Abstract
Background
The suppressors of cytokine signalling (SOCS) are inhibitors of cytokine signalling; methylation of SOCS‐3 has been implicated in the tumorigenesis of liver as well as head and neck cancer.
Aims
This study was performed to elucidate the role of SOCS‐1 and SOCS‐3 in Barrett's adenocarcinoma and its precursor lesions.
Methods
DNA of specimens from 19 Barrett's adenocarcinomas, 56 Barrett's intraepithelial neoplasias (n = 29 low grade and n = 27 high grade), 30 Barrett's mucosa without neoplasia, 20 samples of normal squamous and gastric epithelium and four cell lines were studied using methylation specific PCR for the SOCS‐1 and SOCS‐3 promoter following microdissection. The presence of SOCS‐3 mRNA transcripts was confirmed by semiquantitative real time PCR, and the SOCS‐3 protein was analysed immunohistochemically.
Results
In normal squamous epithelium and normal gastric mucosa, neither SOCS‐3 nor SOCS‐1 methylation was observed. In Barrett's mucosa without intraepithelial neoplasia, SOCS‐3 methylation occurred in 4/30 cases (13%) whereas SOCS‐1 was unmethylated. A hypermethylated SOCS‐3 promoter was found in 14/19 Barrett's adenocarcinomas (74%) and in 20/29 high and 6/27 low grade intraepithelial neoplasias (69% and 22%, respectively). SOCS‐1 promoter hypermethylation occurred in 8/19 adenocarcinomas (42%) and in 6/29 high grade and 1/27 low grade intraepithelial neoplasias (21% and 4%, respectively). Methylation of the SOCS‐3 promoter correlated with downregulation of SOCS‐3 transcripts and protein expression in these tumours and various cell lines. In the cell lines tested, SOCS‐3 and SOCS‐1 transcripts increased after treatment with the demethylation compound 5‐aza‐2‐deoxycytidine.
Conclusions
These data indicate that promoter methylation and subsequent transcript downregulation of SOCS‐3 transcripts and, to a much lesser extent, SOCS‐1 are involved in the multistep carcinogenesis of Barrett's adenocarcinoma.
Barrett's adenocarcinoma arises from Barrett's oesophagus in which an intestinal‐type epithelium (specialised intestinal metaplasia) replaces oesophageal squamous epithelium damaged by gastro‐oesophageal reflux disease. The development of cancer in Barrett's oesophagus follows a multistep pathway. Histologically, there is a progression from intestinal metaplasia (Barrett's mucosa) through low and high grade intraepithelial neoplasia to adenocarcinoma.1,2
To date, the exact cellular and molecular mechanisms leading to neoplastic progression in Barrett's epithelium are still not fully understood.3,4,5 However, the initial step in the carcinogenic process is thought to be an intermediate step in the progression from reflux oesophagitis to oesophageal adenocarcinoma.
Many signalling pathways, such as cellular growth, differentiation and also inflammation, involve the Janus kinases (JAKs), the signal transducers and activators of transcription (STATs), and their endogenous inhibitors of suppressors of cytokine signalling (SOCS) as important players in transmitting external signals from surface membrane to target genes in the nucleus.6
Cancer related defective JAK/STAT/SOCS pathways may not only pertubate cell growth or differentiation, but may also negatively affect tumour response to the cytokine based immunotherapy. The cytokine inducible SH2 domain containing protein and SOCS‐1–7 have been identified in the SOCS family to date.7,8 SOCS proteins act as negative regulators of JAK/STAT pathways and may represent tumour suppressor genes.8 The finding of oncogenic partners in this signalling pathway, especially in human epithelial malignant tumours, may support a prominent role of deregulated pathways in the pathogenesis of diseases. Another possible mechanism, by which SOCS proteins restrict signalling, is to promote protein degradation or interfering with the turnover of certain substrates (eg, activating an E3 ubiquitin ligase).8
Aberrant hypermethylation of CpG islands in promoter regions has been shown to be associated with transcriptional suppression of various genes in several types of epithelial as well as haematopoietic malignancies.9,10,11
SOCS‐1 appears to have tumour suppressor activity as restoration of SOCS‐1 gene expression in hepatocellular carcinoma cells caused growth suppression and induction of apoptosis.12,13 Recently, SOCS‐3 was found to be frequently silenced by hypermethylation in gastrointestinal cancers (eg, in hepatocellular carcinoma,14,15 pancreatic carcinoma16 or hepatoblastomas17). Silencing of SOCS‐3 by promoter methylation in human lung and head and neck cancer has also recently been reported.18,19
Therefore, in the present study, we analysed the status of SOCS‐1 and SOCS‐3 in Barrett's adenocarcinoma and its precursor lesions to elucidate a possible role of these genes in the stepwise carcinogenic process of these tumours.
