Abstract
Insulators, first identified in Drosophila, are DNA sequence elements that shield a promoter from nearby regulatory elements. We have previously reported that a DNA sequence at the 5′ end of the chicken β-globin locus can function as an insulator. It is capable of shielding a reporter gene from the activating effects of a nearby mouse β-globin locus control region element in the human erythroleukemic cell line K562. In this report, we show that most of the insulating activity lies in a 250-bp CpG island (core element), which contains the constitutive DNase I-hypersensitive site (5′HS4). DNA binding assays with the core sequence reveal a complex protein binding pattern. The insulating activity of the core element is multiplied when tandem copies are used. Although CpG islands are often associated with promoters of housekeeping genes, we find little evidence that the core element is a promoter. Furthermore, the insulator differs from a promoter in its ability to block the locus control region effect directionally.
Keywords: chromatin, domain, locus control region
Insulators are DNA sequence elements that block activation of a promoter by an enhancer, but only when placed between them (1). Known insulators include the scs (2, 3) and gypsy (4) elements in Drosophila. We have previously reported that a DNA sequence at the 5′ end of the chicken β-globin locus, containing a constitutive DNase I-hypersensitive site (5′HS4), can also function as an insulator (5). When two copies of this 1.2-kb fragment were introduced between a human Aγ-globin promoter and an upstream enhancer/locus control region (LCR) element from the mouse β-globin locus (6–8), the expression of a reporter gene for neomycin resistance was greatly reduced. Other experiments showed that the insulator sequence could shield the white minigene from position effect in Drosophila.
The hypersensitive site 5′HS4 marks the 5′ end of the chicken β-globin domain (9, 10). Unlike the erythroid-specific hypersensitive sites further 3′ in the domain, 5′HS4 is DNase I-hypersensitive in all tissue types (9). Upstream of 5′HS4, there is an abrupt change in higher order chromatin structure in chicken erythroid cells (10). The chromatin within the β-globin domain (i.e., downstream of 5′HS4) displays the uniform moderate sensitivity to DNase I characteristic of transcriptionally active chromatin domains, whereas the chromatin upstream of 5′HS4 is DNase I-resistant and may be thought of as a kind of “heterochromatic” region. Consistent with this view, the N-terminal tail of histone H4 within the β-globin domain is acetylated at elevated levels when compared with histone H4 in chromatin upstream of the 5′HS4 (10). Elevated histone acetylation has been found in transcriptionally active and decondensed regions, such as the active X chromosome, relative to acetylation in transcriptionally inactive and compacted regions, such as the inactive X chromosome (11).
In this paper we describe in more detail the properties of the chicken insulator. We show by dissection of the 1.2-kb fragment that much of the activity is contained in a 250-bp fragment that includes 5′HS4. The DNA of this core region is G+C-rich and has the properties of a “CpG island,” but does not appear to function as a promoter. The sequence contains multiple small internal repeats, which may explain, in part, the complex footprint results obtained when the DNA is incubated with nuclear extracts. Although these results indicate that multiple factors bind within the core region, the sequence itself contains only a small number of canonical binding sites for transcription factors. One of these is an Sp1 (12) site, another a nearly perfect match to the binding site for the yeast α2 repressor (13), which overlaps with a region homologous to the binding sites for a number of other factors.
MATERIALS AND METHODS
Plasmid Construction.
