The pCaSpeR (1) and pUAST vectors (2) are two of the most commonly used Drosophila transformation vectors. However, although they have great utility in their current form, their multiple cloning sequences (MCSs) have a limited number of unique restriction sites (Figure 1). This is particularly true for the pUAST vector, whose MCS has only five unique sites. Further, neither of the MCSs in pCaSpeR or pUAST are present in small shuttle or cloning vectors, which is problematic, because the large size (>8 kb) of the transgenesis vectors requires sequence manipulations such as site-directed mutagenesis or deletion dropouts to be done in small plasmid vectors and the modified DNA to be moved to the transgenesis vectors. The lack of matching shuttle vectors further constrains the usable cloning sites and can make moving large genomic fragments between a cloning vector and a transgenesis vector problematic.
Figure 1. Comparison of the pCaSpeR5, pUAS-C5, pC5-Kan vectors with the pCaSpeR4 and pUAST vectors.
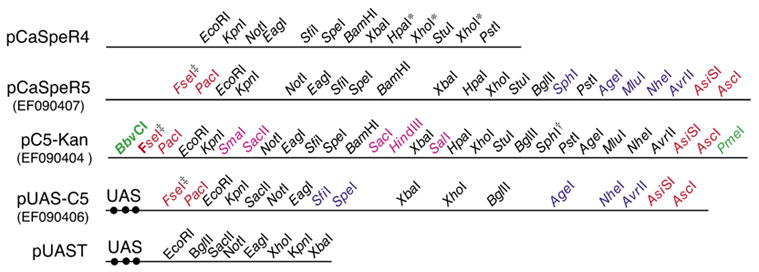
Maps comparing the restriction sites in the new and standard transgenesis vectors are shown. GenBank accession nos. for the CaSper5 vectors are shown in parenthesis below the vector names. UAS, upstream activating sequences containing the binding sites (filled black circles) for the GAL4 transcription factor; red text, new 8-cutter sites for moving fragments from pC5-Kan or pBS-C5 to pCaSper5 or pUAS-C5; blue text, 6-cutter sites present in pCaSpeR5 but not pCaSpeR4 or in pUAS-C5 but not pUAST (however, note there is a different order of sites common to pUAS-C5 and pUAST); bold text, 6-cutter sites present in pC5-Kan but not pCaSpeR4; pink text, 6-cutter sites present in pCaSpeR5 or pUAS-C5 but only unique in pC5-Kan and pBS-C5; green text, blunt or imperfect 8- and 7-cutter sites in pC5-Kan and pBS-C5 that can be used to prevent ligation of the pC5-Kan or pBS-C5 into the target vector in ligations done without gel purification; ◇, 95% of BbvCI ends cannot be religated (New England Biolabs product information); *, multiple sites for these restriction enzymes are present in the multiple cloning sequences (MCSs), but these sites are still pseudo-unique because they are not present in the vector backbone; †, the SphI recognition site contains an ATG that has been eliminated in pC5-KanΔSphI (EF090405) and pBS-C5ΔSphI (EF090403); ‡, FseI is not stable at −20°C, necessitating storage at −70°C for periods >30 days. Inserts into the MCSs can be sequenced using the following primers: pC5-Kan, pC5-Kan FseI 5′-GCCATCACGAGATTTCGATTCC-3′ and pC5-Kan AscI 5′-AAGCAGCAGATTACGCGCAG-3′; pCaSpeR5, pCasper-MCS-PstI 5′-CACGGACATGCTAAGGGTTA-3′ and pCasper-MCS-EcoRI 5′-AGTTCAATGATATCCAGTGCAG-3′; pUAS-C5 pUAST-5737 5′-CCAGCAACCAAGTAAATCAACTGC-3′ and pUAST-5432 5′-CCCATTCATCAGTTCCATAGGTTG-3′; pBS-C5, standard T3, T7, and M13 primers.
To overcome the above limitations, we engineered a new MCS based on the pCaSpeR4 MCS that adds five new 6-base cutter sites, but most importantly flanks the entire MCS by two 8-base cutters on each side (Figure 1). We call this improved vector pCaSpeR5, since the new vector retains all the restriction sites of the pCaSpeR4 MCS in their original order. We also created pUAS-C5 by replacing the MCS of pUAST with a modified version of the pCaSpeR5 MCS (C5 MCS) that lacked the ATG-containing SphI site (Figure 1). Although the pUAST has many common 6-cutter sites in its backbone, the C5 MCS nonetheless adds five new 6-cutter and the flanking four 8-cutter sites to the pUAS expression vector. The pCasper5 and pUAS-C5 vectors were shown to be functional by cloning a GMR-nvYFP transgene (3) into pCaSper5 and nvYFP (3) into pUAS-C5 (Figure 2). Injection of these constructs into embryos yielded typical numbers of transgenic offspring (>33 and >46 transgenic lines per 200 injected embryos for pCaSpeR5 and pUAS-C5, respectively) that expressed yellow fluorescent protein (YFP) strongly in eye discs (pCaSpeR5:: GMR-nvYFP) or in the tracheal system [pUAS-C5:: nvYFP lines crossed to the tracheal btl-gal4 driver (4)] at levels at least comparable or stronger than to similar constructs in other transgenesis vectors (Figure 2).
Figure 2. The pCaSpeR5 and pUAS-C5 vectors are functional.