Materials and methods
Cell lines
Squamous carcinoma cell lines (OE21), adenocarcinoma cell lines (OE19, OE33) and normal lung fibroblasts (CCL‐75 cells) were obtained from the American Type Culture Collection (ATCC, Rockville, Maryland, USA) and from the European Collection of Cell Cultures, respectively. All cell lines were grown in RPMI 1640 medium or Dulbecco (Gibco BRL, Gaithersburg, Maryland, USA) supplemented with 10% fetal bovine serum (Gibco BRL). All cell lines were kept at 37°C in a humidified incubator with 5% CO2 in air.
Patients and tissue samples
Between February 2000 and September 2001, 19 patients with well differentiated Barrett's adenocarcinoma, 56 patients with Barrett's epithelium and intraepithelial neoplasia (n = 29 with low grade (LGIN) and n = 27 with high grade (HGIN) intraepithelial neoplasia) and 30 patients with Barrett's mucosa without neoplasia were selected from the archives to obtain an equal representation of different grades of dysplasia for molecular analysis. Ten normal squamous cell epithelium samples as well as 10 normal gastric mucosa specimens from the cardia region were used as controls. All patients with Barrett's neoplasia received endoscopic mucosal resection. Barrett's mucosa without neoplasia was obtained from biopsies of patients without neoplasia. The present study was in accordance with the ethical standards of the Committee on Human Experimentation of the University of Leipzig and Bochum. All samples were taken during treatment procedures with therapeutic intent. The inclusion criterion for this study was the availability of good quality, paraffin embedded tissue after initial clinical diagnosis. Each tumour was re‐evaluated with regard to typing.20 In all cases, haematoxylin‐eosin stained slides were re‐examined independently by four experienced gastrointestinal pathologists (IT, MV, MS, AT) without knowledge of the clinical data. In the case of conflicting results of grading intraepithelial neoplasia, microscopic re‐evaluation was obtained until concordance of opinion was obtained.
Microdissection and sample processing
For each tumour sample, the histopathological lesions of interest were first identified on routinely stained sections, as described previously,19,21,22 resulting in a nearly complete separation of the target population from neighbouring tissues. In the case of intraepithelial neoplasia, only clearly identifiable neoplastic cells were microdissected. The approximate number of cells was estimated to be at least 1200 per sample for PCR analysis. After microdissection, the tissue samples were put into Eppendorf tubes and standard methods for DNA and RNA extraction were used.22
Methylation analysis
For each tumour sample, the histopathological lesions of interest were identified on routinely stained sections, as described previously.19,21,22 Next, microdissection was performed on formalin fixed, paraffin embedded tissue. Sections (12 μm) cut from paraffin blocks were mounted on glass slides with a thickness of 0.17 mm (very thin glass slides are needed to prevent laser energy from being dispersed before reaching the section of tissue). An ultraviolet laser microscope system was used to remove as much stromal tissue as possible (UV‐laser microbeam; PALM, Bernried, Germany), resulting in a nearly complete separation of the target population from neighbouring tissue. After microdissection,19,22 methylation specific PCR (MSP) was applied to investigate the methylation status of the promoter regions of the SOCS‐1 and SOCS‐3 genes. After an initial bisulfite treatment to modify the DNA, PCR was performed to distinguish methylated from unmethylated DNA, as described by Herman et al.23 According to our previously published protocols,19,21 2 μg of genomic DNA were denatured with 0.3 M NaOH. Hydroquinone 10 mM and 3 mM sodium bisulfite were added and incubated at 50°C for 16 h. Modified DNA was purified using the Wizard DNA purification resin (Qiagen, Hilden, Germany), followed by desulphonating in 0.3 M NaOH, subsequent ethanol precipitation and resuspension in 30–50 μl of water. MSP was performed using specific primers and conditions previously described.19,23 Briefly, a 20 μl reaction volume containing 150 ng of bisulfite modified DNA, 1× PCR buffer, 1.5 mM MgCl2, 0.16 μM dNTPs, 0.25 μM specific primer mix (forward and reverse primers; table 1) and 1 unit of Taq enzyme (Roche, Hamburg, Germany) were used. The primers were designed according to a previously published protocol and adopted to the specific conditions of our tumour samples (table 1).18,19 Placental DNA treated with methyltransferase was used as a positive control for methylation.
Table 1 Primer sequences for methylation specific PCR analysis.
| Gene | Primer | Sequence | Product size |
|---|---|---|---|
| SOCS‐1 | UmspF | Tgaagatggttttgggatttatga | 184 bp |
| UmspR | cacaactcctacaacaaccacacac | ||
| MspF | Tgaagatggtttcgggatttacga | 183 bp | |
| MspR | Acaactcctacaacgaccgcacg | ||
| SOCS‐3 | UmspF | tagtgtgtaagttgtaggagagtgg | 134 bp |
| UmspR | Ctaaacataaaaaaataacactaatccaaa | ||
| MspF | Gtagtgcgtaagttgtaggagag | 139 bp | |
| MspR | Gtaaaaaaataacgctaatccgaa |
MSP, methylation specific PCR; SOCS, suppressors of cytokine signalling.