Plasmids p137, p138, and p168 were derived from pJC5-4 by replacing the 1.2-kb SacI–SacI and XbaI–XbaI insulator fragments with the HindIII–SspI fragment (C1), BglII–SspI fragment (C2), and SacI–BglII fragment (C3), respectively, of the intact 1.2-kb insulator fragment (5). SacI and XbaI linkers were ligated to these fragments after the ends were filled in before being cloned into the SacI and XbaI sites, respectively, of pJC5-4. Plasmids p176 and p179 were constructed by first cloning two (p116) and three copies (p117), respectively, of the 250-bp SacI–HindIII fragment of the 1.2-kb insulator fragment into the pBSII SK+ (Stratagene) BamHI site after the ends were filled in and BamHI linker was ligated. p116 was cut with EcoRI and the 947-bp HindIII–EcoRI fragment from the 2.3-kb HindIII–HindIII fragment of the λ phage DNA was inserted after the ligation of EcoRI linker to the filled in HindIII end. After an SpeI linker was ligated to the EcoRV site, the resulting SpeI–SpeI fragment was inserted into the SpeI site (at the end of the LCR) of pJC5-4 to make p176. The 1.2-kb insulator fragment in the SacI site was then deleted. To construct p179, the three copies of the 250-bp fragment in p117 were doubled to six copies by ligating EcoRI linkers to the filled in ends of the SpeI–EcoRV fragment and inserting back into the EcoRI site of p117. The SpeI–EcoRV fragment of p117, which contains six copies of the insert, was cut out and an SpeI linker was ligated to the EcoRV site. The resulting SpeI–SpeI fragment was ligated into the SpeI site of pJC5-4. The 1.2-kb insulator fragment in the SacI site was then deleted to make p179. To make p231, the LCR was cut out of pJC5-4 with EcoRI and religated. The ends of the EcoRI–EcoRI LCR fragment was then filled in and ligated to KpnI linkers and inserted into the KpnI site of pJC16 (5). To make p233 and p239, the 2.7-kb BamHI–BamHI fragment containing the Aγ-globin promoter linked to the neomycin (G418) resistance gene (5) was first cloned into the BamHI site of pGEM-4Z (Promega) and a 559-bp PvuII fragment containing the Aγ-globin promoter (−299 to +260) was isolated and ligated to SacI linkers and inserted into the SacI site of p231 (for p233) and pJC5-4 (for p239) after deleting the 1.2-kb insulator fragment. Plasmid p180 was constructed by first isolating the 1.7-kb BamHI–BglII fragment containing the Drosophila scs element (2) and ligating either SpeI or XbaI linkers to the ends of the fragment after filling in. The XbaI fragment containing the scs element was inserted into the XbaI site of pJC5-4 replacing the 1.2-kb chicken insulator fragment. Then the other chicken insulator fragment was deleted by cutting with SacI and religating. The scs fragment with SpeI linker was then inserted into the SpeI site placing it between the LCR and the Aγ-globin promoter/neo reporter gene. Plasmids pGL-promoter and pGL2-basic were purchased from Promega. p265 was constructed by inserting the 250-bp BamHI–BamHI fragment from p116 (see above) into the BglII site of pGL2-basic. The plasmid pUPS-1.2 was constructed by removing the 1.2-kb chicken insulator fragment from the SacI site of pJC5-4 and placing it in the NdeI site upstream of the LCR.
DNase I Footprint.
The probes for DNase I in vitro footprinting were prepared by first cloning the SacI–HindIII fragment from pJC5-4, which contains the chicken β-globin 5′HS4, into the BamHI site of pBS II SK+ after the ends were filled in and ligated to BamHI linkers. To label the coding and noncoding strand probes, this plasmid was cut with either HindIII or XbaI, respectively, followed by end-filling with Klenow enzyme and [α-32P]dATP (DuPont/NEN; 6000 Ci/mmol; 1 Ci = 37 GBq). The labeled fragment was liberated by cleavage with XbaI (for coding strand probe) or HindIII (for noncoding strand probe) and was purified on a 5% polyacrylamide gel. DNA binding reactions contained 10 mM Hepes (pH 7.8), 60 mM KCl, 10% glycerol, 1 mM MgCl2, 5 mM DTT, 10 μg of d(A·T), 2.5 μg of BSA, 5–8 μl of K562 nuclear extract (2.2 mg/ml) or HeLa nuclear extract (2.5 mg/ml), and 40,000 cpm of 32P-labeled probe in a final volume of 50 μl. Where indicated, 2 pmol of competitor oligonucleotide was added with the probe. After 20 min at 25°C, 3 μl of 10 mM CaCl2 was added and gently mixed. The binding reaction was digested with DNase I (Worthington–DPRF) from freshly prepared stock solutions of 0.9 and 9.0 ng/μl for free and protein-bound DNA, respectively. Samples were mixed and incubated for 2 min at 25°C. Reactions were stopped with 0.2 ml of 10 mM Tris·HCl (pH 8.0), 25 mM EDTA, 1% SDS, 10 μg of tRNA, and rapid vortexing. DNA was purified by two extractions with phenol/chloroform and ethanol precipitation. DNA fragments were resolved on 8% polyacrylamide/urea gels.
Nuclear Extract Preparation.
The protocol for the nuclear extract preparation was described by Bresnick and Felsenfeld (14).
DNA Sequencing.
The SacI–HindIII fragment containing the chicken β-globin 5′HS4 was first cloned into the BamHI site of pBS II SK+, and the sequencing reaction was performed using the Sequenase Kit (United States Biochemical) with T7 and T3 primers.
Methylation Analysis.
