Abstract
Background
Elevated circulating free fatty acids (FFA) contribute to the development of hepatic steatosis and promote hepatocyte apoptosis by incompletely defined mechanisms. Although the death ligand TRAIL has been implicated in a variety of pathological liver diseases, the role of TRAIL in mediating apoptosis of FFA induced steatotic hepatocytes is unknown.
Aim
We examined TRAIL cytotoxicity in an in vitro model of hepatocyte steatosis induced by FFA.
Methods
Hepatocytes (Huh 7 cells, HepG2 cells, and primary rat hepatocytes) were rendered steatotic by incubation with oleic acid. Apoptosis was assessed morphologically and biochemically by caspase activity. TRAIL receptor regulation was examined using immunoblot analysis and siRNA for targeted knockdown. c‐jun N‐terminal kinase (JNK) inhibition was attained with SP600125.
Results
Oleic acid sensitised the cells to TRAIL but not TNF‐α cytotoxicity. FFA sensitisation to TRAIL occurred at much lower concentrations than required for FFA mediated sensitisation to Fas, or FFA induced lipoapoptosis. Oleic acid treatment led to upregulation of the cognate TRAIL receptor death receptor 5 (DR5) but not death receptor 4 (DR4). The upregulation of DR5 was JNK dependent. siRNA targeted knockdown of either DR5 or DR4 demonstrated that DR5 was responsible for FFA sensitisation to TRAIL killing. DR5 expression was enhanced in steatotic human liver samples.
Conclusion
Our results suggest that FFA induced hepatocyte steatosis sensitises to TRAIL by a DR5 mediated JNK dependent mechanism.
Keywords: cellular steatosis, death receptor, SP600125
Steatosis is a common histological feature of several liver disorders including non‐alcoholic steatohepatitis (NASH), alcohol induced liver disease (ALD), hepatitis C, and drug mediated liver injuries.1,2,3,4 Multiple metabolic derangements lead to the development of hepatocyte steatosis.5 Insulin resistance and elevated circulating free fatty acids (FFA) have been implicated in the development of steatosis in NASH and other forms of hepatic steatosis.6 FFA are injurious to many cells by inducing apoptosis, so called lipoapoptosis.7,8 Indeed, hepatocyte apoptosis is a prominent feature of steatotic liver diseases and serum biomarkers of apoptosis are elevated in patients with NASH.9 Hepatocyte apoptosis is commonly mediated by the death receptors tumour necrosis factor alfa (TNF‐α), tumour necrosis factor related apoptosis inducing ligand (TRAIL), and Fas ligand (FasL). Although TNF‐α and FasL are both implicated in the pathogenesis of NASH,3,6 the role of TRAIL in the pathogenesis of steatotic liver injury is unexplored.
Of five known TRAIL receptors, death receptor 4 (DR4) and death receptor 5 (DR5) are classic death receptors, expressed in fetal and adult liver.10 Enhanced expression of these death receptors is one mechanism by which cells can be sensitised to TRAIL. DR4 and DR5 expression can be upregulated by p53 dependent and p53 independent pathways.11,12,13,14 In particular, DR5 is also regulated by a c‐jun N‐terminal kinase (JNK) dependent manner.15 On ligand binding, DR5 and DR4 are able to activate canonical death signalling pathways leading to activation of caspases culminating in the classic morphological changes of apoptosis.10 Based on these concepts, the objective of this study was to ascertain if steatotic hepatocytes are sensitised to TRAIL cytotoxicity.
Experimental procedures
Cells and fatty acid treatment
Huh 7 , HepG2, and primary rat hepatocytes were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing high glucose (25 mM), 100 000 U/l penicillin, 100 mg/l streptomycin, and 10% fetal bovine serum. Rat hepatocytes were isolated from Sprague‐Dawley rats as previously described.16 Cells were treated with fatty acids as previously described in detail17 and intracellular neutral lipid accumulation assessed by Nile red staining.
Quantitation of apoptosis
Morphological changes of apoptosis were assessed by staining nuclei with 4′,6‐diamidino‐2‐phenylindole (DAPI) as previously described by us.17 Caspase 3/7 activity was quantified utilising a commercially available assay (Apo‐ONE homogeneous Caspase‐3/7 assay, Promega Corporation, Madison, WI, USA).
Immunoblot analysis
Cell lysates were electrophoretically resolved by SDS‐PAGE and immobilised on poly(vinylidene) fluoride (PVDF) membrane. Non‐fat dairy milk, 5%, in TRIS buffered saline (20 mM TRIS, 150 mM NaCl, pH 7.4) with 0.1% Tween‐20 was used to block non‐specific binding sites. Blots were incubated with primary antibodies at 4°C overnight and with secondary antibodies at room temperature for 1 hour. The following primary antibodies were used: rabbit polyclonal antibody (Ab) to DR4 (PSC‐1139‐C100, 1:1000, Axxora LLC, San Diego, CA, USA); polyclonal Ab to DR4 (catalogue No 1139, rabbit, 1:500, ProSci Incorporated, Poway, CA, USA); DR5 (catalogue No 2019, rabbit, 1:500, ProSci Incorporated, Poway, CA, USA); goat polyclonal Ab to DR5 (ALX‐210‐743‐C200, 1:1000); rat monoclonal Ab to cFLIP (Dave 2, ALX‐804‐127‐C100, 1:1000, Alexis Biochemicals, Lausen, Switzerland); rabbit polyclonal Ab to Caspase 8 (551234, 1:2000, BD Pharmingen, San Jose, CA, USA); rabbit polyclonal Ab to Bim (559685, 1:1000, BD Pharmingen); goat polyclonal Ab to Bid (C‐20, sc‐6538, 1:200). Rabbit polyclonal Ab to Mcl‐1 (S‐19, sc‐819, 1:200), mouse monoclonal Ab to Bax (B‐9, sc‐7480, 1:200), goat polyclonal Ab to Bak (G‐23, sc‐832‐G, 1:500), rabbit polyclonal Ab to Bcl‐2 (sc‐492, 1:500), and rabbit polyclonal Ab to Bcl‐xl (sc‐634, 1:500) were from Santa Cruz Biotechnology, Santa Cruz, CA, USA; mouse monoclonal Ab to γ‐tubulin (T6557, 1:5000) from Sigma Pharmaceuticals, St Louis, MO, USA. Peroxidase conjugated secondary antibodies (1:3000, Biosource International, Camarillo, CA, USA) were used to detect antigen‐antibody complexes. Immune complexes were visualised using a chemiluminescent substrate (ECL, Amersham, IL, USA), and Kodak XOMAT film (Eastman Kodak, Rochester, NY, USA).