Table 2 Results of mRNA, methylation specific PCR analysis and immunohistochemistry of suppressors of cytokine signalling‐3 in Barrett's lesions.
| mRNA reduced | MSP methylated bands | Protein (immunohistochemistry) | |
|---|---|---|---|
| Barrett's adenocarcinoma | 14/19 | 14/19 | 6/19* |
| Barrett's HGIN | 20/29 | 20/29 | 9/29 |
| Barrett's LGIN | 6/27 | 6/27 | 21/27 |
| Barrett's mucosa | 4/30 | 4/30 | 26/30 |
HGIN, high grade intraepithelial; LGIN, low grade intraepithelial neoplasia; MSP, methylation specific PCR; SOCS, suppressors of cytokine signalling.
*SOCS‐3 protein was detected in one case with reduced mRNA expression as well as MSP detectable methylated bands.
Bisulfite sequencing for SOCS‐3 and SOCS‐1
Bisulfite treated genomic DNA was amplified by using primers (5′‐GTG‐TAG‐AGT‐AGT‐GAT‐TAA‐ATA‐3′ (forward) and 5′‐TCC‐TTA‐AAA‐CTA‐AAC‐CCC‐CTC‐3′ (reverse)) designed to amplify nucleotides −1084 to −671 of the SOCS‐3 promoter region (the start codon ATG of SOCS‐3 is defined as +1), adopting the protocols published recently by He et al,18 and from our previously published protocol.19 For SOCS‐1, three sets of primers were used.14,19,24 Primers for region 1 were 5‐GAG GAG GGA GGG GAG TTT AGG GTA GTT‐3 (sense) and 5‐TTC AAC CTC AAT AAA CAC AAC TAA AAA A‐3 (antisense). Primers for region 2 were 5‐TTT TTT AGT TGT GTT TAT TGA GGT TGA A‐3 (sense) and 5‐CCA CCT AAT TAT ATA CTA CCA TCC TAC AA‐3 (antisense). Primers for region 3 were 5‐TGT AGG ATG GTA GTA TAT AAT TAG GTG GT‐3 (sense) and 5‐TAA TAC TCC AAC AAC TCT AAA AAA CAA TC‐3 (antisense). The PCR products were cloned into a Topo TA cloning kit (Invitrogen, Carlsbad, California, USA). Two to five randomly picked clones were sequenced on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, California, USA).
Table 3 Results of mRNA, methylation specific PCR analysis and immunohistochemistry of suppressors of cytokine signalling‐1 in Barrett's lesions.
| mRNA reduced | MSP methylated bands | Protein (immunohistochemistry) | |
|---|---|---|---|
| Barrett's adenocarcinoma | 8/19 | 8/19 | 11/19 |
| Barrett's HGIN | 6/29 | 6/29 | 23/29 |
| Barrett's LGIN | 1/27 | 1/27 | 26/27 |
| Barrett's mucosa | 0/30 | 0/30 | 30/30 |
HGIN, high grade intraepithelial; LGIN, low grade intraepithelial neoplasia; MSP, methylation specific PCR; SOCS, suppressors of cytokine signalling.
RT‐PCR
The presence of SOCS‐3 mRNA transcripts was analysed by semiquantitative PCR (LightCycler; Roche) as described previously.19 RNA (200 ng) extracted from approximately 50–60 mg paraffin embedded tissue sample using the RNeasy Mini kit (Qiagen) was reverse transcribed with the primer sequences for a 579 bp fragment of the human SOCS‐3 cDNA (5′ ‐ TTC TAC TGG AGC GCA GTG AC ‐3′ (forward) and 5′‐ACT GGG TCT TGA CGC TGA G‐3′ (reverse)) in 20 μl of RT mix with a QuantiTect SYBR Green RT‐PCR kit (Qiagen) in accordance with the manufacturer's instructions.14,15,18
Demethylation
For expression induction of SOCS‐3 after exposure to 5‐aza‐2‐deoxycytidine (5‐AZA‐DC), a drug that inhibits DNA methylation, subconfluent cultures of the SOCS‐3 non‐expressing cell lines of oesophageal adenocarcinoma (OE19‐obtained from the European Collection of Cell Cultures), was selected. The cell lines were exposed to 1 μM 5‐AZA‐DC for 4 days. After isolation of total RNA using the RNeasy extraction kit (Qiagen), multiplex reverse transcription‐PCR was performed for SOCS‐3 as described above.19
Results
To examine the expression status of SOCS‐3 and SOCS‐1 in cell culture, three cell lines, derived from oesophageal squamous epithelium (OE21) and from Barrett's adenocarcinoma (OE19, OE33), were analysed.
