Abstract
In the budding yeast, Saccharomyces cerevisiae, DNA damage or ribonucleotide depletion causes the transcriptional induction of an array of genes with known or putative roles in DNA repair. The ATM-like kinase, Mec1, and the serine/threonine protein kinases, Rad53 and Dun1, are required for this transcriptional response. In this paper, we provide evidence suggesting that another kinase, Hrr25, is also involved in the transcriptional response to DNA damage through its interaction with the transcription factor, Swi6. The Swi6 protein interacts with Swi4 to form the SBF complex and with Mbp1 to form the MBF complex. SBF and MBF are required for the G1-specific expression of G1 cyclins and genes required for S-phase. We show that Swi6 associates with and is phosphorylated by Hrr25 in vitro. We find that swi4, swi6, and hrr25 mutants, but not mbp1 mutants, are sensitive to hydroxyurea and the DNA-damaging agent methyl methanesulfonate and are defective in the transcriptional induction of a subset of DNA damage-inducible genes. Both the sensitivity of swi6 mutants to methyl methanesulfonate and hydroxyurea and the transcriptional defect of hrr25 mutants are rescued by overexpression of SWI4, implicating the SBF complex in the Hrr25/Swi6-dependent response to DNA damage.
Keywords: DNA repair, transcription, cell cycle
In budding yeast and other eukaryotic cells, exposure to DNA-damaging agents invokes both a checkpoint response and a repair response. Checkpoint pathways delay cell division to allow the repair of damaged DNA prior to proceeding through the cell cycle; in general, checkpoints serve to ensure the integrity of the genome (reviewed in refs. 1 and 2). Three checkpoint responses to DNA damage have been defined in yeast. First, a G2/M checkpoint acts to prevent mitosis in the presence of broken or damaged chromosomes (3–5). Second, an S-phase checkpoint prevents entry into mitosis in the presence of unreplicated DNA (4, 6–8). Finally, a G1 checkpoint acts to delay S-phase entry in response to DNA lesions incurred early in the cell cycle (6, 9).
During checkpoint-induced cell cycle arrest, some genes involved in DNA repair are transcriptionally induced. These genes include RNR1, RNR3 (large subunit of ribonucleotide reductase, ref. 10), RNR2 (small subunit of ribonucleotide reductase, ref. 11), RAD54 (recombinational repair, ref. 12), POL1 (DNA polymerase 1, ref. 13), and CDC9 (DNA ligase, refs. 14 and 15). The importance of the transcriptional activation of repair genes became evident with the isolation of a mutant, dun1 (16), which is defective for DNA damage-induced transcription and is hypersensitive to DNA damaging agents such as methyl methanesulfonate (MMS) and UV irradiation. Recent studies have delineated a pathway by which the damage signal is transduced to the checkpoint and transcriptional response apparatus. The kinases, Mec1 (4, 17) and Rad53 (6, 17), are required for both responses, whereas the Dun1 kinase, believed to act downstream of Mec1 and Rad53, is only required for the transcriptional induction response (16).
Mutations in another kinase, Hrr25, were identified as causing hypersensitivity to double-stranded DNA breaks induced by endonuclease expression, x-irradiation, or continuous exposure to MMS (18). Hrr25 is a casein kinase I (CKI) isoform that has dual-specificity protein kinase activity in vitro (19). In addition to having defects in DNA double-strand break repair, hrr25 mutant cells sporulate poorly, grow very slowly, and show a cell cycle delay in G2 (18). Kinase assays carried out with Hrr25 immunoprecipitates from yeast extracts show phosphorylation of Hrr25 itself as well as many coimmunoprecipitated proteins (20), suggesting that Hrr25 may have multiple substrates in vivo. The potential role of Hrr25 in the transcriptional or checkpoint response to DNA damage has not been investigated.
In this study, we show that Hrr25 interacts with and phosphorylates the Swi6 protein in vitro. Swi6 is a cell cycle-regulatory transcription factor that activates gene expression late in the G1 phase of the cell cycle at START (reviewed in refs. 21 and 22). Swi6 does not bind DNA specifically (23), but functions as a transcription factor through its interaction with different DNA-binding partners (23–27). Swi6 interacts with the Swi4 protein to form the SBF complex (SCB-binding factor), which activates transcription of some G1 cyclin genes and the HO gene through a cis-acting element called the SCB (SWI4/6 cell cycle box; consensus CACGAAA). When bound to the Mbp1 protein, Swi6 forms a second transcription factor, MBF (MCB-binding factor, also known as DSC1), which acts through a distinct upstream sequence element, the MCB [MluI cell cycle box, consensus ACGCGTNA (see refs. 21 and 22)]. The SCB and MCB elements are each sufficient to confer START-specific transcription on heterologous promoters (28–30). MCB elements are found in the promoters of many cell cycle-regulated genes involved in DNA replication such as CDC9, POL1, and the RNR genes (reviewed in ref. 31). In addition to being cell cycle regulated, the expression of some MCB-controlled genes is also induced by DNA damage (e.g., CDC9, POL1, RNR1, RAD51, RAD54, UNG1; refs. 10, 12, and 32–34). Although a role for MCB elements in controlling cell cycle-regulated transcription has been established, their role in DNA damage-induced transcription is unclear.
We find that hrr25 mutants are defective in the transcriptional induction of the RNR2 and RNR3 genes in response to ribonucleotide depletion caused by HU (hydroxyurea) treatment. In addition to defining a biochemical interaction between Hrr25 and Swi6, we show that, like hrr25 mutants, both swi6 and swi4 mutants are sensitive to DNA-damaging agents and defective in the damage-induced transcription of RNR2 and RNR3. Our observations lead us to propose a novel role for the SBF complex (Swi4/Swi6), through its interaction with the Hrr25 protein kinase, in the transcriptional response to DNA damage.