JNK activity assay
JNK induced phosphorylation of c‐Jun was measured utilising a Phospho‐c‐Jun (Ser63) specific antibody using a commercially available in vitro kinase assay according to the manufacturer's directions (Cell Signaling Technology, catalogue No 9810). Briefly, whole cell lysate was immunoprecipitated with immobilised c‐Jun fusion protein. Kinase reaction was performed in the presence of 200 μM ATP, and phospho‐c‐Jun and total c‐Jun detected by immunoblot analysis. An image densitometer was used to quantitate the area of immunoreactivity and the Molecular Analyst software program used to calculate the relative increase in phospho‐c‐jun (BioRad).
Quantitative real time PCR
Polymerase chain reaction (PCR) primers used for human DR5 were as follows: forward 5′‐TGCAGCCGTAGTCTTGATTG‐3′ and reverse 5′‐GCACCAAGTCTGCAAAGTCA‐3′. PCR primers used for human DR4 were as follows: forward 5′‐TGTCAGTGCAAACCAGGAAC‐3′ and reverse 5′‐TGCTCAGAGACGAAAGTGGA‐3′. Commercially available primers for ribosomal 18S RNA (Ambion, Austin, TX, USA) were used as controls. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and reverse transcribed to cDNA using random primers and murine leukaemia virus reverse transcriptase (Invitrogen). The cDNA was amplified using Taq DNA polymerase (Invitrogen) and standard PCR. Quantitative real time PCR (LightCycler, Roche Molecular Biochemicals, Mannheim, Germany) was performed using SYBR green (Molecular probes) as the fluorophore as previously described by us.17
Luciferase reporter assay
Huh 7 cells were co‐transfected with TK‐Renilla‐CMV and firefly luciferase based DR5 reporter plasmid. The firefly luciferase based DR5 reporter plasmid has been previously described.15 Cell lysates were prepared by adding lysis buffer directly to the cells. Firefly luciferase and Renilla luciferase activities in cell lysates were determined using a dual luciferase kit (Promega, Madison, WI, USA).
Small interfering RNA (siRNA) experiments
RNA interference was used to silence DR4 and DR5 gene expression in Huh 7 cells. The sequences of siRNAs used were as follows: siDR4 (TRAIL‐R1) sense 5′‐CACCAAUGCUUCCAACAAUdTdT‐3′18 and siDR5 (TRAIL‐R2) sense 5′‐AAGACCCTTGTGCTCGTTGTC‐3′.19 TRAIL‐R1 siRNA was purchased from Dharmacon Research (Lafayette, CO, USA) and TRAIL‐R2 siRNA was purchased from Qiagen (Valencia, CA, USA). Transient gene silencing was attained by transfection of siRNA into cells using siPORT lipid transfection reagent (Ambion) according to the manufacturer's instructions. Scrambled siRNA was used as a control. Cells were transiently transfected with 2 μg/ml siRNA by using 8 μl of siPORT lipid (Ambion) in a total transfection volume of 0.25 ml of OPTIMEM (Gibco‐Invitrogen Corporation, Carlsbad, CA, USA).
Human studies
After approval by the Mayo Clinic institutional review board immunohistochemistry for DR5 was performed using goat anti‐DR5 (N‐19) and goat ABC Staining System (Santa Cruz Biotechnology) on normal liver samples (n = 5) and liver samples from patients with NASH (n = 10) retrieved from our pathology files.2 Formalin fixed, paraffin embedded liver biopsy tissue was cut into 4‐μm thick sections. Sections were deparaffinised, rehydrated through graded alcohols, and antigen retrieval was performed by heating in ethylenediaminetetraacetic acid (1 mmol/l, pH 8.0) in a vegetable steamer for 40 minutes followed by cooling for 20 minutes. Endogenous peroxidase was quenched with 3% H2O2 in methanol, followed by blocking for 1 hour with 1.5% blocking serum in PBS followed by a 1:40 dilution of primary antibody or normal goat immunoglobulin G (IgG; Sigma, St Louis, MO, USA) overnight at 4°C. Sections were incubated with a second biotinylated antibody, followed by avidin‐biotin‐peroxidase complex for 30 minutes. Sections were developed with 3,3′‐diaminobenzidine and counterstained with haematoxylin. Semi‐quantitative DR5 expression was evaluated using a 4‐point scoring system (0 = no staining, 1 = positive staining in less than 30% hepatocytes per high power field, 2 = positive staining in 30–70% hepatocytes per high power field, and 3 = positive staining in greater than 70% hepatocytes per high power field. All slides were scored by two independent, blinded observers.
Reagents
DAPI (4′,6‐diamidino‐2‐phenylindole), isopropanol, bovine serum albumin, stearic acid, ecosapentaenoic acid, and oleic acid were from Sigma Pharmaceuticals (St Louis, MO, USA). Recombinant human TRAIL was from R&D Systems (Minneapolis, MN, USA). SP‐600125 (JNK inhibitor) was from Calbiochem (San Diego, CA, USA). Z‐VAD‐fmk was from Enzyme Systems (Livermore, CA, USA).