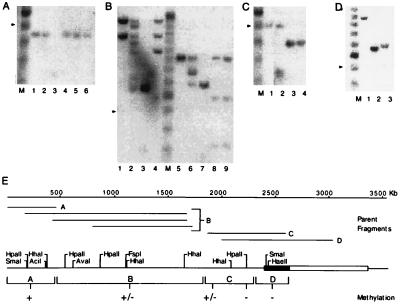
To generate the “parent fragments” shown in Fig. 5, adult chicken red blood cell DNA was first digested for 16 hr with 40 units/μg each of either BamHI and SacI (see Fig. 5A), HaeIII (see Fig. 5B, lanes 1–4), SacI (see Fig. 5 B, lanes 5–9, and C), or StyI (see Fig. 5D). Each sample was phenol/chloroform-extracted and ethanol-precipitated, then divided evenly and either mock-digested or incubated for 16 hr with 40 units/μg of the methylation-sensitive enzyme indicated in the legend to Fig. 5. The DNA in each sample was quantified, and equal amounts of total DNA (8 μg) were loaded onto each lane of a 1.8% agarose gel, electrophoresed, blotted, and hybridized with an appropriate 32P-labeled probe by standard methods.
Figure 5.
Analysis of DNA methylation in the insulator region. In each of the panels (A–D), the sensitivity of the parent fragments illustrated in E to digestion by methylation-sensitive restriction enzymes is shown. In A, a 454-bp parent fragment was mock-digested (lane 1), digested with the methylation-sensitive enzymes HpaII (lane 2), SmaI (lane 4), HhaI (lane 5), and AciI (lane 6), or digested with the methylation-insensitive isoschizomer of HpaII–MspI (lane 3). In lanes 1–4 of B, 1469- and 1219-bp parent fragments were mock-digested (lane 1), digested with the methylation-sensitive enzymes HpaII (lane 2) and AvaI (lane 4), or digested with the methylation-insensitive isoschizomer of HpaII–Msp I (lane 3). In lanes 5–9 of B, an 893-bp parent fragment was mock-digested (lane 5), digested with the methylation-sensitive enzymes HpaII (lane 6), FspI (lane 8), and HhaI (lane 9), or digested with the methylation-insensitive isoschizomer of HpaII–MspI (lane 3). In C, a 503-bp parent fragment was mock-digested (lane 1), digested with the methylation-sensitive enzymes HhaI (lane 2) and HpaII (lane 4), or digested with the methylation-insensitive isoschizomer of HpaII–MspI (lane 3). In D, a 1017-bp parent fragment was mock-digested (lane 1) or digested with the methylation-sensitive enzymes SmaI (lane 2) and HaeII (lane 3). In A–D, the arrowhead indicates the position of the 500-bp fragment of the 100-bp ladder in lane M. The results of these experiments are summarized qualitatively in E. +, Full methylation; +/−, partial methylation; and −, absence of detectable methylation. The boxed region delineates the position of the 1.2-kb insulator; stippling indicates the position of the core.
DNA Transfection of K562 Cells.
The culture conditions and the stable transfection technique were described by Chung et al. (5). Transient transfection was performed in the same way as stable transfection, except that 3 × 107 K562 cells and 10 μg of circular plasmid DNA were used. Data shown in Figs. 1, 2, 7, and 9 are the results of duplicate experiments and are typical of results obtained with the constructions used.
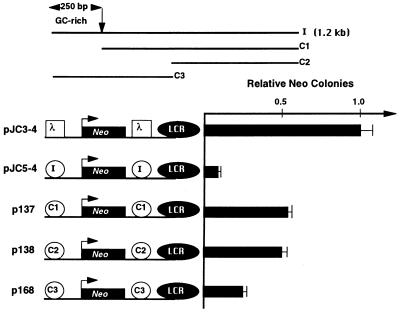
Figure 1.
Localization of the insulator activity. Three DNA fragments containing various regions of the intact 1.2-kb fragment (I, intact fragment; C1, 1 kb of the 3′ region; C2, 600 bp of the 3′ region; and C3, 650 bp of the 5′ region) were inserted on both sides of the Aγ-neo reporter gene. These constructs were stably transfected into K562 cells and G418-resistant colonies were selected and counted 2 to 3 weeks later as described previously (5). The number of G418-resistant colonies for pJC3-4 was set to 1.0.
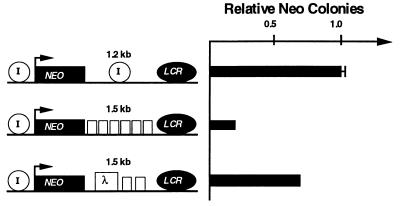
Figure 2.