MATERIALS AND METHODS
Yeast Strains.
All yeast strains used for plating assays and Northern blot analysis were isogenic to strain JO34 (S288C derivative, MATa, ura3-52, lys2-801a, ade2-1070, his3Δ200, leu2-Δ1, SCB-LacZ) with the exceptions noted. The swi4Δ (swi4ΔHIS3, JO57-6B) and swi6Δ (swi6ΔHIS3, JO42-7C) strains have been described (35). The mbp1Δ (mbp1ΔTRP1) allele was constructed using PCR to replace the entire MBP1 coding sequence with the TRP1 gene. The mbp1ΔTRP1 disruption cassette was used to transform strain BY263 (trp1Δ63, GAL2+, otherwise isogenic to JO34). The hrr25Δ deletion strain was made by transformation of a diploid derivative of strain BY263 with an hrr25ΔURA3 disruption allele (18). The diploid was sporulated and meiotic progeny deleted for HRR25 recovered by tetrad dissection. For plating assays and Northern blot analyses, yeast strains were transformed with either vector Yep24 or with a high-copy SWI4 plasmid, pBA314. Other yeast strains are described in the relevant sections below.
Protein Affinity Chromatography and Microsequencing of p54.
Swi6 protein was expressed and purified essentially as described (23). The protein was coupled to AffiGel-10 resin (Bio-Rad) according to the manufacturer’s recommendations. The concentration of coupled protein on the resin was calculated to be 40 μM. For the preparation of yeast extracts, yeast cells (strain BJ2168, a ura3-52 leu2 trp1 prb1-122 pep4-3 prc1-407) were grown to mid-logarithmic phase in YPD medium (36). The cells were then lysed in lysis buffer [100 mM Tris, pH 8.0/100 mM NaCl/10 mM MgCl2/1 mM EDTA/10% glycerol/1 mM DTT/20 mM NaF/50 mM β-glycerophosphate/2 mM benzamidine/2 mg/ml aprotinin/2 mg/ml leupeptin/1 mg/ml pepstatin/1 mM phenylmethylsulfonyl fluoride (PMSF)] using agitation in the presence of glass beads. For small scale experiments, cells from 0.5–1 liter cultures were either vortexed with glass beads in 15-ml Sarstedt tubes or lysed in a Biospec mini-bead beater (Biospec Products, Bartlesville, OK). For preparative scale chromatography, 1.6 g of protein extract was prepared from 30 g of wet cell pellet by lysing with 10 × 20 sec bursts in the mid-sized chamber of a Biospec Beadbeater. After lysis, extracts were centrifuged at 100,000 × g for 1 hr and passed over Swi6 affinity columns. In a typical analytical experiment, approximately 4 mg of protein extract were loaded onto 20 μl micro-columns. For preparative chromatography, the clarified supernatant was loaded onto a 0.5-ml column of the Swi6-coupled resin that had been sequentially washed and equilibrated in SB buffer (20 mM Hepes, pH 7.2/10% glycerol/0.1 mM DTT/0.1 mM PMSF) with 1 M NaCl (SB-1000) and 100 mM NaCl (SB-100). The column was then washed in 10 column volumes of SB-100, and eluted with SB-1000. Analytical affinity chromatography experiments with hrr25 deletion strains were done with strain 7D (hrr25Δ, described in ref. 18) and an isogenic wild-type strain (W303). p54 purified by Swi6 protein affinity chromatography was digested with Achromobacter protease and the peptides prepared for microsequencing as described (37). Peptides were sequenced using an automated protein sequencer (Applied Biosystems models 470, 473, and 477).
Kinase Assays with Affinity Column Eluates.
Hrr25 kinase assays using column eluates were performed in a reaction buffer containing 15 mM Hepes (pH 7.5), 200 mM NaCl, 10 mM MgCl2, 1 mM DTT, 1 μM ATP, and 10 μCi of [γ-32P]ATP (DuPont) (1 Ci = 37 GBq). One-fiftieth of the eluate from a micro-column was added per reaction. Kinase reactions also contained 100 ng Swi6 protein (see above), myelin basic protein (Sigma), histone H1 (Boehringer Mannheim), or casein (Sigma), as indicated. Reactions were stopped after 15 min at 30°C with SDS sample buffer and boiled before electrophoresis on SDS/polyacrylamide gels. The gels were dried and exposed to XAR-5 film (Kodak).
HA–Hrr25 Immunoprecipitation Kinase Assay.
Immunoprecipitation kinase assays with hemagglutinin (HA)-tagged Hrr25 were done essentially as described (38), with modifications. Strain JO34 (wild type) was transformed with a plasmid containing HRR25 tagged at the C terminus with a single HA epitope tag (38) or vector pRS316 (39) and grown in selective medium (36) to maintain the plasmid. Cells were harvested in early logarithmic phase and lysed in IPK buffer (50 mM Tris, pH 7.5/1% Nonidet P-40/0.05% SDS/0.05% sodium deoxycholate/5 mM EDTA/5 mM DTT/100 mM NaCl with protease/phosphatase inhibitors as in lysis buffer). HA–Hrr25 was immunoprecipitated from the extracts with monoclonal antibody 12CA5, washed twice in IPK buffer, twice in IPK buffer with 1 M NaCl without inhibitors, and twice in kinase buffer (38). Where indicated, Swi6 and casein were added to 100 ng per kinase reaction.
Plating Assay for Sensitivity to DNA-Damaging Agents.