Statistical analysis
All data are expressed as the mean (SEM) unless otherwise indicated, from three or more experiments. Differences between groups were compared using Student's t test.
Results
Free fatty acid treatment induces cellular steatosis and sensitisation to TRAIL cytotoxicity
Huh‐7 cells were treated with stearic acid, a saturated fatty acid, and oleic acid, a monounsaturated fatty acid. Each individual FFA induced cellular steatosis (fig 1A) consistent with out earlier observations.17 Next, Huh‐7 cells were treated with stearic acid and oleic acid for 16 hours followed by the addition of recombinant human TRAIL for an additional 4 hours. Stearic acid, oleic acid, or TRAIL treatment alone induced minimal apoptosis (fig 1B). However, the combination of FFA and TRAIL lead to a significant augmentation of cell death as assessed morphologically (fig 1B) and by caspase 3/7 activity (fig 1C). FFA sensitisation to TRAIL cytotoxicity was concentration and time dependent (fig 1D and E), and even concentrations as low as 50 μM sensitised to TRAIL cytotoxicity. In contrast, FFA did not sensitise Huh‐7 cells to TNF‐α cytotoxicity (data not shown). Finally, we determined if oleic acid and stearic acid induced TRAIL sensitisation was caspase dependent. Huh 7 cells treated with the pan‐caspase inhibitor zVAD‐fmk followed by oleic acid or stearic acid and TRAIL were resistant to apoptosis (fig 1F); as an internal control, zVAD‐fmk also inhibited TRAIL killing alone, as well as free fatty acid induced sensitisation to TRAIL. Thus, the TRAIL mediated cytotoxicity was caspase dependent. We next analysed eicosapentaenoic acid (EPA), a polyunsaturated fatty acid. EPA induced cellular steatosis (fig 1G), similar to previously examined fatty acids, and sensitised to TRAIL killing. Furthermore, EPA did not protect against oleic acid toxicity (fig 1H).
Figure 1 Free fatty acid (FFA) treatment induces steatosis and sensitises Huh 7 cells to TRAIL cytotoxicity. (A) Huh 7 cells were treated with 200 μM each of oleic acid and stearic acid to induce steatosis for 4 hours and 16 hours (top and bottom panels, respectively). Nile red staining was utilised to demonstrate steatosis. Both oleic acid and stearic acid induced cellular steatosis following 4 hours and 16 hours of treatment. (B–F) Huh 7 cells were treated with 200 μM each of oleic acid and stearic acid for 16 hours, except for the time course experiment. Steatotic cells were challenged with recombinant human TRAIL (2 ng/ml). Apoptosis was assessed morphologically and biochemically by measuring caspase 3/7 activity. (B) Oleic acid and stearic acid sensitise Huh 7 cells to TRAIL cytotoxicity (p<0.001, FFA + TRAIL vs free fatty acid alone). (C) Caspase 3/7 activity showing sensitisation to TRAIL toxicity in oleic acid and stearic acid treated cells (p<0.05, FFA + TRAIL vs FFA alone). (D) Sensitisation to TRAIL cytotoxicity is concentration dependent, occurring at concentrations that are not toxic, per se (p<0.05 for each concentration, FFA + TRAIL vs FFA alone, except 50 μM oleic acid, where the trend was consistent with sensitisation though statistical significance was not attained). (E) Oleic acid and stearic acid sensitisation to TRAIL cytotoxicity is time dependent. The earliest time point at which fatty acid treatment sensitised to TRAIL cytotoxicity was observed was 4 hours of pre‐incubation with each FFA (p<0.05 compared to each FFA alone). (F) Oleic acid and stearic acid induced TRAIL cytotoxicity is caspase dependent. Pan‐caspase inhibitor zVAD‐fmk significantly attenuated FFA induced TRAIL sensitisation. TRAIL induced cell death (*p<0.05, TRAIL vs zVAD‐fmk+TRAIL), and FFA induced sensitisation to TRAIL (**p<0.001, FFA+TRAIL vs FFA+zVAD‐fmk+TRAIL) was caspase dependent. (G) Eicosapentaenoic acid (EPA) also induces steatosis in Huh 7 cells treated with 200 μM EPA for 4 hours and 16 hours by Nile red staining. EPA treatment (200 μM for 16 hours) sensitises to TRAIL toxicity, as measured morphologically (open bars) and biochemically by caspase 3/7 activity (shaded bars) (p<0.05, FFA vs FFA+TRAIL). (H) Morphological data measuring apoptosis in Huh 7 cells treated with oleic acid (OA) alone or EPA alone or the two in combination (100–200 μM each, 20 hours) is shown.
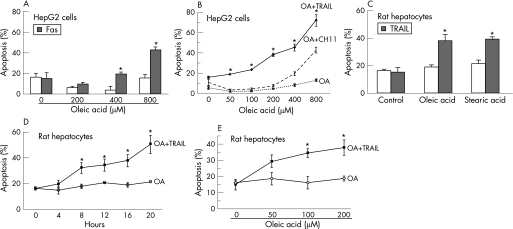
Huh 7 cells do not express Fas, therefore, we utilised HepG2 cells for experiments involving Fas. Consistent with our previous report, oleic acid sensitised HepG2 cells to Fas mediated cell death (fig 2A). However, oleic acid concentrations of 400 μM were necessary to sensitise cells to the Fas agonist (CH11), and no sensitisation was observed at lower concentrations of oleic acid (50–200 μM), despite the observation that these concentrations did sensitise HepG2 cells to TRAIL cytotoxicity (fig 2B). Transformed cells are often preferentially susceptible to TRAIL cytotoxicity, whereas primary hepatocytes may be resistant. Therefore, our next step was to determine if FFA induced steatosis also sensitised to TRAIL killing in primary rat hepatocytes. Minimal basal cytotoxicity was observed in cells treated with oleic acid, stearic acid, or TRAIL alone (fig 2C). However both oleic and stearic acid sensitised hepatocytes to TRAIL cytotoxicity in a time and concentration dependent manner (fig 2D and E). Collectively, these data demonstrate that the FFA, oleic acid and stearic acid, sensitise both transformed and primary hepatocytes to TRAIL cytotoxicity. As oleic acid, a minimally toxic FFA, sensitised to TRAIL cytotoxicity (4–5‐fold increase) to an extent similar to the inherently toxic stearic acid, we utilised oleic acid for the remainder of our studies.