Testing the insulating activity of the core element. Six copies of the 250-bp fragment containing 5′HS4 (p179) or two copies of the fragment plus a 950-bp fragment from λ phage DNA (p176) were inserted between the LCR and the Aγ-neo reporter gene. The number of G418-resistant colonies was determined as in Fig. 1, and the number obtained for pJC5-4, which contains one copy of the intact insulator on both sides of the reporter gene, was set to 1.0.
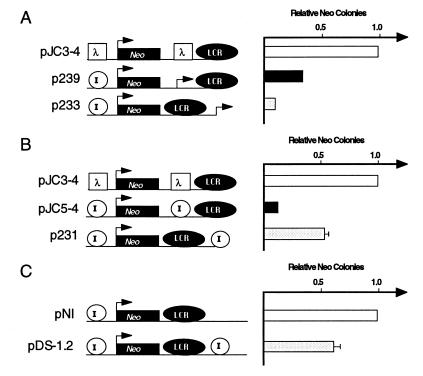
Figure 7.
Comparing the position dependence of insulator activity with that of the Aγ-globin promoter. A 559-bp fragment containing the Aγ-globin promoter (A) or the full-length insulator fragment (B) was inserted either between the LCR and the reporter (p239 and pJC5-4, respectively) or on the outside of the LCR (p233 and p231, respectively). In C, a construct that contains the insulator inserted on the outside of the LCR is compared with an otherwise identical construct in which no insulator has been inserted. For each construct, the number of G418-resistant colonies was determined as in Fig. 1 and normalized to pJC3-4 (A and B) or pNI (C) as indicated. The data presented are the average of two to four independent experiments.
Luciferase Assay.
K562 cells were harvested 48 hr after transfection and washed twice with phosphate-buffered saline (137 mM NaCl/2.7 mM KCl/4.4 mM Na2HPO4/1.4 mM KH2PO4). The cell pellet was lysed with 250 μl of lysis buffer (25 mM Tris-phosphate, pH 7.8/2 mM DTT/2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid/10% glycerol/1% Triton X-100) for 10 min at 25°C. Ten microliters of the extract was mixed with 50 μl of the luciferase assay reagent [20 mM tricine/1.07 mM (MgCO3)4Mg(OH)2·5H2O/2.67 mM MgSO4/0.1 mM EDTA/33.3 mM DTT/270 μM coenzyme A/470 μM luciferin/530 μM ATP]. The samples were placed in an automated luminometer (Berthold Clini-Lumat; London Diagnostics, Eden Prairie, MN) and measured for 10 sec.
RESULTS
In an earlier study (5), we used a colony assay based on G418 resistance to test whether a 1.2-kb fragment containing 5′HS4 can insulate a Aγ-globin promoter/neo reporter gene from a strong β-globin LCR element. We found that a single copy of the l.2-kb fragment was capable of insulating the reporter gene by ≈10-fold and two copies of the fragment insulated by 30-fold.
To define the insulator more precisely, we have now tested a series of deletions of this 1.2-kb fragment for insulator activity (Fig. 1). Deletion of the 250-bp region, which contains 5′HS4 (fragment C1), resulted in a significant, but not complete, loss of insulator activity (5-fold loss). A deletion of an additional 400 bp (C2) had no further effect. The 650-bp fragment containing the 250-bp region (C3) conferred some insulation but less than the original 1.2-kb fragment. This suggested that although there is some weaker activity within the remaining 1 kb, much of the insulator activity is concentrated in the 250-bp region. We will refer to the 250-bp region as the “core” element of the insulator. The sequence of the core element is shown in Fig. 3.
Figure 3.
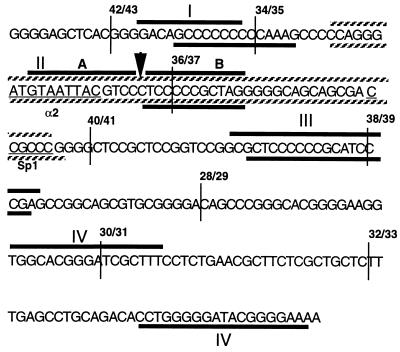
The sequence of the insulator core element. The heavy bars indicate the footprinted regions observed for both the coding (upper) and noncoding (lower) strand. The footprinted regions are numbered (I–IV) according to their positions on each strand; note that the positions of the fourth footprinted regions on the upper and lower strands do not coincide. The 5′ boundaries of the overlapping oligonucleotides used as competitors in DNase I footprint analysis (Fig. 4) are indicated as vertical lines. Oligonucleotide 42/43 is 25 bp long; all of the remaining oligonucleotides are 40 bp long. The extended footprint II and a hypersensitive site seen in the presence of the competing oligonucleotide 34/35 are shown as striped bars and a vertical arrow, respectively. The vertical arrow also indicates the boundary between the footprints IIA and IIB. The potential binding sites for Sp1, the yeast α2 repressor, and suppressor of hairy wing are underlined.