For viability assays, cells were grown to early logarithmic phase in minimal medium. The cells were harvested, washed twice, and resuspended in 100 mM KH2PO4 (pH 7.5). The cell suspension was then briefly sonicated and counted using a hemacytometer. Cells were plated at densities of 100, 500, 5,000, 50,000, and 500,000 (for hrr25 mutants) per plate onto SD minimal plates (36) containing either no drugs, 100 mM HU (Sigma), 200 mM HU, 0.01% MMS (Sigma), or 0.02% MMS. The plates were incubated at 30°C until full-size colonies appeared on the plates with the highest drug concentration. The percent viability (Table 1) was calculated as the percentage of viable colonies on drug-containing plates versus nondrug containing plates. In calculating viability, plates containing at least 200 colonies were used in the calculations where possible.
Table 1.
Survival of hrr25Δ, swi4Δ, swi6Δ, and mbp1Δ mutants in HU and MMS
| Strains | 100 mM HU | 150 mM HU | 0.01% MMS | 0.02% MMS |
|---|---|---|---|---|
| Wild type | 69 | 64 | 63 | 6 |
| swi4Δ | <0.5 | <0.5 | <0.5 | <0.5 |
| swi6Δ | <0.5 | <0.5 | 4 | <0.5 |
| mbp1Δ | 88 | 78 | 42 | 4 |
| swi6Δ 2μSWI4 | 53 | 36 | 22 | <0.5 |
| Wild type | 94 | 92 | 90 | 3 |
| hrr25Δ | <0.5 | <0.5 | <0.5 | <0.5 |
| hrr25Δ 2μSWI4 | <0.5 | <0.5 | <0.5 | <0.5 |
The strains above were grown to early logarithmic phase in minimal media, harvested, washed, and resuspended in phosphate buffer before plating onto minimal plates containing either HU or MMS in the concentrations shown. Numbers represent the percentage of colonies formed on drug plates relative to plating in the absence of drug (see Materials and Methods). The results of two independent experiments are shown (separated by the line space).
Northern Blot Analysis.
Cultures for RNA extraction were grown in minimal media to early logarithmic phase. An aliquot of cells was taken from the culture (0 time point) and HU was then added to a final concentration of 200 mM. For each time point, 15 ml of cells were harvested and total RNA was prepared and Northern blot analysis was performed as described (36). The blots were exposed to Kodak XAR-5 film for autoradiography and quantitated using a PhosphorImager (Molecular Dynamics) and imagequant 3.33 software. The RNR2, RNR3, UBI4, and ACT1 probes used for Northern blot analysis have been described (6, 10, 40).
RESULTS
Binding of Hrr25 to Swi6 Protein Affinity Columns.
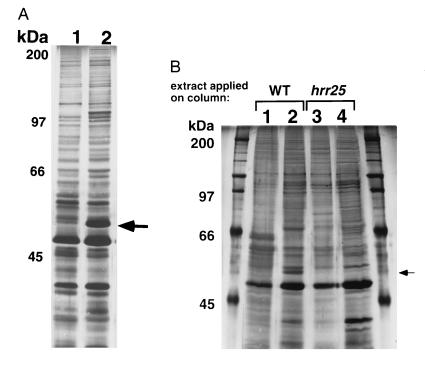
To identify proteins that may regulate Swi6 activity, we used protein affinity chromatography to look for proteins in crude yeast extracts that physically associate with Swi6. We compared the profile of proteins from yeast extracts that were retained on a Swi6 affinity resin to those proteins bound by the resin alone. We detected a 54-kDa protein (p54) that bound specifically to the Swi6 column but not to the control column (Fig. 1A, p54 indicated by arrow). Microsequencing of purified p54 yielded two peptide sequences that showed a perfect or near perfect match with the published amino acid sequence of Hrr25 (18), a dual-specificity CKI isoform (peptide 1, IGSGSFGDIYHGTNLISGEEVAI, amino acids 15–37; peptide 2, DLNANSNAAS?K, amino acids 312–323). We confirmed that p54 was indeed Hrr25 in two ways. First, p54 was absent in eluates from a Swi6 affinity column loaded with extracts from an hrr25Δ strain (Fig. 1B). Second, antibodies raised against Hrr25 (20) recognized a 54-kDa band in Swi6 column eluates but not in control column eluates (data not shown). We conclude that Swi6 and Hrr25 form a specific protein complex in vitro.
Figure 1.
Binding of p54(Hrr25) to Swi6 protein affinity columns. (A) Yeast extracts from BJ2168 were loaded onto either a control column (no coupled protein, lane 1) or a Swi6-coupled column (lane 2) and eluted with 1 M NaCl. (B) Extracts made from either a wild-type (lanes 1 and 2) or an hrr25Δ strain (lanes 3 and 4) were loaded onto either a control column (lanes 1 and 3) or Swi6-coupled columns (lanes 2 and 4) and eluted with 1 M NaCl. Protein molecular weight markers are indicated to the left of the gel photographs and the p54 (Hrr25) is indicated by the arrow on the right.
Swi6 Is a Substrate for the Hrr25 Protein Kinase in Vitro.