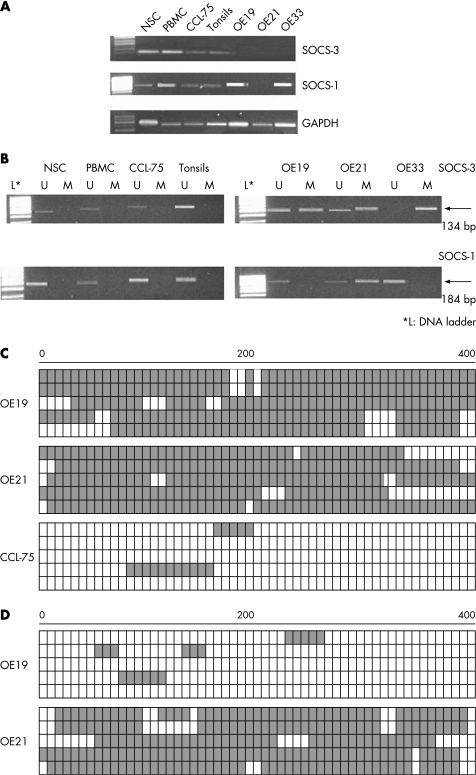
SOCS‐3 transcripts were dramatically decreased or absent in all three cell lines (fig 1A). In contrast, SOCS‐3 expression was detectable in all normal control cell lines, including primary cell cultures (“NSC” normal squamous epithelial cells; see fig 1A), peripheral blood mononuclear cells of healthy donors and CCl‐75 cells (fibroblasts) (fig 1A). SOCS‐1 transcript was decreased or absent in OE21 cells, but detectable in normal epithelial cells (NSC), peripheral blood mononuclear cells of normal donors, in tonsils, OE19 and OE33 (fig 1A).
Figure 1 (A) mRNA expression analysis of suppressors of cytokine signalling (SOCS)‐3 and SOCS‐1 in normal squamous epithelial cells (NSC), peripheral blood mononuclear cells (PBMC) of healthy volunteers, CCL‐75 cells (lung fibroblasts), tonsils, OE19, OE21 and OE33 cells. SOCS‐3 mRNA was absent in OE19, OE21 and OE33 cells. In normal tissue as well as in CCL‐75, SOCS‐3 expression was observed. SOCS‐1 mRNA was detectable in NSC, PBMC, CCL‐75 and tonsils. In contrast with SOCS‐3, SOCS‐1 was detectable in all cell lines, except OE21. (B) Methylation specific PCR analysis of SOCS‐3 and SOCS‐1 in NSC, PBMC of healthy volunteers as well as in CCL‐75 cells, tonsils and various tumour cell lines (OE19, OE21 and OE33). Bands (134 bp for SOCS‐3 and 184 bp for SOCS‐1, respectively) in lanes labelled “U” represent unmethylated DNA products amplified with non‐methylation specific primers. Bands in lanes labelled “M” refer to methylated DNA products amplified with methylation specific primers (134 and 183 bp respectively). L, DNA ladder. GAPDH, glyceraldehyde phosphate dehydrogenase. (C, D) Bisulfite sequencing analysis of cell lines. Open and filled squares represent unmethylated and methylated CpG islands, respectively. We sequenced five clones of PCR products amplified from bisulfite treated genomic DNA for each cell line. OE19 and OE21 exhibited heavily methylated CpG islands of SOCS‐3 (C). Methylation of the SOCS‐1 gene was examined with three primer sets. OE21 exhibited a heavily methylated promoter region whereas OE19 lacked methylation (D).
To analyse the possible causal mechanism of the decrease or lack of transcripts, the CpG islands of the SOCS‐1 and SOCS‐3 promoters were analysed, using MSP. We found that in those cell lines with undetectable SOCS‐3 transcripts (OE21, OE19 and OE33), promoter methylation occurred (fig 1B). In those cells with detectable SOCS‐3 transcripts, no methylated bands were observed. MSP for SOCS‐1 promoter revealed a hypermethylated promoter only for OE21 cells, the cell line with absent SOCS‐1 transcripts (fig 1A, 1B).