Figure 2 Free fatty acid (FFA) treatment sensitises HepG2 cells and primary hepatocytes to TRAIL cytotoxicity. (A) Sensitisation to Fas (using agonistic antibody CH11, 150 ng/ml) in HepG2 cells is concentration dependent (p<0.05 compared to each FFA alone). (B) Oleic acid (OA) induces greater sensitisation to TRAIL compared to Fas in HepG2 cells. At each concentration of oleic acid statistically significant sensitisation was observed to TRAIL cytotoxicity compared to Fas cytotoxicity (p<0.01). (C) Primary rat hepatocytes treated with oleic acid or stearic acid are sensitised to TRAIL cytotoxicity (p<0.05 compared to FFA alone). (D) Oleic acid induced sensitisation to TRAIL cytotoxicity in time dependent in primary rat hepatocytes, statistically significant from 8 hours onwards (p<0.05). (E) Oleic acid induced sensitisation to TRAIL cytotoxicity in concentration dependent in primary rat hepatocytes, (p<0.05 for 100 μM and 200 μM oleic acid).
FFA induced TRAIL sensitisation is JNK dependent
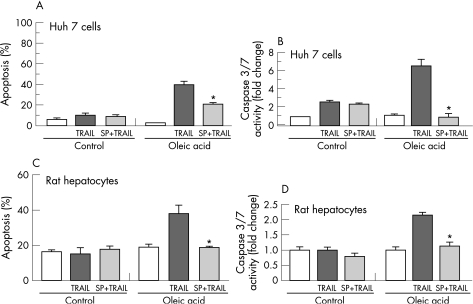
Because FFA induce JNK activation and JNK regulates the expression of TRAIL and its receptors15 we next ascertained if JNK activation is associated with sensitisation of oleic acid treated Huh 7 cells to TRAIL. JNK 1/2 inhibition by SP600125 reduced TRAIL toxicity by 50% in oleic acid treated cells as determined by morphological criteria (fig 3A) and caspase 3/7 activity (fig 3B). Similarly, in primary rat hepatocytes JNK 1/2 inhibition reduced TRAIL toxicity in oleic acid treated cells by 50% as assessed either by morphological criteria (fig 3C) or caspase 3/7 activity (fig 3D). Taken together these data indicate that oleic acid induced sensitisation to TRAIL is JNK 1/2 dependent.
Figure 3 Oleic acid induced TRAIL sensitisation is JNK dependent. Huh 7 cells and primary rat hepatocytes were each treated with 200 μM oleic acid. SP600125 a pharmacological JNK 1/2 inhibitor was used (10 μM, 10 minutes pretreatment). Recombinant human TRAIL was used at a concentration of 2 ng/ml. Apoptosis was assessed by morphological criteria and by caspase 3/7 activity. (A) Oleic acid treated Huh 7 cells are sensitised to TRAIL cytotoxicity in a JNK dependent manner as measured morphologically. In the presence of SP600125 oleic acid induced TRAIL sensitisation was reduced significantly (p<0.05). (B) Oleic acid treated Huh 7 cells are sensitised to TRAIL cytotoxicity in a JNK dependent manner as measured biochemically by caspase 3/7 activity. In the presence of SP600125 oleic acid induced TRAIL sensitisation was reduced significantly (p<0.05). (C) Oleic acid treated primary rat hepatocytes are sensitised to TRAIL cytotoxicity in a JNK dependent manner as measured morphologically. In the presence of SP600125 oleic acid induced TRAIL sensitisation was reduced significantly (p<0.05). (D) Oleic acid treated primary rat hepatocytes are sensitised to TRAIL cytotoxicity in a JNK dependent manner as measured biochemically by caspase 3/7 activity. In the presence of SP600125 oleic acid induced TRAIL sensitisation was reduced significantly (p<0.05).