Since a single copy of the core element was not a very strong insulator, we asked whether its strength was copy number-dependent by replacing the 1.2-kb fragment with either six copies of the core element or two copies of the core element and a spacer DNA from λ phage (Fig. 2). The construct with two copies of the core element (p176) was as active as a single copy of the original 1.2-kb fragment (pJC5-4). The construct with six copies of the core element had an insulator activity that was 3-fold greater than the activity of the two-copy core element construct (p179). Thus the insulator activity of the core element, like that of the original 1.2-kb fragment, is dependent on the number of copies.
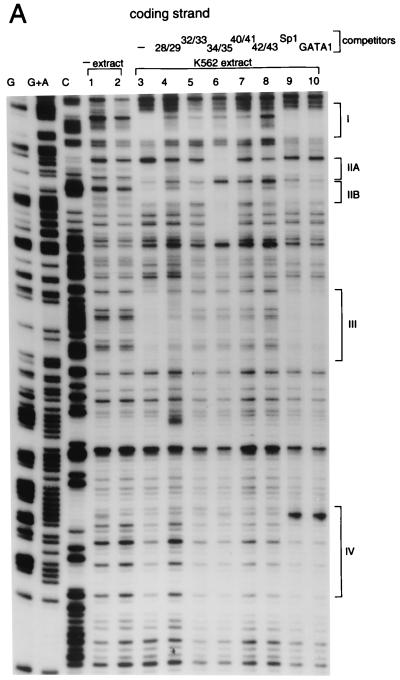
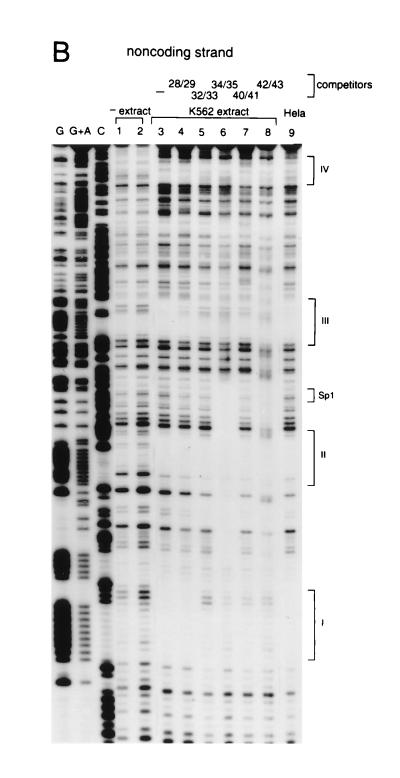
Binding of proteins to the core DNA was studied by in vitro footprinting, which shows at least four protected regions with K562 nuclear extracts. These are marked on the sequence (Fig. 3) and on the autoradiogram (Fig. 4). Footprint patterns obtained with HeLa extract were not significantly different (see Fig. 4B). The footprint experiments were repeated in the presence of some of the unlabeled competing oligomers indicated in Fig. 3. In some cases, different oligomers crosscompete for the same footprints. For example, footprint III is disrupted not only by self-competition (probe 40/41) but also by the other probes tested. This is likely to be a reflection of the base sequence similarities of different regions of the core. In gel-shift experiments, while each of the oligomers gave rise to more than one complex, only some of these complexes were competed by non-self-oligomers (data not shown). Thus, while some of the same nuclear factors may bind to different regions of the core, additional as yet unidentified proteins are capable of forming specific complexes with these regions.
Figure 4.
DNase I footprint analysis of the core element. DNA binding reactions were incubated for 20 min at 25°C with K562 nuclear extract. (A) Coding strand analysis. The four most prominent footprints are labeled I–IV. The oligonucleotide competitor, where used, is indicated above the lanes. See Fig. 3 for the sequence compositions of the competitors as well as a summary of the footprint analysis. To control for specificity, oligonucleotide competitors with GATA1 and Sp1 binding sites were also used (lanes 9 and 10, respectively). (B) Noncoding strand analysis. In addition to the four observed footprints, the location of the canonical Sp1 binding site is also indicated.
