presence of M-CSF (5 ng/ml) for 12–18 h. Nonadherent cells were cultured for a further 2 days in the presence of M-CSF (30 ng/ml), starved for 2 h, and then treated with RANKL (50 ng/ml) with or without sCD200 (0.5 μg/ml) for 30 min. The cells were then subjected to Western blot analysis with the indicated antibodies against IkBα and JNK and their phosphorylated forms. Numbers represent relative expression of P-IκB and P-JNK over JNK. The addition of sCD200 restored the activation of JNK but not of IκBα. (C) Bone-marrow-derived macrophages from 6-week-old wild-type mice were cultured in the presence of M-CSF (30 ng/ml) and RANKL (50 ng/ml) with or without the recombinant extracellular domain of the CD200 receptor (sCD200R). sCD200R blocked the fusion of macrophages (SD; n = 5), whereas recombinant prostate-specific antigen had no effect (osteoclast surface, 0.38 ± 0.1; osteoclast number, 170.0 ± 23.3). (D) Bone-marrow-derived macrophages from 6-week-old wild-type mice were cultured in the presence of M-CSF (30 ng/ml) for 2 days before being transduced with the retroviral vector MigR1, which encoded, or not, shRNAs designed after the CD200R1 cDNA. A construct encoding random (rdm) oligonucleotides was used as a control. Each of the three targeting retroviral constructs, namely shRNAi1, shRNAi2, and shRNAi3, abolished the expression of CD200R1 (see Western blot, Bottom Left) and prevented the formation of multinucleate osteoclasts (Top and Right). These experiments were reproduced several times with similar results.
We postulated next that, if the CD200-CD200R interaction plays a role in fusion, interference with this interaction should block fusion. We engineered a histidine-tagged soluble recombinant protein that included the extracellular domain of mouse CD200R (sCD200R). We cultured bone marrow cells from wild-type mice in the presence of M-CSF (30 ng/ml) and RANKL (50 ng/ml) in the absence and presence of sCD200R (50–400 ng/ml) and soluble histidine-tagged recombinant prostate-specific antigen (400 ng/ml). As anticipated, osteoclastogenesis was blocked in the presence of sCD200R in a dose-dependent manner (Fig. 4C).
We next asked whether osteoclasts express other CD200R-like molecules (19). We found that mouse osteoclasts expressed transcripts that encoded CD200R and also CD200RLc (data not shown). To date, the function of this additional receptor remains unclear. Because CD200 could activate alternative receptors, such as CD200Lc, and because sCD200R could block the interaction of CD200 with alternative receptors, we questioned whether CD200 signals specifically through CD200R to modulate fusion. To do so, we attempted to silence the expression of CD200R in fusing macrophages by RNAi with short-hairpin RNA (shRNA). We generated three retrovirus-based shRNA constructs that targeted mouse CD200R (shRNAi1, shRNAi2, and shRNAi3), as well as a construct that encoded random sequences (MigR1rdm). We transduced bone marrow macrophages isolated from wild-type mice with these constructs, as well as the empty vector (MigR1). Each one of the shRNA constructs (shRNAi1, shRNAi2, and shRNAi3) interfered with the expression of CD200R and prevented the fusion of osteoclasts (Fig. 4D). By contrast, neither MigR1 nor MigR1rdm affected the expression of CD200R and the differentiation of osteoclasts. Together, these results confirmed the proposed central role for the CD200-CD200R axis in the fusion of macrophages and in osteoclastogenesis.
The Absence of CD200 Does Not Impair the Differentiation of Osteoblasts.
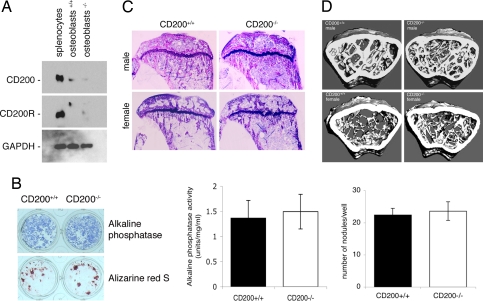
To determine whether osteoblasts express CD200 and its receptor, CD200R, we cultured bone marrow cells for 9 days in the presence of ascorbic acid (50 μg/ml) and β-glycerophosphate (10 mM). We then subjected these bone marrow-derived osteoblasts to Western blot analysis. Such an approach confirmed the relatively low-level expression of CD200 (12) and the absence of CD200R in osteoblasts (Fig. 5A). To determine whether the absence of CD200 affects the differentiation of osteoblasts, we compared the alkaline phosphatase (ALP) activity and the ability to form bone-like nodules of bone marrow-derived osteoblasts from CD200-deficient and wild-type mice. The absence of CD200 had no effect on ALP activity and on the formation of nodules by osteoblasts (Fig. 5B).