Oleic acid induces JNK dependent DR5 expression
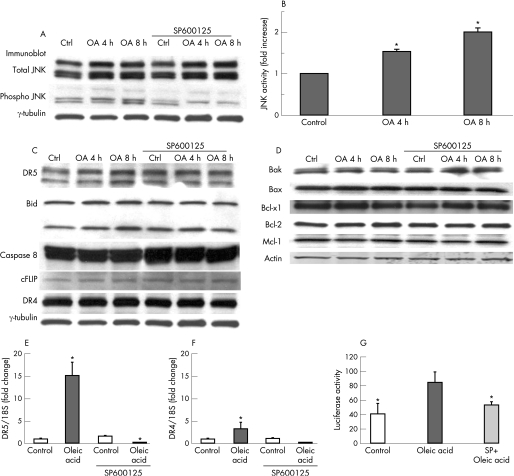
JNK 1/2 can modulate apoptosis in several ways, including activation of pro‐apoptotic Bcl‐2 family proteins, inactivation of anti‐apoptotic Bcl‐2 family proteins, induction of BH‐3 only proteins, degradation of cFLIP, or modulation of death receptor expression.15,17,20,21,22,23 To define how JNK 1/2 modulated TRAIL sensitivity in oleic acid treated cells, we examined the possibility that JNK 1/2 activation induced TRAIL receptor DR4 and/or DR5 expression. Oleic acid treatment led to a time dependent increase in JNK 1/2 phosphorylation, and c‐Jun phosphorylation in a JNK activity assay indicative of its activation (fig 4A and 4B). As expected, this activation was blocked by SP600125, a JNK inhibitor. Furthermore, oleic acid treatment led to a time dependent increase in DR5 protein expression; which was also attenuated by SP600125 (fig 4C). In contrast, DR4 protein expression was unchanged. Death signals emanating from TRAIL ligation with DR5 are processed at the TRAIL death inducing signalling complex (DISC). This activation culminates in the activation of caspase 8 and cleavage of Bid. cFLIP is an endogenous inhibitor of death receptor signalling at the level of the DISC, while the antiapoptotic proteins, Bcl‐xl and Mcl‐1, block TRAIL signalling at the level of the mitochondria, via inhibitory interactions with proapoptotic proteins, Bak and Bax. Furthermore, JNK can regulate anti‐apoptotic Bcl‐2 and pro‐apoptotic Bax, controlling susceptibility to apoptotic stimuli.24 Therefore, we examined if oleic acid induced TRAIL sensitisation was contingent upon alterations in expression of these cell death effectors. Cellular protein levels of caspase 8, cFLIP, and Bid were unchanged in oleic acid treated cells. Expression of the antiapoptotic Bcl‐2 family proteins, Bcl‐xl, Bcl‐2, and Mcl‐1, and the pro‐apoptotic proteins Bak and Bax, were also unchanged (fig 4D). Therefore, oleic acid sensitisation to TRAIL cytotoxicity appeared to be specific for DR5 upregulation. To further confirm the induction of DR5 by FFA, DR5 mRNA expression was assessed by real time PCR. Huh 7 cells treated with oleic acid exhibited a 15‐fold increase in DR5 expression compared to control (p<0.001) (fig 4E). SP600125 decreased DR‐5 transcription by 90% compared to oleic acid alone (p<0.05). In contrast a significant increase was not observed for DR4 mRNA expression (fig 4F), consistent with the protein expression data. Furthermore, in a luciferase reporter gene assay, oleic acid treatment led to a twofold increase in luciferase activity (fig 4G); this increase was also reduced by SP600125. These data indicate JNK mediated DR5 upregulation is a potential underlying mechanism for oleic acid induced TRAIL sensitivity.
Figure 4 Oleic acid (OA) treatment sensitises to TRAIL cytotoxicity via JNK dependent upregulation of DR5. Huh 7 cells were treated with 200 μM oleic acid, with and without a pharmacological JNK 1/2 inhibitor, SP600125. (A) JNK activation, evidenced by phosphospecific immunoblotting, is seen in oleic acid treated cells, and is prevented in the presence of SP600125. (B) JNK activity assay data in oleic acid treated Huh 7 cells are shown. JNK activity assay (described in methods) product, phospho‐c‐jun, was quantitated and normalised to total c‐jun expression for each experimental condition (p<0.001 compared to control). (C) Immunoblot analysis shows selective upregulation of DR5 in oleic acid treated cells, in a time dependent manner. The DR5 upregulation is abrogated in the presence of SP600125. DR4, Bid, caspase 8, and cFLIP levels are unchanged. (D) Immunoblot analysis for pro‐apoptotic and anti‐apoptotic members of the Bcl‐2 family demonstrates that expression of Bak, Bax, Bcl‐xl, Bcl‐2, and Mcl‐1 was unchanged. (E) Quantitative real time PCR was performed for DR5 expression in oleic acid treated Huh 7 cells, in the absence and presence of SP600125. Oleic acid treatment induced a 15‐fold increase in DR5 expression (p<0.05, compared to control); this was completely abrogated in the presence of SP600125 (p<0.05, compared to oleic acid alone). (F) Quantitative real time PCR was performed for DR4 expression in oleic acid treated Huh 7 cells, in the absence and presence of SP600125. A slight increase was observed in DR4 expression, this did not attain statistical significance (p = 0.2, compared to control). (G) Reporter gene assay using a DR5 luciferase plasmid demonstrates that 200 μM oleic acid treatment induced an increase in DR5 expression (p<0.05); this is JNK dependent as it is abrogated in the presence of SP 600125 (p<0.05).
DR5 mediates oleic acid induced TRAIL sensitisation
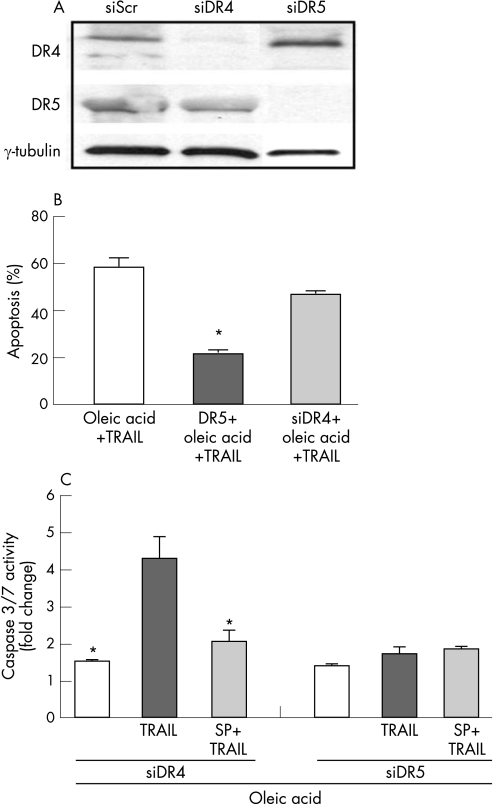
To confirm that DR5 indeed mediates oleic acid induced sensitisation to TRAIL, DR5 expression was knocked down using siRNA. We first verified the efficacy and specificity of the respective siRNAs targeting DR5 and DR4 (fig 5A) in Huh 7 cells. Next, cells targeted with DR5 or DR4 siRNA were treated with oleic acid and TRAIL. Oleic acid treated DR5 siRNA transfected cells demonstrated greater than a 60% reduction in TRAIL cytotoxicity by morphological criteria (fig 5B) and caspase 3/7 activity (fig 5C). In contrast, oleic acid treated DR4 siRNA transfected cells did not demonstrate any protection from TRAIL cytotoxicity. These data demonstrate that oleic acid induced sensitisation to TRAIL is indeed DR5 dependent.