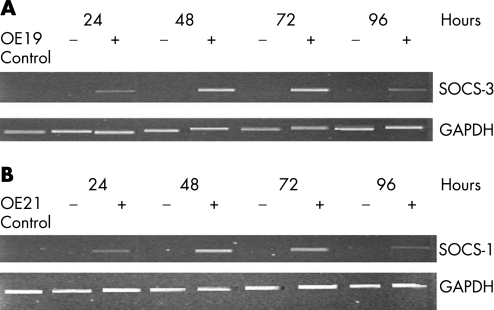
To confirm that promoter hypermethylation was responsible for the lack of SOCS‐3 as well as SOCS‐1 expression in the cell lines tested, 5‐AZA‐DC treatment was performed. After exposure of the SOCS‐3 non‐expressors OE19 cells to 5‐AZA‐DC, a drug that inhibits DNA methylation, for 3 days, re‐expression of SOCS‐3 was detected (fig 2A), with little or no change in the expression of the housekeeping gene, glyceraldehyde phosphate dehydrogenase. Treatment of the SOCS‐1 non‐expressor OE21 also exhibited re‐expression after demethylation treatment (fig 2B).
Figure 2 Expression of suppressors of cytokine signalling (SOCS)‐3 and SOCS‐1 transcripts after treatment with 5‐aza‐2‐deoxycytidine (5‐AZA‐DC). Expression of SOCS‐3 before (−) and after (+) treatment of OE19 and OE21 cells as well as with 5‐AZA‐DC. GAPDH, glyceraldehyde phosphate dehydrogenase.
To analyse the CpG islands in detail, bisulfite sequencing was performed for the detection of the extent of CpG site methylation of the SOCS‐3 and SOCS‐1 promoter regions, respectively. Consistent with the MSP results, we found that the SOCS‐3 CpG islands in OE21, OE19 as well as in OE33 were heavily methylated (fig 1C). Strong methylation of the SOCS‐1 promoter was detected in OE21 cells, but not in OE19 and OE33 cells (fig 1D).
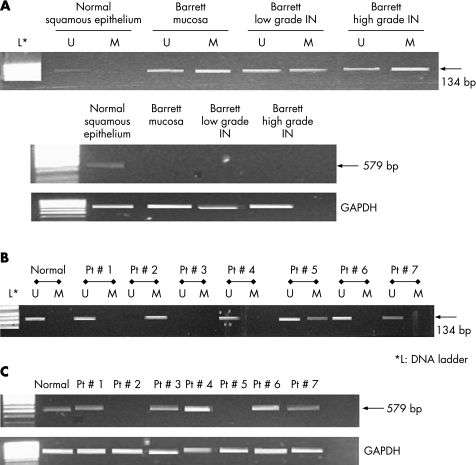
To assess the methylation status of the SOCS‐3 and SOCS‐1 promoter region in human tumours, 19 Barrett's adenocarcinomas, 29 high grade and 27 low grade intraepithelial neoplasia samples and 30 specimens with Barrett's mucosa were analysed after microdissection (fig 3; tables 2, 3). In (normal) Barrett mucosa without intraepithelial neoplasia, SOCS‐3 methylation was detected in four cases (13%). In LGIN, SOCS‐3 methylation occurred in 6/27 cases (22%) and in 20/29 cases (69%) of HGIN. Fourteen of 19 Barrett's adenocarcinoma samples (74%) showed decreased or even absent mRNA expression for SOCS‐3, as indicated by RT‐PCR (fig 3C). In association with this, we found hypermethylation in these 14 tumour samples by MSP, but not in their matched non‐neoplastic normal tissue samples.
Figure 3 Correlation of methylation in the promoter region with silencing of the suppressors of cytokine signalling (SOCS)‐3 gene of corresponding normal epithelium and Barrett's mucosa with and without low and high grade intraepithelial neoplasia (IN). These samples were from the same patient, using microdissection to analyse different parts of the mucosectomy specimen (A). Seven patients with Barrett's adenocarcinoma are also shown (B). Bands (134 bp) in lanes labelled “U” were unmethylated DNA products amplified with non‐methylation‐specific primers. Bands (134 bp) in lanes labelled “M” were methylated DNA products amplified with methylation specific primers. L, DNA ladder. The upper lanes with SOCS‐3 expression were analysed by reverse transcription‐PCR. Glyceraldehyde phosphate dehydrogenase (GAPDH) was used as an internal control.
The SOCS‐1 promoter region was also examined in all 105 specimens. SOCS‐1 methylation was observed in 1/27 (4%) LGIN and in 6/29 (21%) HGIN. A hypermethylated SOCS‐1 promoter with reduced mRNA transcripts was detected in 8/19 (42%) Barrett's adenocarcinomas. In a few cases, unmethylated bands (U) were also visible in the tumour tissue, which may result from admixed normal cells within the tumour specimens (eg, granulocytes, fibroblasts), even though microdissection was applied. In Barrett's mucosa without neoplasia, the SOCS‐1 transcript was detectable in all 30 cases. In 20 normal squamous epithelium or gastric mucosa, neither SOCS‐3 nor SOCS‐1 methylation occurred.