Fig. 5.
The absence of CD200 increases bone density. (A) Bone marrow cells from 6- to 8-week-old CD200-deficient and wild-type mice were plated in 24-well plates (5 × 106 cells/well) and cultured for 9–11 days in α-MEM supplemented with ascorbic acid (50 μg/ml) and β-glycerophosphate (10 mM) to acquire the osteoblast phenotype. Osteoblast lysates were analyzed for protein concentration and subjected to Western blot analysis with antibodies directed against mouse CD200, CD200R, and GAPDH. (B) Bone-marrow-derived osteoblasts were examined for ALP activity and stained for calcium with alizarin red S to allow quantitation of the number of nodules per well (SD; n = 6). Cell lysates were analyzed for ALP activity (Left; SD; n = 6). These experiments were repeated three times with similar results. (C) Toluidine blue-stained sections of proximal tibiae from 2-month-old CD200-deficient male and female mice and wild-type mice. (Scale bar, 1 mm.) (D) Microcomputed tomography analysis of distal femurs from 6-month-old male and female CD200-deficient mice. Note the increased density of trabeculae inside the distal femur of CD200-deficient male and female mice as compared with wild types. The widest diameter of the bone sections correspond ≈3 mm.
CD200-Deficient Mice Had Higher Bone Density and Fewer Osteoclasts than Wild-Type Mice.
Our in vitro data clearly indicate that CD200 and its receptor CD200R play a positive role in the differentiation of osteoclasts but not of osteoblasts. If this is true, then mice that lack CD200 should have a lower number of osteoclasts and a higher bone mass.
To address this question, we first subjected 2-month-old male and female CD200−/− and wild-type mice to DEXA analysis (see Material and Methods). Consistent with our hypothesis, both male and female CD200−/− mice had higher spinal bone densities than corresponding wild-type mice (SI Fig. 7A). Peripheral quantitative tomography analysis of the femurs from these mice revealed that CD200 deficiency was associated with an increase in the total density of the shaft in both males and females and of the distal femur in females, as compared with age- and sex-matched wild-type mice (SI Fig. 7B). In CD200-deficient female mice, there was a decrease in the trabecular area of the shaft and the distal part of the femur, whereas in CD200-deficient male mice, there was an increase in the trabecular area of the distal femur only, as compared with the respective wild-type mice. In CD200-deficient male mice, there was an increase and in CD200-deficient female mice a decrease in periosteal circumference, in both the shaft and the distal femur, as compared with corresponding wild-type mice. It appeared, therefore, that CD200 deficiency has led to the enhanced accumulation of bone.
To confirm that the increase in total bone density was the result of a decrease in the number of osteoclasts, we subjected the distal femurs from CD200-deficient and age- and sex-matched wild-type mice to histomorphometric analysis. Our results confirmed that CD200-deficient mice, both males and females, had an increase in trabecular bone volume when compared with wild types (Fig. 5C and SI Fig. 7C). We also found a decrease in the relative bone surface area that was occupied by osteoclasts in both male and female CD200-deficient mice. To our surprise, despite the increase in bone density in CD200-deficient female mice, we found a decrease in the relative surface area of bone that was covered by osteoblasts (SI Fig. 7C). This result further suggested that it was, indeed, the osteoclasts that were responsible for the higher bone volume in CD200-deficient mice.
To determine whether the increase in bone volume persisted with aging, we subjected the distal femurs from both CD200-deficient and wild-type 6-month-old mice to high-resolution microcomputed tomography analysis. Both male and female CD200-deficient mice had accumulated more trabecular bone than the corresponding wild types (Fig. 5D and SI Table 2). This observation was supported by peripheral quantitative tomography analysis of the same bones, which showed that trabecular density was higher in the CD200-deficient mice than in the corresponding wild types (SI Fig. 7D).
Discussion
The CD200-CD200R axis appears to be a central player in the fusion and/or multinucleation of macrophages, which is required for the differentiation of osteoclasts, and the regulation of bone mass. Although our results confirm that mononucleate macrophages do not express CD200 (14), they reveal that their fusion is accompanied by strong and de novo expression of CD200. Not only is the expression of CD200 abruptly induced in fusing osteoclasts, but absence of CD200 impairs osteoclastogenesis, with a subsequent increase in bone volume and, hence, a form of osteosclerosis.