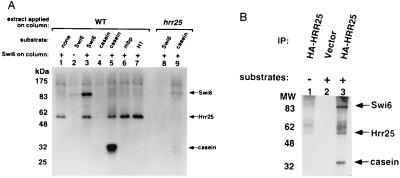
Although HRR25 shows strong homology to known CKI isoforms and has been shown to have casein kinase activity in vitro (20), biologically relevant substrates have not been identified. We tested whether the interaction between Hrr25 and Swi6 might reflect the fact that Swi6 is a substrate for the Hrr25 kinase by using Swi6 affinity column eluates as a source of the Hrr25 protein for in vitro kinase assays. A kinase activity that was specific to the Swi6 column eluates could efficiently phosphorylate recombinant Swi6 protein in vitro (Fig. 2A, lane 3). The Swi6 kinase activity was absent in eluates from a resin-only control column and absent in column eluates derived from hrr25Δ extracts (Fig. 2A, lanes 2 and 8). The kinase activity seen in the Swi6 column eluates phosphorylated casein but not myelin basic protein or histone H1 (Fig. 2A, lanes 5–7). Phosphorylation of a 54-kDa protein that was absent in hrr25Δ extracts was also seen in the Swi6 column eluates and most likely corresponds to autophosphorylation of Hrr25 (20) (Fig. 2A, lanes 1, 3, and 5–7). Furthermore, both the phosphorylation of casein and Swi6 were inhibited by CKI-7, a specific inhibitor of human CKI (data not shown, ref. 41). These observations show that Swi6 is phosphorylated by a kinase with the expected properties of Hrr25. Swi6 was also phosphorylated by Hrr25 kinase immunoprecipitated from yeast extracts with a HA tag (Fig. 2B). These data demonstrate that Swi6 is a substrate for the Hrr25 protein kinase in vitro.
Figure 2.
In vitro phosphorylation of Swi6 by Hrr25. (A) Kinase assays were done on eluates from Swi6 affinity columns or from control columns (no coupled protein). The presence of Swi6 on the column resin is indicated by a “+” above the lane, whereas “−” denotes the control column with no coupled protein. The columns were loaded with extracts from either a wild-type (lanes 1–7) or hrr25Δ strain (lanes 8 and 9) as indicated above the figure (“extract applied on column”). Exogenous substrate (100 ng) was added to the kinase assays as indicated above the lanes. mbp, myelin basic protein; H1, histone H1. (B) Kinase assays were done with 12CA5 (anti-HA) immunoprecipitates from yeast cells expressing an HA–Hrr25 fusion protein (lanes 1 and 3) or cells transformed with an empty vector (lane 2). In lanes 2 and 3, 100 ng of casein and Swi6 were added to the kinase reaction as indicated by a “+.” The position of migration of phosphorylated Swi6, Hrr25, and casein are indicated on the right. Molecular weight markers are shown on the left.
Both SBF (swi4Δ/swi6Δ) and hrr25Δ Mutants Show Sensitivity to the DNA-Damaging Agent, MMS, and the DNA Synthesis Inhibitor, HU.
Because deletion of HRR25 is known to cause sensitivity to some DNA damaging agents (18), we determined the requirement for Swi6 in the DNA damage response. SWI6 deletion strains have previously been shown to exhibit reduced viability after a transient exposure to MMS (12). We assayed the sensitivity of swi6, swi4, mbp1, and hrr25 mutants to continuous exposure to MMS or HU in a plating assay (Table 1). MMS is a DNA alkylating agent that is known to cause DNA strand breaks (for example, see ref. 42), whereas HU inhibits DNA synthesis through inhibition of ribonucleotide reductases (43). Both agents are known to elicit transcriptional induction of DNA repair genes, most notably the RNR genes (for review, see ref. 44). We found that both the swi6 and hrr25 deletion strains showed a marked decrease in viability versus wild type when plated onto media containing either MMS or HU (Table 1). In addition, we found that swi4 but not mbp1 mutants were sensitive to growth in the presence of MMS and HU. The viability of an mbp1 mutant was previously observed to be unaffected by a transient exposure to MMS (12). The drug sensitivity of the swi4 and swi6 deletion strains is not a secondary consequence of an unusual cell cycle distribution of cells in the culture since, in our strain background, neither mutant showed an abnormal flourescence-activated cell sorter profile when grown in minimal medium (data not shown). As described earlier, Swi6 binds to DNA through either of two DNA-binding subunits forming either the SBF complex with Swi4 or the MBF complex with Mbp1. Our data suggest that the SBF but not the MBF complex is involved in the sensitivity of swi6 deletion strains to MMS and HU.
Although Swi6 is normally essential for transcriptional activation through both the SCB and the MCB elements, overproduction of Swi4 can bypass the Swi6 requirement for activation of an SCB reporter (45). Since our data implicated the SBF complex (Swi4/Swi6) in the sensitivity of an swi6 mutant to MMS and HU, we tested the ability of Swi4 overproduction to rescue the drug sensitivity of an swi6 mutant. We found that a high-copy SWI4 plasmid partially rescued the sensitivity of a swi6 mutant to plating in the presence of HU or MMS (Table 1). Swi4 overproduction failed to suppress the MMS and HU sensitivity of an hrr25 mutant in this assay (Table 1).
The MMS and HU Sensitivity of hrr25Δ and SBF-Deficient Strains May Reflect a Defect in the Transcriptional Induction of DNA Repair Genes.