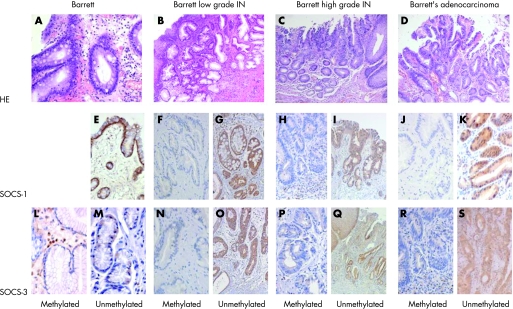
Immunohistochemistry was used to assess SOCS‐3 and SOCS‐1 at the protein level. SOCS‐1 and SOCS‐3 were detected in 11/19 and 6/19 Barrett adenocarcinomas, respectively (fig 4). All of the 11 SOCS‐1 positive tumours contained an unmethylated SOCS‐1 promoter, while SOCS‐3 protein was detected in one specimen with a methylated SOCS‐3 promoter. SOCS‐3 protein expression was undetectable in HGIN and LGIN, which harboured a methylated SOCS‐3 promoter (fig 3). In contrast, SOCS‐1 and SOCS‐3 positivity occurred in normal epithelial and inflammatory cells (granulocytes, lymphocytes) as well as in tumour surrounding fibrous tissue (fig 3). Within a given Barrett's adenocarcinoma, a nearly homogeneous expression was observed.
Figure 4 Immunohistochemical staining of suppressors of cytokine signalling (SOCS)‐1 and SOCS‐3 in Barrett's mucosa, and in specimens with low grade and high grade intraepithelial neoplasia (IN). SOCS‐1 and SCOC‐3 immunoreactivity within epithelial cells and fibrous tissue (including fibroblasts and inflammatory cells), irrespective of the methylation status of the tumour. SOCS‐1 and SOCS‐3 staining of a methylated and unmethylated specimen of patients with Barrett mucosa, IN (low and high grade) as well as Barrett adenocarcinoma. (A) HE: Barrett. (B) HE: Barrett's low grade IN. (C) HE: Barrett's high grade IN. (D) HE: Barrett's adenocarcinoma. (E) SOCS‐1: Barrett unmethylated. (F) SOCS‐1: Barrett's low grade IN methylated. (G) SOCS‐1: Barrett's low grade IN unmethylated. (H) SOCS‐1: Barrett's high grade IN methylated. (I) SOCS‐1: Barrett's high grade IN unmethylated. (J) SOCS‐1: Barrett's adenocarcinoma methylated. (K) SOCS‐1: Barrett's adenocarcinoma unmethylated. (L) SOCS‐3: Barrett methylated. (M) SOCS‐3: Barrett unmethylated. (N) SOCS‐3: Barrett's low grade IN methylated. (O) SOCS‐3: Barrett's low grade IN unmethylated. (P) SOCS‐3: Barrett's high grade IN methylated. (Q) SOCS‐3: Barrett's high grade IN unmethylated. (R) SOCS‐3: Barrett's adenocarcinoma methylated. (S) SOCS‐3: Barrett's adenocarcinoma unmethylated. HE; hematoxylin‐eosin stained sections of the SOCS‐1 and SOCS‐3 expression.
Discussion
There is increasing evidence that abnormalities in STAT/SOCS proteins are involved in the pathogenesis of certain human epithelial and non‐epithelial malignancies.19,25,26,27,28,29,30,31 Cancer associated malfunction of the JAK/STAT/SOCS pathway may negatively influence (tumour and also stromal) cell response to (cytokine based) immunotherapies and innate immunity, as has been demonstrated recently.31,32 Aberrant hypermethylation of CpG islands within promoter regions silencing gene transcription has been recognised as a mechanism for inactivating tumour suppressor genes in cancer. Many recent findings indicate that SOCS proteins act, in addition, as adaptors that regulate the turnover of certain substrates by interacting with and activating an E3 ubiquitin ligase.31,32 Thus SOCS proteins act as negative regulators of JAK/STAT pathways and may represent tumour suppressor genes.
We report that inactivation of SOCS‐3—and to a lesser extent SOCS‐1—is frequently observed in Barrett adenocarcinoma as well as in precursor lesions, mainly due to promoter hypermethylation. A possible mechanism for the involvement of SOCS‐3 in human cancers has been reported recently by He et al18 and our group.19 In lung and also in head and neck cancer, SOCS‐3 functions as a growth suppressor and inducer of apoptosis.18,19 In the present study, we found that frequent hypermethylation of the functional SOCS‐3 promoter region was correlated with silencing of the SOCS‐3 gene in Barrett adenocarcinoma and also in precursor lesions. Normal, non‐neoplastic expressing SOCS‐3, transcripts showed a functionally active promoter. In low and high grade neoplasia as well as in Barrett mucosa, SOCS‐3 was also methylated, showing increasing rates of methylation with higher grades of neoplasia.