Our analysis of the number of bone marrow macrophages/osteoclast precursor cells as a percentage of the total number of bone marrow cells, which was similar in CD200-deficient and wild-type mice, suggests the CD200-CD200R axis does not control the differentiation of premonocytes (17, 18). This is in contrast with the numbers of splenic and mesenteric lymph node macrophages, which are elevated in mice that lack CD200 (14). It is possible that monocytes from bone marrow are less differentiated than those from lymphoid organs, which might express low levels of CD200. Nevertheless, the decreases in the numbers of osteoclasts in CD200-deficient mice cannot be attributed to decreases in numbers of precursor cells.
Of possible relevance to fusion, genes for CD200-like proteins have been identified in the genomes of some, but not all, members of families of double-stranded DNA viruses, such as poxviruses, herpesviruses, and adenoviruses (20, 21). Moreover, the product of the K14 gene of Kaposi's sarcoma-associated herpesvirus is a ligand for CD200R (20, 21). Similarly, M141R is a cell-surface protein encoded by myxoma virus with significant homology at the amino acid level to CD200, required for the full pathogenesis of myxoma virus in the European rabbit (22). Most importantly, both CD200 and its viral homologs activate the CD200R to down-regulate basophil (HHV-8; ref. 23) and macrophage (HHV-8 and M141R; refs. 21 and 22) function. Hence, as might be the case for CD47, which is homologous to proteins encoded by vaccinia and myxoma virus (24, 25), viruses might have “stolen” CD200 to allow them to evade the immune response and to fuse with and infect cells. Of note, in both Drosophila myoblast fusion and mammalian macrophage fusion, members of the IgSF mediate adhesion and also initiate intracellular signaling events (4), suggesting a supporting role for the CD200-CD200R axis in fusion; however, this is highly hypothetical.
Although the CD200-CD200R axis plays an inhibitory role in the immune system (see ref. 11 for a review), it appears to play a supporting role in macrophage fusion via RANK signaling, because the absence of CD200 or the silencing of CD200R slow down the differentiation of osteoclasts. Because it has been proposed that the MFR/SIRPα also transmits an inhibitory signal to myeloid cells via its ITIM domain and facilitates macrophage fusion (1, 2), it is possible that these two axes, namely MFR/SIRPα-CD47 and CD200-CD200R, work in tandem to secure the fusion of osteoclasts while preventing their activation in response to CD200R and MFR/SIRPα ligation. Indeed, macrophages might require “deactivation” or “suppression” to proceed with fusion. This is a concept that necessitates further investigation. In that respect, mice that lack both CD47 and CD200 might provide a model to answer this question. In addition, we cannot exclude the possibility that CD200 and its receptor associate both in cis and in trans via their N-terminal domains, because the fusing partners are both macrophages. Indeed, it will be of interest to determine whether downstream signaling is differentially activated in cis or in trans in future studies.
That a defect in osteoclastogenesis in CD200-deficient mice results from a defect in activation downstream of RANK suggests possible cross-talk between CD200R and RANK. We should note, however, that, although the absence of CD200 slows down osteoclastogenesis, it does not prevent the expression of MFR/SIRPα and CD44, which are candidate members of the fusion machinery in macrophages. It remains to be determined whether the absence of CD200 affects the expression of DC-STAMP, the most recently identified component of the macrophage fusion machinery (2, 26). Together, our results suggest that the machinery for macrophage fusion involves multiple and, possibly, redundant molecules.
The absence of CD200 increased bone mass, and the soluble recombinant extracellular domain of CD200R blocked macrophage fusion in vitro. Thus, CD200 and its receptor might be recently discovered targets in efforts to prevent bone loss. However, even though osteoblasts in culture express low levels of CD200, and the absence of CD200 does not affect their differentiation in vitro, we cannot exclude a possible role for CD200 in these cells in vivo. Further studies involving the treatment of animal models with the soluble recombinant extracellular domain of CD200R will help clarify this issue.
Materials and Methods
Animals and Cells.
CD200−/− mice were produced by homologous recombination, as described (14). Mice whose bones were subjected to histomorphometric analysis received two i.p. injections of calcein (3 μg/g body weight; Merck, Darmstadt, Germany) on days 1 and 6 before death. The Yale Animal Care and Use Committee approved all experiments.
Bone-marrow-derived macrophages and osteoclasts were generated from 6- to 12-week old CD200−/−, and CD200+/+ mice were prepared as before (16). Osteoblast ALP and mineralized nodule formation assays were performed according to ref. 27.