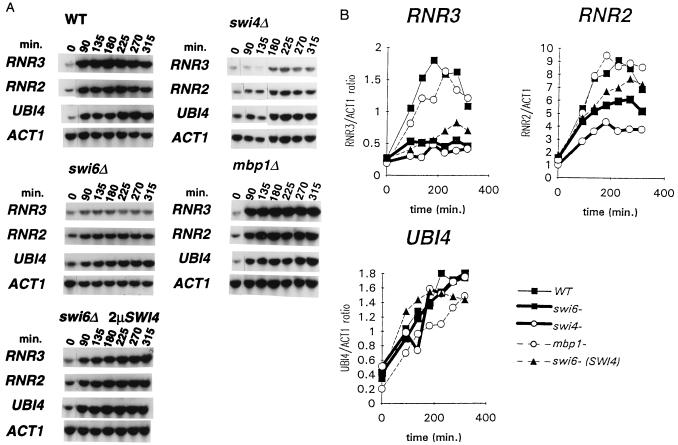
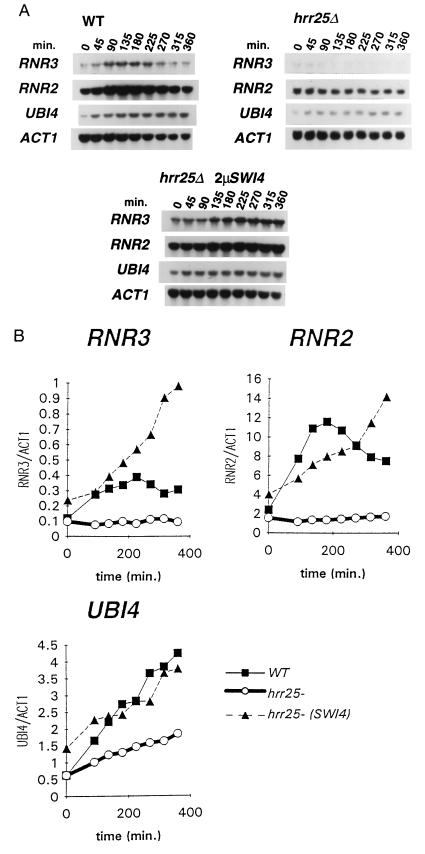
Two classes of HU-sensitive mutants have been characterized: (i) mutants defective in the S-phase checkpoint that are unable to inhibit mitotic division in the presence of unreplicated DNA (6, 7) and (ii) mutants defective in the transcriptional induction of RNR gene expression after HU depletion (7, 16). Because neither swi4, swi6, nor hrr25 cells appeared to show a defective S-phase checkpoint response (data not shown; S. Elledge, personal communication; see Discussion), we used Northern blot analysis to test the induction of RNR gene expression in swi4, swi6, hrr25, mbp1, and wild-type cells upon treatment with HU. Three genes encoding ribonucleotide reductase have been identified in yeast: RNR1 and RNR3 encode the large subunit of the enzyme, whereas RNR2 encodes the small subunit (46). RNR3 transcription was induced 4- to 6-fold following HU treatment of the wild-type strain (Figs. 3 and 4). In contrast, RNR3 transcription was not significantly induced in strains deleted for swi4, swi6, or hrr25 (Figs. 3 and 4). SWI4 and SWI6 were also required for maximal RNR3 transcription in response to MMS treatment (data not shown). hrr25 mutants also failed to induce RNR2 expression (Fig. 4 A and B), whereas the induction of RNR2 was less dramatically affected in the swi4 and swi6 mutant strains (Fig. 3 A and B). The RNR1 gene was not transcriptionally induced in response to HU in our strain background (data not shown). Consistent with the relative resistance of the mbp1 mutant to MMS and HU (Table 1), RNR2 and RNR3 expression was comparable to wild type following HU treatment of an mbp1 deletion strain (Fig. 3 A and B). The failure to induce RNR3 transcription was not due to low viability or slow response of the mutant cells because UBI4 transcription was induced normally by HU treatment in swi4 and swi6 mutants (Fig. 3) and also in hrr25 cells (Fig. 4) although not to wild-type levels. UBI4 encodes polyubiquitin and is transcriptionally induced by a variety of physiological stresses (47) through regulatory mechanisms that appear distinct from those controlling damage induction of RNR genes (16, 17).
Figure 3.
Transcriptional induction of genes after treatment with 200 mM HU in swi4Δ, swi6Δ, and mbp1Δ strains. (A) Yeast strains (indicated at the top of each panel) were grown in minimal media to early logarithmic phase and a 0 time point was taken before HU was added to 200 mM. Aliquots of cells were taken after HU addition at the time points specified (in minutes). Total RNA was extracted and Northern hybridization analysis was performed with the probes indicated to the left of each panel. All time points shown for each probe are from the same exposure of the Northern blot. Blots were sequentially hybridized with the different probes. (B) PhosphorImager analysis of the Northern blots shown in A. The RNA levels of RNR2, RNR3, and UBI4 relative to ACT1 were determined and plotted versus time after HU addition. ACT1 encodes actin and served as a loading control. swi6 (SWI4) indicates an swi6 deletion strain transformed with a high-copy SWI4 plasmid.
Figure 4.
Transcriptional induction of RNR2, RNR3, and UBI4 after treatment with 200 mM HU in an hrr25Δ strain. (A) RNA was isolated from the strains indicated at the top of each panel after treatment with HU as described in the legend to Fig. 3. Total RNA was extracted and Northern hybridization analysis was performed with the probes indicated to the left. (B) PhosphorImager analysis of the Northern blots shown in A. The RNA levels of RNR2, RNR3, and UBI4 relative to ACT1 were determined and plotted versus time after HU addition.
Since overproduction of SWI4 rescued the inviability of swi6 mutants in the presence of HU, we assayed RNR2 and RNR3 gene expression in swi6 mutants transformed with a high-copy SWI4 plasmid (Fig. 3). Overproduction of SWI4 in the swi6 strain partially restored inducibility of both RNR2 and RNR3, although not to wild-type levels. It is possible that the overexpression of SWI4 rescued the inviability of the swi6 mutant in HU by causing a prolonged transcriptional response over a longer period of time than we assayed in our Northern blot analysis. In an hrr25 strain, ectopic expression of SWI4 almost completely rescued RNR2 induction, whereas RNR3 expression was increased relative to the wild-type strain after 200 min in the presence of HU (Fig. 4 A and B). Swi4 overproduction in the hrr25 mutant also increased UBI4 transcription in untreated cells (Fig. 4B, 0 time point), but the induction of UBI4 over time was similar to that seen in the hrr25 mutant (Fig. 4B).