Figure 5 Oleic acid induced TRAIL sensitisation is DR5 dependent. (A) Western blot analysis was performed for DR5 and DR4 expression using whole cell lysates from Huh 7 cells transfected with respective siRNAs. The specificity and effectiveness of each siRNA is demonstrated. In cells treated with siDR4 a decrease in DR4 level was observed, DR5 levels were unchanged. In contrast, in cells treated with siDR5 a decrease in DR5 level was observed, DR4 levels were unchanged. Scrambled siRNA was used as control. (B) Apoptosis was assessed morphologically in cells transfected with siDR5 and siDR4. Oleic acid treatment sensitised to TRAIL cytotoxicity. This was abrogated in siDR5 transfected cells (p<0.05) and unchanged in siDR4 transfected cells. (C) Caspase 3/7 activity was assessed in cells transfected with siDR5 and siDR4. Oleic acid treatment (200 μM) sensitised to TRAIL cytotoxicity; this was JNK dependent, and the sensitisation was unchanged in siDR4 transfected cells (p<0.05). In siDR5 transfected cells TRAIL sensitisation was abolished and predictably unchanged in the presence of JNK inhibitor, SP600125.
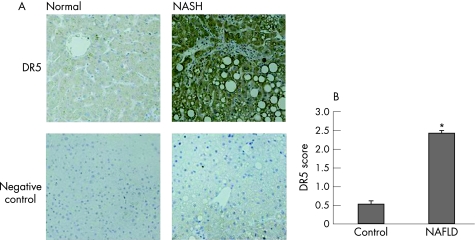
DR5 expression is enhanced in human liver in NASH
To establish a clinical correlation to our in vitro model we performed immunohistochemical analysis for DR5 expression in human liver from patients with NASH. Representative photomicrographs (fig 6A) and quantitative data (fig 6B) demonstrate that DR5 expression was enhanced in NASH.
Figure 6 DR5 expression in human liver samples from patients with NASH. (A) Immunohistochemistry was performed for DR5 expression using liver biopsy samples from patients with NASH. Normal liver biopsy samples were used as controls. Representative photomicrographs (original magnification ×200) demonstrate upregulation of DR5 expression in NASH compared to normal liver. (B) DR5 score was calculated based on the scoring system described in the methods section. Two or more random high power fields were scored per sample by two independent observers. Significant increase in DR5 expression in hepatocytes was observed in liver tissue from patients with NASH (p<0.001).
Discussion
In the current study, FFA directly sensitise hepatocytes to TRAIL induced apoptosis. FFA are elevated in the metabolic syndrome and correlate with fibrosis in patients with NAFLD.25 Furthermore, we have demonstrated that the saturated FFA, stearic acid and palmitic acid, are inherently cytotoxic and induce hepatocyte lipoapoptosis.17 Our earlier study also demonstrated the relative lack of toxicity of the monounsaturated fatty acids, oleic acid and palmitoleic acid. We have now demonstrated that both monounsaturated and saturated FFA sensitise hepatocytes to TRAIL cytotoxicity. Furthermore, polyunsaturated fatty acids fail to protect against oleic acid toxicity and sensitise to TRAIL.
Normal human hepatocytes, in conditions of health, are resistant to TRAIL cytotoxicity.26 In contrast, pathophysiological disturbances can sensitise hepatocytes to TRAIL mediated liver injury. In patients with hepatitis B virus infection, TRAIL expression is increased on CD8 and CD4 T cells and correlated with liver injury.27 In cell culture, HBV replication confers sensitivity to TRAIL induced apoptosis.28 Hepatitis C virus (HCV) core protein sensitises cultured hepatocyte derived cell lines to TRAIL induced apoptosis and TRAIL expression is increased in livers of patients with HCV.1 In experimental adenoviral hepatitis, TRAIL sensitisation occurs in an NF‐κB dependent manner.29 In immune mediated murine hepatitis, Listeria monocytogenes hepatotoxicity and concanavalin A induced hepatitis, liver injury is also TRAIL dependent.30 Furthermore, TRAIL has been shown to induce steatosis and apoptosis in an adenoviral model of hepatitis, while leading to steatosis alone in a model of alcohol induced liver disease.31 The current studies strongly implicate FFA induced steatosis as another pathophysiological perturbance sensitising hepatocytes to TRAIL cytotoxicity.
FFA activate the mitogen activated protein kinase (MAPK) cascades, especially c‐jun N‐terminal kinase (JNK).17 Indeed, FFA mediated lipoapoptosis in hepatocytes and apoptosis in several other cells types is JNK dependent. Two isoforms of JNK, JNK 1 and 2, are expressed in the liver,32,33,34 and a cytotoxic role has been described for each. JNK 1 mediates dietary obesity induced and methionine‐choline deficient dietary murine steatohepatitis.35,36 JNK 2 plays a predominant part in FFA induced, acetaminophen induced, and TNF‐α induced liver injury.17,37,38 JNK mediated direct oleic acid toxicity is minimal in hepatocytes, probably related to the magnitude of JNK activation. Oleic acid activation of JNK 1/2, although definite and sustained, is not of the magnitude observed with saturated FFA. Thus, the specific JNK isoform, duration of activation, and magnitude of activation are all key processes in determining the nature of the cellular toxicity effected by this stress kinase. Oleic acid does sensitise to TRAIL killing, both in steatotic Huh 7 cells and steatotic primary rodent hepatocytes in a JNK 1/2 dependent fashion. JNK isoform specific analysis was not performed in the current study; however, based on our earlier observations, we speculate that JNK 2 may be the specific isoform responsible for sensitising hepatocytes to TRAIL cytotoxicity by FFA.