Curiously, unlike the other footprints, footprint II (IIA and B on the coding strand) is not disrupted by any of the competing probes, including 34/35; despite the fact that 34/35 includes and is larger than footprint II, its addition actually extends the footprint rather than diminishing it. One possible explanation for this behavior is that an abundant or high-affinity factor that binds within footprint II inhibits other factors from binding to the region around it. Probe 34/35 presumably titrates off this inhibitor and allows the other factors to bind, creating a new and extended footprint. This might also explain why even though the ubiquitous factor Sp1 (12) binds in a gel-shift assay (data not shown) to its canonical binding motif (CCGCCC) located in probe 36/37, it does not footprint in the absence of competing probe 34/35. In the presence of 34/35, footprint II extends to the Sp1 binding site (see Fig. 4B). This footprinted region also contains a 9 of 10 base sequence identity to the binding site for a monomer of the α2 repressor from yeast (ref. 13; see also Fig. 3).
It is evident from the sequence shown in Fig. 3 that the core element is G+C-rich. Its GC content is ≈70% with a high density of CpG dinucleotides—22 of 250 bp, with a CpG/GpC ratio of 0.78. The DNA both 5′ and 3′ to the core is not notably G+C-rich, nor is there a preponderance of CpGs; the ≈1 kb of sequence 3′ to the core contains only 15 CpGs (data not shown). Since regions of high CpG density are frequently undermethylated, we used methylation-sensitive restriction enzymes to examine the DNA methylation status of the core and surrounding sequences. Adult chicken red blood cell genomic DNA was first digested with restriction enzymes to generate parent fragments representative of the DNA at varying distances from the core (Fig. 5E). Each of these fragments was then subjected to limit digestion with the methylation-sensitive enzymes indicated in Fig. 5. The core region as well as the DNA within 200 bp 5′ of the core is digested to completion with the methylation-sensitive enzymes HpaII, SmaI, and HaeII, indicating that the CpGs in this region are not methylated (data in Fig. 5 C, lanes 3 and 4, and D, lanes 2 and 3). Varying degrees of partial methylation were observed in the next ≈2 kb of DNA upstream of the core, followed by complete methylation beginning ≈2.2 kb upstream of the core. The absence of a suitable combination of probes and parent fragments in the DNA 3′ to the core made it impossible to assess the methylation status of this region. An HpaII site ≈1.5 kb 3′ to the core, however, is at least partially methylated (data not shown).
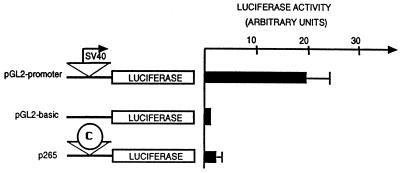
CpG islands have been found in association with the promoters of housekeeping genes (15, 16). For this reason we tested whether the core region has promoter function (Fig. 6). The core sequence was coupled to a luciferase reporter gene and transfected into K562 cells. We compared its activity to a control construction carrying the simian virus 40 (SV40) early promoter and to another control, which lacked any promoter. As shown in Fig. 6, the core sequence showed only marginal (1.8-fold) activity when compared with the promoterless control, and both were far less active than the SV40 promoter (20-fold). Even lower activity was observed with the core element in the opposite orientation (data not shown).
Figure 6.
Testing the core element for promoter activity. Three reporter genes, including pGL2-basic, which has no promoter, pGL2-promoter, which has the SV40 early promoter, and p265, which has the core element inserted in place of the SV40 promoter, were transiently transfected into K562 cells, and luciferase activity was measured 48 hr later.
As a more direct approach, we searched for transcripts in vivo in the neighborhoods 3′ and 5′ of the core element. Northern blot analysis of RNA prepared from 5- and 11-day embryonic erythroid cells, as well as from chicken embryo fibroblasts, showed no detectable transcripts when hybridized with probes to the regions 0–503 bp upstream of the core element and 374–661 bp downstream of the core element, whereas a control histone H5 (17) probe gave a distinct hybridization band in the erythroid cell samples (data not shown).