DISCUSSION
We have made two sets of observations that suggest a functional interaction between the Hrr25 protein kinase and the Swi6 transcription factor. First, Hrr25 interacts with and phosphorylates Swi6 in vitro and second, hrr25 and swi6 mutants share a defect in the induction of RNR gene expression in response to HU in vivo. In addition, we find that swi4 mutants, but not mbp1 mutants, are defective in the induction of RNR genes in response to HU. Overproduction of SWI4 rescues the HU and MMS sensitivity of swi6 mutants and alleviates the transcriptional induction defect of both swi6 and hrr25 mutants (Table 1; Figs. 3 and 4). Thus, although our data suggest that Swi6 is a target of Hrr25, only the SBF complex and not the MBF complex appears to be involved in the RNR transcriptional response.
Two observations support a role for Hrr25 and SBF that is specific to the induction of gene expression in response to DNA damage and not simply a role in providing a basal transcriptional activity. First, we find that uninduced levels of RNR2 and RNR3 expression are not affected by mutation of SWI4, SWI6, or HRR25 (Figs. 3 and 4). Second, we find that the activity of SCB::lacZ and MCB::lacZ reporter genes in untreated cells is not reduced by mutation of HRR25, demonstrating that Swi6 is functional in an hrr25 mutant in the absence of DNA damage (Y.H., unpublished data). We infer that Hrr25 and SBF are specifically involved in mediating the transcriptional induction of RNR2/3 in response to HU and are not providing a basal activity that is modulated by another damage-responsive factor.
Previous studies have shown that RNR2 inducibility is not blocked by protein synthesis inhibitors suggesting that a preexisting factor is likely responsible for DNA damage-induced transcription (48, 49). Our finding that SBF is involved in upregulating RNR gene expression in response to HU suggests that SBF may be one such factor acting directly on the RNR promoters. The RNR3 gene contains three matches to the SCB consensus within 350 bp upstream of the ATG. Likewise, the RNR2 gene contains one near match in its upstream sequences (CTCGAAA). Both promoters also contain matches, or near matches, to the MCB consensus element (46). Although several observations suggest that, at least in certain promoter contexts, the principle binding sites for SBF are SCB sequences, other data suggest that SBF may also act through MCB elements (reviewed in ref. 21).
Alternatively, SBF may be acting through another, unidentified element to mediate the DNA damage response. Dissection of the SWI4 and CLN2 promoters has provided evidence that both Swi4 and Swi6 may act through upstream sequences distinct from SCB or MCB elements (50–52). In support of an alternative element mediating the SBF-dependent DNA damage response, we and others do not see elevated expression from SCB nor MCB::lacZ reporter genes after DNA damage (Y.H., unpublished data; MCB reporter also cited in refs. 12 and 53). Moreover, DNA damage-induced expression of CDC9 was still seen when MCB elements were deleted from its promoter (12) and DNA damage-inducibility of RNR2 is maintained in a promoter deletion mutant lacking the putative SCB sequence (49). The Hrr25 and SBF dependence of RNR promoter mutants lacking SCB and MCB sequences has yet to be assessed.
Promoter analyses of damage-inducible genes have uncovered numerous DNA elements and DNA-binding proteins that are involved in the transcriptional response to DNA damage (reviewed in ref. 44). Also, genetic screens have identified mutants defective in regulation of damage-inducible promoters (16, 54). It will be interesting to assess the relationship between these mutants or any of the unidentified proteins and SWI4, SWI6, or HRR25. It is possible that different elements in the complex promoters of damage-inducible genes may mediate the response to different types of DNA damage. For example, although RNR2 and RNR3 are induced in response to UV or UV-mimetic agents (10, 48, 49), neither hrr25, swi4, or swi6 mutants display any UV sensitivity (refs. 12 and 18; and M.H., unpublished data). This implies that either (i) RNR induction is not required for maximum viability after UV irradiation or (ii) UV irradiation induces transcription of the RNR genes through a pathway independent of Hrr25/SBF. The cellular sensing apparatus for UV irradiation is believed to be different from the sensors for DNA damage and ribonucleotide depletion (17, 55). We are currently testing the UV inducibility of the RNR genes in swi4, swi6, and hrr25 mutants.
We found that Swi4 overproduction rescued the drug sensitivity of the swi6 mutant and induced a modest increase in RNR2/3 gene expression in response to HU (Table 1, Fig. 3). Assays of spindle elongation following HU treatment of an swi4 mutant and viability assays following HU treatment of swi4, swi6, and hrr25 mutants suggested that the S-phase checkpoint is intact in these mutants (data not shown; Steve Elledge, personal communication). However, swi4, swi6, and hrr25 mutants do show defects in RNR2/3 transcriptional induction (Figs. 3 and 4). Therefore, we suggest that overproduction of SWI4 rescues the inviability of the swi6 mutant on HU through (i) its effect on RNR transcription, which may become significant over a prolonged period or (ii) the induction of other genes that may be required to survive HU treatment, whose expression can be stimulated directly or indirectly by SWI4. SWI4 may be important for survival or recovery after prolonged exposure to DNA-damaging agents since the viability of an swi4ts mbp1 mutant was not affected by a transient exposure to MMS (12). Although overexpression of SWI4 only partially rescued the transcriptional defect of the swi6 mutant, it completely rescued the lack of RNR2/3 inducibility in the hrr25 mutant strain (Fig. 4). This observation, together with our finding that overexpression of SWI4 rescued the MMS and HU sensitivity of an swi6 mutant, strongly implicates SBF in the transcriptional induction of RNR genes in response to DNA damage.