Our results suggest that SOCS‐3 silencing results from promoter methylation and may represent an important cause of constitutive activation of the JAK/STAT pathway in the malignant transformation of Barrett's mucosa. It may also act as an important epigenetic event during Barrett carcinogenesis, as SOCS‐3 inactivation was also found in Barrett epithelium as well as intraepithelial neoplasia, a pre‐neoplastic, pre‐malignant lesion.
The significance of SOCS‐3 being inactivated in Barrett mucosa without intraepithelial neoplasia remains unclear. It has recently been described that in a model of chronic inflammation, as it is the case in gastro‐oesophageal reflux disease, the endogenous SOCS‐3 is a critical negative regulator of multiple cell types orchestrating inflammatory disease.33,34 Joint inflammation in SOCS‐3 negative mice was particularly severe and was characterised by increased numbers of neutrophils and macrophages and showed increased production of and enhanced responsiveness to granulocyte‐colony stimulating factor and interleukin 6.35 Gastric refluxate has not been shown to be genotoxic, which opens the possibility that the effect of gastro‐oesophageal reflux on the development of Barrett's mucosa represents an epigenetic effect mediated by methylation of anti‐inflammatory cytokines, such as SOCS‐3.
We speculate that inactivated SOCS‐3 increases the inflammatory process in Barrett mucosa. This might be in agreement with the clinical observation that Barrett's mucosa may progress to the next step of intraepithelial neoplasia, even when the reflux cessates. The rate of SOCS‐3 methylation in 13% of “normal” Barrett's lesions might be an explanation for the epidemiological observation that only 10% of patients with Barrett's mucosa will eventually develop adenocarcinoma.
Therefore, the phenomenon of SOCS‐3, and to a lesser extent, SOCS‐1, silencing as a result of promoter methylation may represent a common event during Barrett's carcinogenesis. SOCS‐3 itself may potentially function as an important tumour suppressor gene.14 The high prevalence of SOCS‐3 promoter hypermethylation also supports targeted therapies of the JAK/STAT pathway or its downstream targets.
Acknowledgements
This paper was supported by the Bundesministerium für Bildung und Forschung (BMB+F), Interdisciplinary Centre for Clinical Research (IZKF) at the University of Leipzig (01KS9504/1, project D01).
Abbreviations
5‐AZA‐DC - 5‐aza‐2‐deoxycytidine
HGIN - high grade intraepithelial
JAK - Janus kinase
LGIN - low grade intraepithelial neoplasia
MSP - methylation specific PCR
SOCS - suppressors of cytokine signalling
STATs - signal transducers and activators of transcription neoplasia
Footnotes
Competing interests: None.
References
- 1.Flejou J F. Barrett's oesophagus: from metaplasia to dysplasia and cancer. Gut 200554i6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitzgerald R C. Barrett's oesophagus and oesophageal adenocarcinoma: how does acid interfere with cell proliferation and differentiation? Gut 200554 (Suppl1) 1, i21–1, i26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn J R, Risk J M, Langan J E.et al Physical and transcript map of the minimally deleted region III on 17p implicated in the early development of Barrett's oesophageal adenocarcinoma. Oncogene 2003224134–4142. [DOI] [PubMed] [Google Scholar]
- 4.Morales C P, Souza R F, Spechler S J. Hallmarks of cancer progression in Barrett's oesophagus. Lancet 20023601587–1589. [DOI] [PubMed] [Google Scholar]
- 5.Tannapfel A. Molecular findings in Barrett's epithelium. Dig Dis 200422126–133. [DOI] [PubMed] [Google Scholar]
- 6.Fujimoto M, Naka T. Regulation of cytokine signaling by SOCS family molecules. Trends Immunol 200324659–666. [DOI] [PubMed] [Google Scholar]
- 7.Cacalano N A, Sanden D, Johnston J A. Tyrosine‐phosphorylated SOCS‐3 inhibits STAT activation but binds to p120 RasGAP and activates Ras. Nat Cell Biol 20013460–465. [DOI] [PubMed] [Google Scholar]
- 8.Ali S, Nouhi Z, Chughtai N.et al SHP‐2 regulates SOCS‐1‐mediated Janus kinase‐2 ubiquitination/degradation downstream of the prolactin receptor. Biol Chem 200327852021–52031. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M, Fraga M F, Paz M F.et al Cancer epigenetics and methylation. Science 20022971807–1808. [DOI] [PubMed] [Google Scholar]
- 10.Herman J G, Baylin S B. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 20033492042–2054. [DOI] [PubMed] [Google Scholar]
- 11.Mirmohammadsadegh A, Marini A, Nambiar S.et al Epigenetic silencing of the PTEN gene in melanoma. Cancer Res 2006666546–6552. [DOI] [PubMed] [Google Scholar]