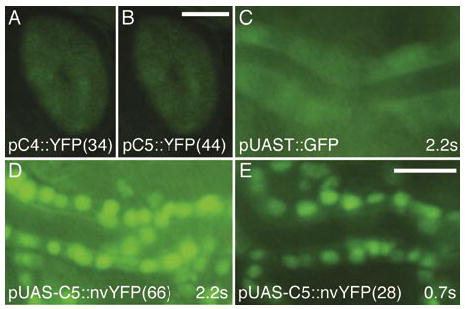
(A and B) Constructs expressing a yellow fluorescent protein (YFP) (3) driven by the glass multimer reporter (GMR) enhancer element (5) produce comparable expression in adult eyes of transgenic animals made using (A) pCaSpeR4 (pC4::YFP) and (B) pCaSpeR5 (pC5::YFP) vectors. Representative lines are shown with line numbers in parenthesis. Exposure times for A and B were 2.2 s and were made using a Model MZFLIII microscope (Leica Microsystems GmbH, Wetzlar, Germany), a Model C4742-95 camera (Hamamatsu Photonics K.K, Hamamatsu, Japan), and a green fluorescent protein (GFP) filter set (Chroma Technology, Rockingham, VT, USA). Scale bar for A and B in panel B, 25 μm. (C E) A nuclear-localized venus-YFP (nvYFP) (3) construct in pUAS-C5 driven by the tracheal-specific btl-Gal4 appears brighter (D and E) than the widely used pUAST-GFP driven by btl-Gal4 (C) (4). Images are of live stage 16 embryos heterozygous for each of the driver and responder constructs. Images were captured using an AxioPlan II microscope, an AxioCam camera (both from Carl Zeiss, Jena, Germany), and a GFP filter set using the indicated exposure times. Scale bar for C E in panel E, 5 μm.
To facilitate clone manipulation, we created a small ampicillin (Amp)-resistant vector named pBS-C5 by replacing the MCS of pBlueScript® (Stratagene, La Jolla, CA, USA) with the C5 MCS. BbvCI and PmeI sites flanking the C5 MCS were also added to facilitate gel-less cloning (see below). pBS-C5 is 3.1 kb compared with the >8 kb of the pUAS or pCaSpeR vectors. Thus, DNA fragments can be cloned into and then manipulated in pBSc5MCS using any of the 6-base cutter sites, and then the modified fragment can be moved to pCaSpeR5 or pUAS-C5 using the flanking 8-base cutter sites. Note that all four 8-base cutters are compatible with buffer 4 (New England Biolabs, Ipswich, MA, USA) to allow straightforward liberation of the insert by a single double digest.
We also created a small kanamycin (Kan)-resistant shuttle vector named pC5-Kan by inserting the C5 MCS into 2.5 kb pHSX vector (Kevin Jones, personal communication) that had been modified to make the C5 MCS cloning sites unique. Because pC5-Kan has a different drug resistance than pCaSpeR5 or pUAS-C5, DNA fragments can be moved from pC5-Kan into pCaSpeR5 or pUAS-C5 without gel purification by digesting pC5-Kan with two appropriate 8-base cutters and either of BbvCI or PmeI and ligating the heat-inactivated mixture into the pCaSpeR5 or pUAS-C5 trans-genesis vectors that have been digested with the corresponding 8-base cutters. Plating the transformation mixture on Amp plates makes purification of the fragment to be cloned unnecessary, because any uncut or recircularized shuttle vector will not grow after plating. The pC5-Kan vector should not productively ligate into the transgenesis vectors because cutting with BbvCI (7-base cutter) or PmeI (8-base cutter) yields incompatible ends. Eliminating the need for gel purification would be particularly useful for manipulation of large genomic clones that are inefficiently recovered from gel purification procedures and for high-throughput site-directed mutagenesis or deletion scanning. In tests of the gel-less cloning approach, the combination of PacI and AscI cut the target vector with variable efficiency, yielding 80% 20% of transformants having the desired insertions, with the remainder of the transformants being empty target vector. Further work will be required to identify restriction enzymes most effective for gel-less cloning. Nevertheless, the availability of transgenesis vectors with improved cloning sites and matching shuttle vectors with flanking 8-cutter sites will be of general utility to the Drosophila community. Therefore, all constructs described in this paper are being made available for nonprofit use via the Drosophila Genome Resource Center (Bloomington, IN, USA).
Acknowledgments
We thank I.T. Helenius for comments on the manuscript and R. Carthew for suggesting the gel-less cloning strategy and the pHSX vector. S.M.P. was supported by National Institutes of Health (NIH) Lung Biology training grant no. 5 T32 HL076139-03. G.J.B. was a recipient of the National Science Foundation (NSF) Career Award IBN-0133411 and NIH R01 GM069540. Support was also provided by a NSF Alliance for Graduate Education and the Professoriate (AGEP) grant. The GenBank® accession nos. for new constructs are pCaSpeR5, EF090407; pUAS-C5, EF090406; pC5-Kan, EF090404; pC5-Kan SphI, EF090405; pBS-C5, EF090402; pBS-C5 SphI, EF090403.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
To purchase reprints of this article, contact: Reprints@BioTechniques.com
References
- 1.Thummel C, Pirrotta V. New pCaSpeR P element vectors. Drosoph Inf Serv. 1992;71:150. [Google Scholar]
- 2.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 3.Le T, Liang Z, Patel H, Yu MH, Sivasubramaniam G, Slovitt M, Tanentzapf G, Mohanty N, et al. A new family of Drosophila balancer chromosomes with a w-dfd-GMR yellow fluorescent protein marker. Genetics. 2006;174:2255–2257. doi: 10.1534/genetics.106.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiga Y, Tanaka-Matakatsu M, Hayashi S. A nuclear GFP/β-galactosidase fusion protein as a marker for morphogenesis in living Drosophila. Dev Growth Diff. 1996;38:99–106. [Google Scholar]
- 5.Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]