Ectopic SWI4 also increased the basal level of UBI4 expression in the hrr25 mutant but had little effect on inducibility consistent with the fact that neither the swi6 nor swi4 mutant showed any defect in UBI4 induction (Fig. 3). In contrast to the swi6 mutant, Swi4 overproduction failed to rescue the inviability of the hrr25 mutant on HU plates (Table 1). The pleiotropic phenotypes of an hrr25 mutant are consistent with the possibility that, in addition to regulating the induction of RNR gene expression, HRR25 is involved in the expression of other stress response genes important for surviving in suboptimal conditions.
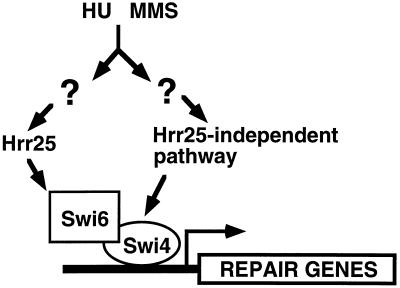
Together, our data support a model whereby the phosphorylation of Swi6 by Hrr25 promotes SBF-dependent induction of DNA repair genes in response to DNA damage or HU-induced depletion of ribonucleotides. Modification of Swi6 by Hrr25 may be necessary to allow SBF to function at times in the cell cycle when it is not normally active. For example, Swi6 protein is present throughout the cell cycle but is largely cytoplasmic from G2 until late mitosis when it enters the nucleus (56, 57). We have found that Swi6 becomes predominantly nuclear following treatment of cells with MMS (Y.H., unpublished data). Phosphorylation of Swi6 by Hrr25 may promote redistribution of Swi6 to the nucleus in response to DNA damage. In addition, our observations suggest that Swi4 may be regulated in response to HU by an HRR25-independent mechanism. First, the transcription induction defect of an hrr25 mutant is completely bypassed by SWI4 overexpression. Second, SWI4 is not phosphorylated by Hrr25 in vitro and it is not required for the association of Swi6 and Hrr25 (Y.H., unpublished data). Taken together, these data suggest that SBF may receive DNA damage signals through both subunits: an Hrr25–Swi6 pathway and an Hrr25-independent pathway through Swi4 (Fig. 5). As discussed earlier, the Mec1, Rad53, and Dun1 kinases are all required for RNR gene induction in response to DNA damage (4, 6, 16, 17). It will be of interest to delineate the relationship between the Hrr25–Swi6, Swi4, and Mec1 pathways in damage-inducible gene expression.
Figure 5.
Model for the role of Hrr25 and SBF (Swi4/Swi6) in the transcriptional response to DNA damage. The Hrr25 protein kinase is proposed to phosphorylate Swi6 in response to DNA damage. An Hrr25-independent pathway may also function through Swi4. These two pathways serve to activate SBF to promote the transcriptional induction of repair genes. See text for details.
Acknowledgments
We thank Barbara Funnell, Lea Harrington, and Mike Tyers for comments on the manuscript. We thank Pascale Rousseau for making the mbp1Δ strain used in this study, Angela Logan for her part in the drug-sensitivity assays, and Stephen Elledge for providing plasmids for the RNR2, RNR1, and UBI4 probes. This work was supported by the Medical Research Council of Canada. B.A. was a scholar of the same agency, S.M. was a Fellow of the National Sciences and Engineering Research Council of Canada, and Y.H. was supported in part by an Ontario Graduate Scholarship.
Footnotes
Abbreviations: MMS, methyl methanesulfonate; HU, hydroxyurea; CKI, casein kinase I; HA, hemagglutinin.
References
- 1.Hartwell L H, Kastan M B. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 2.Weinert T A, Lydall D. Semin Cancer Biol. 1993;4:129–140. [PubMed] [Google Scholar]
- 3.Weinert T A, Hartwell L H. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 4.Weinert T A, Kiser G L, Hartwell L H. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 5.Murray A. Curr Opin Genet Dev. 1993;5:5–11. doi: 10.1016/s0959-437x(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 6.Allen J B, Zhou Z, Side W, Friedberg E C, Elledge S J. Genes Dev. 1994;8:2416–2428. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 7.Navas T A, Zhou Z, Elledge S. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 8.Paulovich A G, Hartwell L H. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 9.Siede W, Friedberg A S, Friedberg E C. Proc Natl Acad Sci USA. 1993;90:7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elledge S J, Davis R W. Genes Dev. 1990;4:740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- 11.Elledge S J, Davis R W. Mol Cell Biol. 1987;7:2783–2793. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston L H, Johnson A L. Nucleic Acids Res. 1995;23:2147–2142. doi: 10.1093/nar/23.12.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston L H, White J H M, Johnson A L, Lucchini G, Plevani P. Nucleic Acids Res. 1987;15:5017. doi: 10.1093/nar/15.13.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson T A, Prakash L, Prakash S, Osley M A, Reed S I. Mol Cell Biol. 1985;5:226–235. doi: 10.1128/mcb.5.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker D G, White J M, Johnston L H. Nucleic Acids Res. 1985;13:8223–8237. doi: 10.1093/nar/13.23.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou A, Elledge S J. Cell. 1993;74:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]
