Abstract
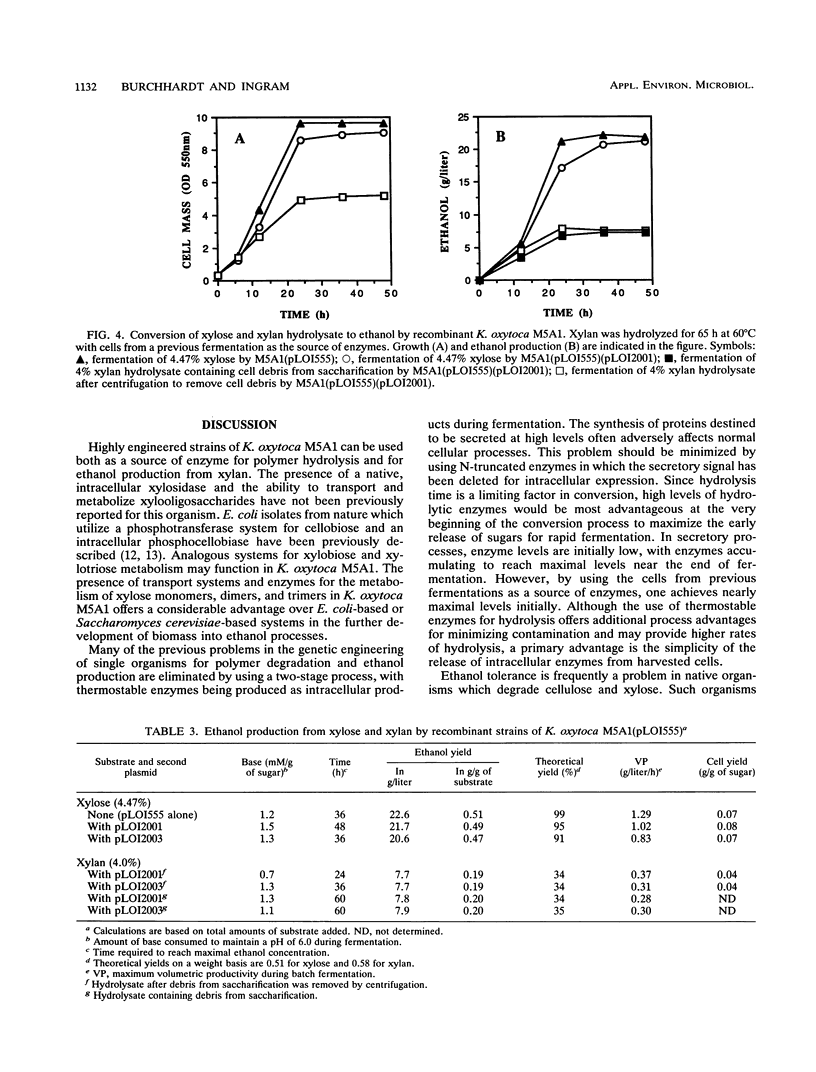
A two-stage process was evaluated for the fermentation of polymeric feedstocks to ethanol by a single, genetically engineered microorganism. The truncated xylanase gene (xynZ) from the thermophilic bacterium Clostridium thermocellum was fused with the N terminus of lacZ to eliminate secretory signals. This hybrid gene was expressed at high levels in ethanologenic strains of Escherichia coli KO11 and Klebsiella oxytoca M5A1(pLOI555). Large amounts of xylanase (25 to 93 mU/mg of cell protein) accumulated as intracellular products during ethanol production. Cells containing xylanase were harvested at the end of fermentation and added to a xylan solution at 60 degrees C, thereby releasing xylanase for saccharification. After cooling, the hydrolysate was fermented to ethanol with the same organism (30 degrees C), thereby replenishing the supply of xylanase for a subsequent saccharification. Recombinant E. coli metabolized only xylose, while recombinant K. oxytoca M5A1 metabolized xylose, xylobiose, and xylotriose but not xylotetrose. Derivatives of this latter organism produced large amounts of intracellular xylosidase, and the organism is presumed to transport both xylobiose and xylotriose for intracellular hydrolysis. By using recombinant M5A1, approximately 34% of the maximal theoretical yield of ethanol was obtained from xylan by this two-stage process. The yield appeared to be limited by the digestibility of commercial xylan rather than by a lack of sufficient xylanase or by ethanol toxicity. In general form, this two-stage process, which uses a single, genetically engineered microorganism, should be applicable for the production of useful chemicals from a wide range of biomass polymers.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bounias M. N-(1-naphthyl)ethylenediamine dihydrochloride as a new reagent for nanomole quantification of sugars on thin-layer plates by a mathematical calibration process. Anal Biochem. 1980 Aug;106(2):291–295. doi: 10.1016/0003-2697(80)90523-0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Béguin P. Molecular biology of cellulose degradation. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- Dombek K. M., Ingram L. O. Determination of the intracellular concentration of ethanol in Saccharomyces cerevisiae during fermentation. Appl Environ Microbiol. 1986 Jan;51(1):197–200. doi: 10.1128/aem.51.1.197-200.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grépinet O., Chebrou M. C., Béguin P. Nucleotide sequence and deletion analysis of the xylanase gene (xynZ) of Clostridium thermocellum. J Bacteriol. 1988 Oct;170(10):4582–4588. doi: 10.1128/jb.170.10.4582-4588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grépinet O., Chebrou M. C., Béguin P. Purification of Clostridium thermocellum xylanase Z expressed in Escherichia coli and identification of the corresponding product in the culture medium of C. thermocellum. J Bacteriol. 1988 Oct;170(10):4576–4581. doi: 10.1128/jb.170.10.4576-4581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G., Faunce W., 3rd Functional genes for cellobiose utilization in natural isolates of Escherichia coli. J Bacteriol. 1987 Jun;169(6):2713–2717. doi: 10.1128/jb.169.6.2713-2717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kricker M., Hall B. G. Biochemical genetics of the cryptic gene system for cellobiose utilization in Escherichia coli K12. Genetics. 1987 Mar;115(3):419–429. doi: 10.1093/genetics/115.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd L. R., Cushman J. H., Nichols R. J., Wyman C. E. Fuel ethanol from cellulosic biomass. Science. 1991 Mar 15;251(4999):1318–1323. doi: 10.1126/science.251.4999.1318. [DOI] [PubMed] [Google Scholar]
- Ohta K., Beall D. S., Mejia J. P., Shanmugam K. T., Ingram L. O. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl Environ Microbiol. 1991 Apr;57(4):893–900. doi: 10.1128/aem.57.4.893-900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Beall D. S., Mejia J. P., Shanmugam K. T., Ingram L. O. Metabolic engineering of Klebsiella oxytoca M5A1 for ethanol production from xylose and glucose. Appl Environ Microbiol. 1991 Oct;57(10):2810–2815. doi: 10.1128/aem.57.10.2810-2815.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailliez P., Girard H., Longin R., Beguin P., Millet J. Cellulose Fermentation by an Asporogenous Mutant and an Ethanol-Tolerant Mutant of Clostridium thermocellum. Appl Environ Microbiol. 1989 Jan;55(1):203–206. doi: 10.1128/aem.55.1.203-206.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utt E. A., Eddy C. K., Keshav K. F., Ingram L. O. Sequencing and expression of the Butyrivibrio fibrisolvens xylB gene encoding a novel bifunctional protein with beta-D-xylosidase and alpha-L-arabinofuranosidase activities. Appl Environ Microbiol. 1991 Apr;57(4):1227–1234. doi: 10.1128/aem.57.4.1227-1234.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]