TRAIL induces apoptosis via ligation with two cognate cell surface receptors, TRAIL‐R1 (DR4) and TRAIL‐R2 (DR5/TRICK‐2/KILLER).39,40 We examined expression of DR5 and DR4 and demonstrated that oleic acid induced TRAIL sensitisation is dependent upon DR5 upregulation. This upregulation of DR5 was JNK dependent as it was blocked by SP 600125. Furthermore, enhanced DR5 expression was observed in liver biopsy samples from patients with NASH. JNK activation leads to phosphorylation and activation of transcription factors, SP1 and AP1. We have previously demonstrated that SP1 mediates JNK dependent DR5 upregulation by toxic bile acids and this transcription factor probably contributes to DR5 upregulation by FFA as well.
In summary, these data implicate a pathogenic role for TRAIL in steatosis associated liver injury. FFA induced sensitisation to TRAIL mediated apoptosis was observed over a physiological range of FFA concentrations, strengthening the premise that TRAIL plays a significant part in steatotic liver injury. Enhanced DR5 expression was observed in liver samples from patients with NASH. The potential role for TRAIL in a wide variety of steatotic liver diseases requires further study and verification. However, if confirmed, these data have potential therapeutic implications (for example, caspase or JNK inhibitors) for the treatment of NAFLD.
Acknowledgements
The authors would like to acknowledge the excellent secretarial support of Ms Erin Nystuen‐Bungum.
Abbreviations
ALD - alcohol induced liver disease
DAPI - 4′,6‐diamidino‐2‐phenylindole
DISC - death inducing signalling complex
DR - death receptor
EPA - eicosapentaenoic acid
FFA - free fatty acid
HCV - hepatitis C virus
JNK - C‐jun N‐terminal kinase
MAPK - mitogen activated protein kinase
NAFLD - non‐alcoholic fatty liver disease
NASH - non‐alcoholic steatohepatitis
OA - oleic acid
PCR - polymerase chain reaction
PVDF - poly(vinylidene) fluoride
siRNA - small interfering RNA
TNF‐α - tumour necrosis factor alfa
TRAIL - tumour necrosis factor related apoptosis inducing ligand
Footnotes
Competing interests: None.
This work was supported by NIH grant DK 41876 and the Mayo and Palumbo.
References
- 1.Chou A H, Tsai H F, Wu Y Y.et al Hepatitis C virus core protein modulates TRAIL‐mediated apoptosis by enhancing Bid cleavage and activation of mitochondria apoptosis signaling pathway. J Immunol 20051742160–2166. [DOI] [PubMed] [Google Scholar]
- 2.Feldstein A E, Canbay A, Angulo P.et al Hepatocyte apoptosis and fas expression are prominent features of human nonalcoholic steatohepatitis. Gastroenterology 2003125437–443. [DOI] [PubMed] [Google Scholar]
- 3.Ribeiro P S, Cortez‐Pinto H, Sola S.et al Hepatocyte apoptosis, expression of death receptors, and activation of NF‐kappaB in the liver of nonalcoholic and alcoholic steatohepatitis patients. Am J Gastroenterol 2004991708–1717. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Danielsson A, Zern M A. Toxicity of hepatotoxins: new insights into mechanisms and therapy. Expert Opin Invest Drugs 19998585–607. [DOI] [PubMed] [Google Scholar]
- 5.Sanyal A J. Mechanisms of disease: pathogenesis of nonalcoholic fatty liver disease. Nat Clin Pract Gastroenterol Hepatol 2005246–53. [DOI] [PubMed] [Google Scholar]
- 6.Feldstein A E, Canbay A, Guicciardi M E.et al Diet associated hepatic steatosis sensitizes to Fas mediated liver injury in mice. J Hepatol 200339978–983. [DOI] [PubMed] [Google Scholar]
- 7.Unger R H. Minireview: weapons of lean body mass destruction: the role of ectopic lipids in the metabolic syndrome. Endocrinology 20031445159–5165. [DOI] [PubMed] [Google Scholar]
- 8.Listenberger L L, Schaffer J E. Mechanisms of lipoapoptosis: implications for human heart disease. Trends Cardiovasc Med 200212134–138. [DOI] [PubMed] [Google Scholar]
- 9.Wieckowska A, Zein N N, Yerian L M.et al In vivo assessment of liver cell apoptosis as a novel biomarker of disease severity in nonalcoholic fatty liver disease. Hepatology 20064427–33. [DOI] [PubMed] [Google Scholar]
- 10.Kimberley F C, Screaton G R. Following a TRAIL: update on a ligand and its five receptors. Cell Res 200414359–372. [DOI] [PubMed] [Google Scholar]
- 11.Wu G S, Burns T F, McDonald E R., 3rdet al Killer/DR5 is a DNA damage‐inducible p53‐regulated death receptor gene. Nat Genet 199717141–143. [DOI] [PubMed] [Google Scholar]
- 12.Sheikh M S, Burns T F, Huang Y.et al p53‐dependent and ‐independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor alpha. Cancer Res 1998581593–1598. [PubMed] [Google Scholar]
- 13.El‐Deiry W S. Regulation of p53 downstream genes. Semin Cancer Biol 19988345–357. [DOI] [PubMed] [Google Scholar]
- 14.Takimoto R, El‐Deiry W S. Wild‐type p53 transactivates the killer/DR5 gene through an intronic sequence‐specific DNA‐binding site. Oncogene 2000191735–1743. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi H, Grambihler A, Canbay A.et al Bile acids up‐regulate death receptor 5/TRAIL‐receptor 2 expression via a c‐Jun N‐terminal kinase‐dependent pathway involving Sp1. J Biol Chem 200427951–60. [DOI] [PubMed] [Google Scholar]