- 17.Kiser G, Weinert T A. Mol Biol Cell. 1996;7:703–718. doi: 10.1091/mbc.7.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoekstra M F, Liskay R M, Ou A C, DeMaggio A J, Burbee D G, Heffron F. Science. 1991;253:1031–1034. doi: 10.1126/science.1887218. [DOI] [PubMed] [Google Scholar]
- 19.Hoekstra M F, Dhillon N, Carmel G, DeMaggio A J, Lindberg R A, Hunter T, Kuret J. Mol Biol Cell. 1994;5:877–886. doi: 10.1091/mbc.5.8.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeMaggio A J, Lindberg R A, Hunter T, Hoekstra M F. Proc Natl Acad Sci USA. 1992;89:7008–7012. doi: 10.1073/pnas.89.15.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breeden L. Curr Top Microbiol Immunol. 1995;208:95–127. doi: 10.1007/978-3-642-79910-5_5. [DOI] [PubMed] [Google Scholar]
- 22.Koch C, Nasmyth K. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 23.Sidorova J, Breeden L. Mol Cell Biol. 1993;13:1069–1077. doi: 10.1128/mcb.13.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews B J, Moore L. Proc Natl Acad Sci USA. 1992;89:11852–11856. doi: 10.1073/pnas.89.24.11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Primig M, Sockanathan S, Auer H, Nasmyth K. Nature (London) 1992;358:593–597. doi: 10.1038/358593a0. [DOI] [PubMed] [Google Scholar]
- 26.Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- 27.Siegmund R F, Nasmyth K. Mol Cell Biol. 1996;16:2647–2655. doi: 10.1128/mcb.16.6.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breeden L, Nasmyth K. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 29.McIntosh E M, Atkinson T, Storms R K, Smith M. Mol Cell Biol. 1991;11:329–337. doi: 10.1128/mcb.11.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowndes N F, Johnson A L, Johnston L H. Nature (London) 1991;350:247–250. doi: 10.1038/350247a0. [DOI] [PubMed] [Google Scholar]
- 31.Toyn J H, Toone W M, Morgan B A, Johnston L H. Trends Biochem Sci. 1995;20:70–73. doi: 10.1016/s0968-0004(00)88960-4. [DOI] [PubMed] [Google Scholar]
- 32.Johnson A L, Barker D G, Johnston L H. Curr Genet. 1986;11:107–112. doi: 10.1007/BF00378201. [DOI] [PubMed] [Google Scholar]
- 33.Johnston L H, White J H M, Johnson A L, Lucchini G, Plevani P. Nucleic Acids Res. 1987;15:5017–5030. doi: 10.1093/nar/15.13.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basile G, Aker M, Mortimer R K. Mol Cell Biol. 1992;12:3235–3246. doi: 10.1128/mcb.12.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogas J, Andrews B J, Herskowitz I. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 36.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 37.Collins K, Kobayashi R, Greider C W. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 38.Tyers M, Tokiwa G, Nash R, Futcher B. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski R S, Hieter P. Genetics. 1989;12:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrington L A, Andrews B J. Nucleic Acids Res. 1996;24:558–565. doi: 10.1093/nar/24.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chijiwa T, Hagiwara M, Hidaka H. J Biol Chem. 1989;264:4924–4927. [PubMed] [Google Scholar]
- 42.Dhillon N, Hoekstra M. EMBO J. 1994;13:2777–2788. doi: 10.1002/j.1460-2075.1994.tb06571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harder J, Follmann H. Free Radical Res Commun. 1990;10:281–286. doi: 10.3109/10715769009149896. [DOI] [PubMed] [Google Scholar]
- 44.Bachant J B, Elledge S J. In: DNA Damage and Repair: Biochemistry, Genetics and Cell Biology. Nickoloff J A, Hoekstra M, editors. Clifton, NJ: Humana; 1996. in press. [Google Scholar]
- 45.Breeden L, Nasmyth K. Nature (London) 1987;329:651–654. doi: 10.1038/329651a0. [DOI] [PubMed] [Google Scholar]
- 46.Elledge S H, Zhou Z, Allen J B. Trends Biochem Sci. 1992;17:119–123. doi: 10.1016/0968-0004(92)90249-9. [DOI] [PubMed] [Google Scholar]
- 47.Treger J M, Heichman K A, McEntee K. Mol Cell Biol. 1988;8:1132–1136. doi: 10.1128/mcb.8.3.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elledge S J, Davis R W. Mol Cell Biol. 1989;9:4932–4940. doi: 10.1128/mcb.9.11.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurd H K, Roberts J W. Mol Cell Biol. 1989;9:5359–5371. doi: 10.1128/mcb.9.12.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cross F, Hoek M, McKinney J D, Tinkelenberg A H. Mol Cell Biol. 1994;14:4779–4787. doi: 10.1128/mcb.14.7.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stuart D, Wittenberg C. Mol Cell Biol. 1994;14:4788–4801. doi: 10.1128/mcb.14.7.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foster R, Mikesell G E, Breeden L. Mol Cell Biol. 1993;13:3972–3801. doi: 10.1128/mcb.13.6.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elledge S J, Zhou Z, Allen J B, Navas T A. BioEssays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- 54.Zhou Z, Elledge S J. Genetics. 1992;131:851–866. doi: 10.1093/genetics/131.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Devary Y, Rosette J, DiDonata J, Karin M. Science. 1993;261:1442–1445. doi: 10.1126/science.8367725. [DOI] [PubMed] [Google Scholar]
- 56.Sidorova J M, Mikesell G E, Breeden L. Mol Biol Cell. 1995;6:1641–1658. doi: 10.1091/mbc.6.12.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taba M R, Muroff I, Lydall G, Tebb G, Nasmyth K. Genes Dev. 1991;5:2000–2013. doi: 10.1101/gad.5.11.2000. [DOI] [PubMed] [Google Scholar]