Reagents.
Recombinant mouse RANKL and M-CSF were obtained from R&D Systems (Minneapolis, MN). A mouse monoclonal antibody directed against rat CD200 and rat monoclonal antibodies directed against mouse CD200 and its receptor CD200R were purchased from Serotec (Raleigh, NC). Mouse anti-rat CD200R antibody was kindly provided by A. N. Barclay (Oxford University, Oxford, U.K.). A polyclonal antibody directed against the intracellular domain of MFR has been published (10). Rabbit polyclonal antibodies directed against p38, phosphorylated-p38 (P-p38), ERK1/2, P-ERK1/2, JNK, and mouse monoclonal antibodies directed against IκB, P-IκB, and P-JNK were obtained from Cell Signaling Technology (Beverly, MA). A monoclonal antibody directed against mouse CD44 was obtained from BD Bioscience (Franklin Lakes, NJ). A mouse monoclonal antibody directed against GAPDH was purchased from Novus Biologicals (Littleton, CO). Horseradish peroxidase-conjugated F(ab′)2 directed against rabbit and mouse IgG were purchased from Jackson ImmunoResearch (West Grove, PA). Rat anti-mouse monoclonal antibodies used for flow cytometry included anti-Mac1 (CD11b) conjugated to fluorescein (Mac1-FITC; M1/70) and rat FITC-IgG2b (PharMingen, San Diego, CA); anti-c-fms conjugated to phycoerythrin (c-fms-PE; IgG2b) and anti-c-Kit conjugated to allophycocyanin (c-kit-APC; IgG2b); and isotype matching antibodies (eBioscience, San Diego, CA). Secondary antibody anti-rat IgG2a conjugated to FITC was purchased from PharMingen. Phalloidin-Alexa fluor 568 was purchased from Invitrogen (Carlsbad, CA) and dentin discs from IDS (Bolton, U.K.). All supplies and reagents for tissue culture were endotoxin-free. Some bone marrow cells were treated with polymyxin B sulfate for 24 h to avoid the effects of the endotoxin before treatment. Dentin discs were obtained from IDS and osteologic slides from BD Bioscience.
Flow Cytometry.
Cells were stained with the primary antibody, incubated for 30 min on ice, and washed twice with washing buffer (5% FCS/PBS). The secondary antibody was added, and the cells were incubated for 30 min on ice. After incubation, cells were washed twice with washing buffer and suspended in washing buffer for FACS analysis, which was performed by using a FACScalibur (BD Bioscience).
RT-PCR.
See SI Text.
Real-Time PCR.
See SI Text.
Generation of the Soluble Extracellular Domain of Mouse CD200 (sCD200) and CD200R (sCD200R).
See SI Text.
Retrovirus-Mediated shRNAi.
See SI Text.
Bone Radiography, Microcomputed Tomography, Bone Densitometry, and Histomorphometry.
See SI Text.
Statistical Analysis.
Statistically significant differences among experimental groups were evaluated by the analysis of variances (28). The significance of mean changes was determined by an unpaired Student's two-tailed t test, and significance was recognized when P < 0.05.
Supplementary Material
Acknowledgments
We thank Dr. N. A. Barclay for his generous gift of the anti-rat CD200R antibody and Dr. Hua Zhu Ke for his help with pQCT and microCT analyses. We thank Dr. Ann Altman for careful editing of this manuscript. J.K. was the recipient of a Boehringer Ingelheim Fellowship. This work was supported by funds from the National Institutes of Health (Grant DE12110, to A.V.).
Abbreviations
- IgSF
Ig superfamily
- CD200R
CD200 receptor
- ALP
alkaline phosphatase
- MFR/SIRPα
macrophage fusion receptor/signal regulatory protein α
- MEM
minimum essential medium
- RANK
receptor activator of NF-κB
- RANKL
RANK ligand
- M-CSF
macrophage colony-stimulating factor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702811104/DC1.