- 16.Spivey J R, Bronk S F, Gores G J. Glycochenodeoxycholate‐induced lethal hepatocellular injury in rat hepatocytes. Role of ATP depletion and cytosolic free calcium. J Clin Invest 19939217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malhi H, Bronk S F, Werneburg N W.et al Free fatty acids induce JNK‐dependent hepatocyte lipoapoptosis. J Biol Chem 200628112093–12101. [DOI] [PubMed] [Google Scholar]
- 18.Ren Y G, Wagner K W, Knee D A.et al Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol Biol Cell 2004155064–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou W, Liu X, Yue P.et al c‐Jun NH2‐terminal kinase‐mediated up‐regulation of death receptor 5 contributes to induction of apoptosis by the novel synthetic triterpenoid methyl‐2‐cyano‐3,12‐dioxooleana‐1, 9‐dien‐28‐oate in human lung cancer cells. Cancer Res 2004647570–7578. [DOI] [PubMed] [Google Scholar]
- 20.Harris C A, Johnson E M., Jr BH3‐only Bcl‐2 family members are coordinately regulated by the JNK pathway and require Bax to induce apoptosis in neurons. J Biol Chem 200127637754–37760. [DOI] [PubMed] [Google Scholar]
- 21.Kharbanda S, Saxena S, Yoshida K.et al Translocation of SAPK/JNK to mitochondria and interaction with Bcl‐x(L) in response to DNA damage. J Biol Chem 2000275322–327. [DOI] [PubMed] [Google Scholar]
- 22.Deng X, Xiao L, Lang W.et al Novel role for JNK as a stress‐activated Bcl2 kinase. J Biol Chem 200127623681–23688. [DOI] [PubMed] [Google Scholar]
- 23.Chang L, Kamata H, Solinas G.et al The E3 ubiquitin ligase itch couples JNK activation to TNFalpha‐induced cell death by inducing c‐FLIP(L) turnover. Cell 2006124601–613. [DOI] [PubMed] [Google Scholar]
- 24.Kim B J, Ryu S W, Song B J. JNK‐ and p38 kinase‐mediated phosphorylation of Bax leads to its activation and mitochondrial translocation and to apoptosis of human hepatoma HepG2 cells. J Biol Chem 200628121256–21265. [DOI] [PubMed] [Google Scholar]
- 25.Nehra V, Angulo P, Buchman A L.et al Nutritional and metabolic considerations in the etiology of nonalcoholic steatohepatitis. Dig Dis Sci 2001462347–2352. [DOI] [PubMed] [Google Scholar]
- 26.Ashkenazi A, Pai R C, Fong S.et al Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest 1999104155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han L H, Sun W S, Ma C H.et al Detection of soluble TRAIL in HBV infected patients and its clinical implications. World J Gastroenterol 200281077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssen H L, Higuchi H, Abdulkarim A.et al Hepatitis B virus enhances tumor necrosis factor‐related apoptosis‐inducing ligand (TRAIL) cytotoxicity by increasing TRAIL‐R1/death receptor 4 expression. J Hepatol 200339414–420. [DOI] [PubMed] [Google Scholar]
- 29.Mundt B, Kuhnel F, Zender L.et al Involvement of TRAIL and its receptors in viral hepatitis. Faseb J 20031794–96. [DOI] [PubMed] [Google Scholar]
- 30.Zheng S J, Wang P, Tsabary G.et al Critical roles of TRAIL in hepatic cell death and hepatic inflammation. J Clin Invest 200411358–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mundt B, Wirth T, Zender L.et al Tumour necrosis factor related apoptosis inducing ligand (TRAIL) induces hepatic steatosis in viral hepatitis and after alcohol intake. Gut 2005541590–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiao L, Han S I, Fang Y.et al Bile acid regulation of C/EBPbeta, CREB, and c‐Jun function, via the extracellular signal‐regulated kinase and c‐Jun NH2‐terminal kinase pathways, modulates the apoptotic response of hepatocytes. Mol Cell Biol 2003233052–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Schattenberg J M, Rigoli R M.et al Hepatocyte resistance to oxidative stress is dependent on protein kinase C‐mediated down‐regulation of c‐Jun/AP‐1. J Biol Chem 200427931089–31097. [DOI] [PubMed] [Google Scholar]
- 34.Bruckner S R, Estus S. JNK3 contributes to c‐jun induction and apoptosis in 4‐hydroxynonenal‐treated sympathetic neurons. J Neurosci Res 200270665–670. [DOI] [PubMed] [Google Scholar]
- 35.Hirosumi J, Tuncman G, Chang L.et al A central role for JNK in obesity and insulin resistance. Nature 2002420333–336. [DOI] [PubMed] [Google Scholar]
- 36.Schattenberg J M, Singh R, Wang Y.et al JNK1 but not JNK2 promotes the development of steatohepatitis in mice. Hepatology 200643163–172. [DOI] [PubMed] [Google Scholar]
- 37.Gunawan B K, Liu Z X, Han D.et al c‐Jun N‐terminal kinase plays a major role in murine acetaminophen hepatotoxicity. Gastroenterology 2006131165–178. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Singh R, Lefkowitch J H.et al Tumor necrosis factor‐induced toxic liver injury results from JNK2‐dependent activation of caspase‐8 and the mitochondrial death pathway. J Biol Chem 200628115258–15267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan G, O'Rourke K, Chinnaiyan A M.et al The receptor for the cytotoxic ligand TRAIL. Science 1997276111–113. [DOI] [PubMed] [Google Scholar]
- 40.Sheridan J P, Marsters S A, Pitti R M.et al Control of TRAIL‐induced apoptosis by a family of signaling and decoy receptors. Science 1997277818–821. [DOI] [PubMed] [Google Scholar]