Because we saw some weak promoter activity in the above transient transfection experiments, we wished to compare the activity of a known promoter with that of the insulator in our colony assay. We therefore compared the effect of the human Aγ-globin promoter when inserted on either side of the LCR to the effect of the insulator (Fig. 7 A and B). We made constructions in which either the Aγ-globin promoter or the insulator was placed between the LCR and the reporter (p239 and pJC5-4) or on the other side of the LCR (p233 and p231). We find that the activity of the Aγ-globin promoter is unlike that of the insulator: whether it is placed between the LCR and the reporter gene (p239) or on the outside of the LCR (p233), the Aγ-globin promoter blocks the activation of the reporter gene (Fig. 7A). In fact, the Aγ-globin promoter blocks more effectively when it is placed on the outside of the LCR. In contrast, the chicken insulator is much more effective in blocking the LCR when it is placed between the LCR and the promoter than when it is placed on the outside of the LCR (Fig. 7B and see below). These results demonstrate that although a promoter can compete for LCR action, it represses from either side of the LCR, and, therefore, its action is unlike that of the chicken β-globin insulator.
In the previous experiments, the LCR is ≈5 kb from the reporter in the control construct (pJC3-4), whereas this distance is only ≈2.5 kb in the construct with the insulator outside the LCR (p231). To account for this difference, we examined the effect of an outside copy of the insulator in an otherwise identical construct (Fig. 7C). With these constructs, the results are identical to those presented in Fig. 7B; when placed outside the LCR, the insulator has only a mild (1.7-fold) inhibitory effect on reporter activity. Thus, regardless of the distance between the LCR and the reporter, we consistently observe a 1.5- to 2-fold reduction in LCR activity when the insulator is placed on the outside of the LCR. This is significantly less than the 8- to 12-fold reduction routinely observed when the insulator is inserted between the LCR and the promoter. The small inhibitory effect of an outside insulator observed in these experiments contrasts with the slight stimulatory effect that we reported in an earlier study (figure 4 of ref. 5). We have found that this discrepancy results from an error in one of the constructs (pJC19) used in the earlier study. This difference does not affect the conclusion that the effect of the insulator is clearly dependent on its position relative to the LCR.
DISCUSSION
We have begun a dissection of the chicken β-globin insulator element to determine with greater precision which DNA sequences and factors confer the insulating activity. The activity of the 1.2-kb fragment containing the constitutively hypersensitive element, 5′HS4, appears to be distributed throughout the fragment, but about half the activity appears to reside in a 250-bp core segment. Two copies of this segment are sufficient to give activity equivalent to that of the entire 1.2-kb fragment. The core DNA is rich in G and C residues, and there are many short repeated sequence motifs within it.
Because the high CpG density of the core region is commonly associated with regions of unmethylated cytosine, we investigated the methylation status of the insulator region. We find that an ≈500-bp region, which includes the core is unmethylated in adult red blood cells. This pattern of methylation was also observed in adult liver DNA (data not shown), indicating that like the DNase I hypersensitivity of this region, the demethylation is maintained independent of the tissue and/or globin expression. While full methylation is not observed until ≈2.2 kb upstream of the core, partial methylation occurs over the intervening ≈2 kb. It is worth noting that the region over which this transition in DNA methylation status extends coincides with the region in which a transition in the degree of DNase I sensitivity and histone hyperacetylation was observed for the globin locus (10). We are currently investigating whether the methylation status of the core plays any role in its ability to act as an insulator.
The core has the properties of a CpG island, and in this respect it resembles the promoter of a housekeeping gene. However, the core appears to have very little promoter activity when assayed by transient transfection in the same cell line used for the insulation studies. Furthermore, there is no evidence in vivo of a defined RNA transcript corresponding to the DNA sequences on either side of 5′HS4. As shown in Fig. 7, LCR inhibition by the insulator is position dependent in contrast with the bidirectional inhibitory effect of the Aγ-globin promoter.
Along with the canonical Sp1 (12) binding site and four other potential Sp1 sites, we have also found an almost perfect match to a single copy of the yeast α2 repressor (13) binding site in the core. This sequence also shares an 8 of 11 base pair homology with the binding site for the suppressor of hairy wing protein. It has been shown that multiple copies of this site exhibit insulating properties in Drosophila (18). More generally, the site is reminiscent of several homeodomain protein binding sites, and the footprint data (Fig. 4) show that it is occupied in the presence of nuclear extracts. We note that binding sites in yeast for the α2/mcm1 complex are capable of organizing nucleosome placement over a considerable distance (13). Although it would be attractive to suppose that this is somehow related to insulator function in the globin locus, preliminary experiments suggest that the homology to this site is not necessary for the chicken β-globin insulator action (A.C.B. and G.F., unpublished work).