References
- 1.Vignery A. Int J Exp Pathol. 2000;81:291–304. doi: 10.1111/j.1365-2613.2000.00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vignery A. Trends Cell Biol. 2005;15:188–193. doi: 10.1016/j.tcb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Vignery A. J Exp Med. 2005;202:337–340. doi: 10.1084/jem.20051123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen EH, Grote E, Mohler W, Vignery A. FEBS Lett. 2007;581:2181–2193. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 6.Dalgleish AG, Beverly P, Clapham P, Crawford D, Greaves M, Weiss R. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 7.Klatzmann D, Champagne E, Chamaret S, Grust J, Guetard D, Hercent T, Gluckmann JC, Montagnier L. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 8.Saginario C, Qian H-Y, Vignery A. Proc Natl Acad Sci USA. 1995;92:12210–12214. doi: 10.1073/pnas.92.26.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saginario C, Sterling H, Beckers C, Kobayashi R, Solimena M, Ullu E, Vignery A. Mol Cell Biol. 1998;18:6213–6223. doi: 10.1128/mcb.18.11.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han X, Sterling H, Chen Y, Saginario C, Brown EJ, Frazier WA, Lindberg FP, Vignery A. J Biol Chem. 2000;275:37984–37992. doi: 10.1074/jbc.M002334200. [DOI] [PubMed] [Google Scholar]
- 11.Minas K, Liversidge J. Crit Rev Immunol. 2006;26:213–230. doi: 10.1615/critrevimmunol.v26.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee L, Liu J, Manuel J, Gorczynski RM. Immunol Lett. 2006;105:150–158. doi: 10.1016/j.imlet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Hatherley D, Cherwinski HM, Moshref M, Barclay AN. J Immunol. 2005;175:2469–2474. doi: 10.4049/jimmunol.175.4.2469. [DOI] [PubMed] [Google Scholar]
- 14.Hoek RM, Ruuls SR, Murphy CA, Wright GJ, Goddard R, Zurawski SM, Blom B, Homola ME, Streit WJ, Brown MH, et al. Science. 2000;290:1768–1771. doi: 10.1126/science.290.5497.1768. [DOI] [PubMed] [Google Scholar]
- 15.Sterling H, Saginario C, Vignery A. J Cell Biol. 1998;143:837–847. doi: 10.1083/jcb.143.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Cuartas C, Cui W, Choi Y, Crawford DT, Ke H.-Z., Kobayashi KS, Flavell RA, Vignery A. J Exp Med. 2005;201:1169–1177. doi: 10.1084/jem.20041444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arai F, Miyamoto T, Ohneda O, Inada T, Sudo T, Brasel K, Miyata T, Anderson DM, Suda T. J Exp Med. 1999;190:1741–1754. doi: 10.1084/jem.190.12.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimi E, Aoki K, Saito H, D'Acquisto F, May MJ, Nakamura I, Sudo T, Kojima T, Okamoto F, Fukushima H, et al. Nat Med. 2004;10:617–624. doi: 10.1038/nm1054. [DOI] [PubMed] [Google Scholar]
- 19.Wright GJ, Puklavec MJ, Willis AC, Hoek RM, Sedgwick JD, Brown MH, Barclay AN. Immunity. 2000;13:233–242. doi: 10.1016/s1074-7613(00)00023-6. [DOI] [PubMed] [Google Scholar]
- 20.Chung YH, Means RE, Choi JK, Lee BS, Jung JU. J Virol. 2002;76:4688–4698. doi: 10.1128/JVI.76.10.4688-4698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster-Cuevas M, Wright GJ, Puklavec MJ, Brown MH, Barclay AN. J Virol. 2004;78:7667–7676. doi: 10.1128/JVI.78.14.7667-7676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cameron CM, Barrett JW, Liu L, Lucas AR, McFadden G. J Virol. 2005;79:6052–6067. doi: 10.1128/JVI.79.10.6052-6067.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiratori I, Yamaguchi M, Suzukawa M, Yamamoto K, Lanier LL, Saito T, Arase H. J Immunol. 2005;175:4441–4449. doi: 10.4049/jimmunol.175.7.4441. [DOI] [PubMed] [Google Scholar]
- 24.Parkinson JE, Sanderson CM, Smith GL. Virology. 1995;214:177–188. doi: 10.1006/viro.1995.9942. [DOI] [PubMed] [Google Scholar]
- 25.Cameron CM, Barrett JW, Mann M, Lucas A, McFadden G. Virology. 2005;337:55–67. doi: 10.1016/j.virol.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 26.Yagi M, Miyamoto T, Sawatani Y, Iwamoto K, Hosogane N, Fujita N, Morita K, Ninomiya K, Suzuki T, Miyamoto K, et al. J Exp Med. 2005;202:345–351. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morinobu M, Nakamoto T, Hino K, Tsuji K, Shen ZJ, Nakashima K, Nifuji A, Yamamoto H, Hirai H, Noda M. J Exp Med. 2005;201:961–970. doi: 10.1084/jem.20041097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zar JH. Biostatistical Analysis. Englewood Cliffs, NJ: Prentice-Hall; 1984. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.