- 12.Miyoshi H, Fujie H, Shintani Y.et al Hepatitis C virus core protein exerts an inhibitory effect on suppressor of cytokine signaling (SOCS)‐1 gene expression. J Hepatol 200543757–763. [DOI] [PubMed] [Google Scholar]
- 13.Calvisi D F, Ladu S, Gorden A.et al Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 20061301117–1128. [DOI] [PubMed] [Google Scholar]
- 14.Yoshikawa H, Matsubara K, Qian G S.et al SOCS‐1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth‐suppression activity. Nat Genet 20012829–35. [DOI] [PubMed] [Google Scholar]
- 15.Ogata H, Kobayashi T, Chinen T.et al Deletion of the SOCS3 gene in liver parenchymal cells promotes hepatitis‐induced hepatocarcinogenesis. Gastroenterology 2006131179–193. [DOI] [PubMed] [Google Scholar]
- 16.Fukushima N, Sato N, Sahin F.et al Aberrant methylation of suppressor of cytokine signalling‐1 (SOCS‐1) gene in pancreatic ductal neoplasms. Br J Cancer 200389338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang B, Guo M, Herman J G.et al Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol 20031631101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He B, You L, Uematsu K.et al SOCS‐3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc Natl Acad Sci USA 200310014133–14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber A, Hengge U R, Bardenheuer W.et al SOCS‐3 is frequently methylated in head and neck squamous cell carcinoma and its precursor lesions and causes growth inhibition. Oncogene 2005246699–6708. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton S R, Aaltonen L A. eds. WHO classification of tumours: pathology and genetics of tumours of digestive system Lyon: IARC, 2002
- 21.Weber A, Langhanki L, Sommerer F.et al Mutations of the BRAF gene in squamous cell carcinoma of the head and neck. Oncogene 2003224757–4759. [DOI] [PubMed] [Google Scholar]
- 22.Sommerer F, Vieth M, Markwarth A.et al Mutations of BRAF and KRAS2 in the development of Barrett's adenocarcinoma. Oncogene 200423554–558. [DOI] [PubMed] [Google Scholar]
- 23.Herman J G, Graff J R, Myohanen S.et al Methylation‐specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996939821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshimo Y, Kuraoka K, Nakayama H.et al Epigenetic inactivation of SOCS‐1 by CpG island hypermethylation in human gastric carcinoma. Int J Cancer 20041121003–1009. [DOI] [PubMed] [Google Scholar]
- 25.Larsen L, Röpke C. Suppressors of cytokine signalling: SOCS. APMIS 2002110833–844. [DOI] [PubMed] [Google Scholar]
- 26.Niu G, Heller R, Catlett‐Falcone R.et al Gene therapy with dominant‐negative Stat3 suppresses growth of the murine melanoma B16 tumor in vivo. Cancer Res 1999595059–5063. [PubMed] [Google Scholar]
- 27.Grandis J R, Drenning S D, Zeng Q.et al Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc Natl Acad Sci U S A 2000974227–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai H, Kim Y S, Konishi N.et al Combined hypermethylation and chromosome loss associated with inactivation of SSI‐1/SOCS‐1/JAB gene in human hepatocellular carcinomas. Cancer Lett 200218659–65. [DOI] [PubMed] [Google Scholar]
- 29.Galm O, Yoshikawa H, Esteller M.et al SOCS‐1, a negative regulator of cytokine signaling, is frequently silenced by methylation in multiple myeloma. Blood 20031012784–2788. [DOI] [PubMed] [Google Scholar]
- 30.Nagai H, Naka T, Terada Y.et al Hypermethylation associated with inactivation of the SOCS‐1 gene, a JAK/STAT inhibitor, in human hepatoblastomas. J Hum Genet 20034865–69. [DOI] [PubMed] [Google Scholar]
- 31.Valentino L, Pierre J. JAK/STAT signal transduction: regulators and implication in hematological malignancies. Biochem Pharmacol 200671713–721. [DOI] [PubMed] [Google Scholar]
- 32.Kortylewski M, Kujawski M, Wang T.et al Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med 2005111314–1321. [DOI] [PubMed] [Google Scholar]
- 33.Terrell A M, Crisostomo P R, Wairiuko G M.et al JAK/STAT/SOCS signalling circuits and associated cytokine‐mediated inflammation and hypertrophy in the heart. Shock 200626226–234. [DOI] [PubMed] [Google Scholar]
- 34.Mitsuyama K, Matsumoto S, Rose‐John S.et al STAT3 activation via interleukin‐6 trans‐signalling contributes to ileitis in SAMP1/Yit mice. Gut 2006551263–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong P K, Egan P J, Croker B A.et al SOCS‐3 negatively regulates innate and adaptive immune mechanisms in acute IL‐1‐dependent inflammatory arthritis. J Clin Invest 20061161571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]