The results presented here in some ways resemble a recent analysis by Vazquez and Schedl (19) of the functional domains of the Drosophila scs insulator element. They showed that a relatively small portion of the element could function as an insulator, that the effect was increased by using multiple copies, and that the active portion was relatively G+C-rich. We have found, however, that the full-length scs element has no effect in our LCR blocking assay (data not shown). The inability of the scs element to function as an insulator in our assay may reflect the absence in K562 cells of some critical protein found in Drosophila or a different mechanism of insulation. We have not tested scs′ in our assay. Schreiber and Schaffner (20) have shown that certain plasmid sequences are able to block the action of the SV40 enhancer when placed between it and a promoter. They found that these sequences were G+C-rich and had the composition of a CpG island, and they suggested that such sequences might serve in vivo as inhibitors of enhancer action. We note also that Raina et al. (21) have observed in the maize Spm promoter a G+C-rich sequence 576 bp long that appears to have insulator properties. These results are entirely consistent with the properties of the β-globin insulator.
A number of models have been proposed for the mode of action of insulators (although insulators do not necessarily share a single mechanism). We and others have suggested that they might work by preventing “tracking” of a distant enhancer complex along the chromatin, by serving as a signal for sequestering the region in the nuclear matrix so as to physically isolate the region, or by interacting with a second such insulator to form an isolated loop, which blocks access to any factors outside the loop. It is also possible that the chicken β-globin insulator functions in vivo simply as a barrier against the incursion of the heterochromatic region into the β-globin locus. Our analysis of the core reveals that it shares very few properties with enhancers or LCR elements. It does not have any of the familiar binding sites associated with the globin promoters or LCR elements, perhaps not surprising since 5′HS4 is not erythroid-specific. One of the striking properties of the core is its G+C-rich content and absence of CpG methylation, which qualifies it as a CpG island. The absence of promoter activity from the core, or of detectable transcription from sequences surrounding it, suggests that this CpG island may serve to mark an important regulatory sequence that is not a promoter. The reduction in the size of the active element to a 250-bp region should allow us to identify the protein factors important for the insulation function. These may be novel proteins, or they may be familiar transcription factors acting in new ways.
Acknowledgments
We are grateful to Drs. Julio Vazquez and Paul Schedl for providing the Drosophila scs element and to members of the Felsenfeld laboratory for critical reading of this manuscript.
Footnotes
References
- 1.Eissenberg J C, Elgin S C R. Trends Genet. 1991;7:335–340. doi: 10.1016/0168-9525(91)90424-o. [DOI] [PubMed] [Google Scholar]
- 2.Kellum R, Schedl P. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 3.Kellum R, Schedl P. Mol Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roseman R R, Pirrotta V, Geyer P K. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung J H, Whitely M, Felsenfeld G. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 6.Crossley M, Orkin S. Curr Opin Genet Dev. 1993;3:232–237. doi: 10.1016/0959-437x(93)90028-n. [DOI] [PubMed] [Google Scholar]
- 7.Engel J D. Trends Genet. 1993;9:304–309. doi: 10.1016/0168-9525(93)90248-g. [DOI] [PubMed] [Google Scholar]
- 8.Moon A M, Ley T J. Proc Natl Acad Sci USA. 1990;87:7693–7697. doi: 10.1073/pnas.87.19.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reitman M, Felsenfeld G. Mol Cell Biol. 1990;10:2774–2786. doi: 10.1128/mcb.10.6.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeppesen P, Turner B M. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 12.Briggs M R, Kadonaga J T, Bell S P, Tjian R. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 13.Roth S Y, Dean A, Simpson R T. Mol Cell Biol. 1990;10:2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bresnick E H, Felsenfeld G. J Biol Chem. 1993;268:18824–18834. [PubMed] [Google Scholar]
- 15.Bird A P. Trends Genet. 1987;3:342–347. [Google Scholar]
- 16.Larsen F, Gundersen G, Lopez R, Prydz H. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 17.Affolter M, Cote J, Renaud J, Ruiz-Carillo A. Mol Cell Biol. 1987;7:3663–3672. doi: 10.1128/mcb.7.10.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spana C, Harrison D A, Corces V G. Genes Dev. 1988;2:1414–1423. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- 19.Vazquez J, Schedl P. EMBO J. 1994;13:5984–5993. doi: 10.1002/j.1460-2075.1994.tb06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schreiber E, Schaffner W. Somatic Cell Mol Genet. 1989;15:591–603. doi: 10.1007/BF01534920. [DOI] [PubMed] [Google Scholar]
- 21.Raina R, Cook D, Federoff N. Proc Natl Acad Sci USA. 1993;90:6355–6359. doi: 10.1073/pnas.90.13.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]