Abstract
The fungus Ustilago maydis is a biotrophic pathogen of maize (Zea mays). In its genome we have identified an ortholog of YAP1 (for Yeast AP-1-like) from Saccharomyces cerevisae that regulates the oxidative stress response in this organism. yap1 mutants of U. maydis displayed higher sensitivity to H2O2 than wild-type cells, and their virulence was significantly reduced. U. maydis yap1 could partially complement the H2O2 sensitivity of a yap1 deletion mutant of S. cerevisiae, and a Yap1-green fluorescent protein fusion protein showed nuclear localization after H2O2 treatment, suggesting that Yap1 in U. maydis functions as a redox sensor. Mutations in two Cys residues prevented accumulation in the nucleus, and the respective mutant strains showed the same virulence phenotype as Δyap1 mutants. Diamino benzidine staining revealed an accumulation of H2O2 around yap1 mutant hyphae, which was absent in the wild type. Inhibition of the plant NADPH oxidase prevented this accumulation and restored virulence. During the infection, Yap1 showed nuclear localization after penetration up to 2 to 3 d after infection. Through array analysis, a large set of Yap1-regulated genes were identified and these included two peroxidase genes. Deletion mutants of these genes were attenuated in virulence. These results suggest that U. maydis is using its Yap1-controlled H2O2 detoxification system for coping with early plant defense responses.
INTRODUCTION
To survive, plants have developed efficient defense systems against pathogenic microbes. One of the most rapid plant defense reactions after pathogen attack is the so-called oxidative burst, which constitutes the production of reactive oxygen species (ROS), primarily superoxide and H2O2, at the site of attempted invasion (Apostol et al., 1989). ROS is primarily generated by plasma membrane–localized NADPH oxidases (Doke et al., 1996). Apoplastic peroxidases bound to cell wall polymers use the generated H2O2 or phenolic substrates in a peroxidation cycle, leading to the synthesis of lignin and other phenolic polymers, which provide additional plant barriers against pathogen attack (Chen and Schopfer, 1999). The produced ROS activate plant defense responses, including programmed cell death, or function as second messenger in the induction of various plant defense-related genes (Torres and Dangl, 2005). Due to the toxicity of ROS molecules and their importance in plant defense responses, plants and plant pathogens have developed strategies for ROS detoxification (see Apel and Hirt, 2004). As one strategy, nonenzymatic antioxidants like ascorbate, GSH, tocopherol, flavonoids, alkaloids, and carotenoids are produced. The second strategy is enzymatic ROS scavenging through superoxide dismutase, ascorbate peroxidase, cytochrome C-peroxidase, glutathione peroxidase, and catalases, generally using NAD(P)H as reducing equivalents (Asada, 1999; Campos et al., 2005).
One of the central regulators whose action provides protection against oxidative stress in Saccharomyces cerevisiae is Yap1p (encoded by YAP1). This transcription factor is a basic domain/leucine zipper (bZIP) protein of the AP-1 family that was originally identified by its ability to recognize the mammalian AP-1 binding site (Moye-Rowley et al., 1989). Other functional domains include two Cys-rich domains (CRDs) (C-terminal domain designated c-CRD and the N-terminal domain termed n-CRD) (Delaunay et al., 2000; Toone et al., 2001), which are critical for the YAP1-mediated resistance to oxidative stress and for the appropriate subcellular localization of Yap1p (Coleman et al., 1999; Kuge et al., 2001). In response to H2O2, Yap1p is oxidized by a glutathione peroxidase-like protein (Gpx3/Opr1) and changes its conformation by forming two intramolecular disulfide bonds that activate the protein and mask a nuclear export sequence (Delaunay et al., 2000). This compromises binding of the nuclear exportin Crm1p, leading to nuclear retention of the active form (Kuge et al., 1997; Wood et al., 2003). Proteins related to Yap1p from S. cerevisiae have been found in Candida albicans (Alarco and Raymond, 1999), Schizosacharomyces pombe (Toone et al., 1998), Kluyveromyces lactis (Billard et al., 1997), and Cochliobolus heterostrophus (Lev et al., 2005). In these microorganisms, Yap1p is involved in activating genes involved in oxidative stress tolerance, drug tolerance, and heavy metal resistance (Wu et al., 1993; Gounalaki and Thireos, 1994; Hirata et al., 1994; Lee et al., 1999; Dumond et al., 2000; Wysocki et al., 2004). Upon H2O2 stress, ∼500 genes are upregulated in S. cerevisiae and many of them have Yap1p binding sites in their promoters (Harshman et al., 1988; Kuge and Jones, 1994; Wu and Moye-Rowley, 1994). Among the Yap1p-activated genes, a significant number is directly involved in the detoxification of ROS, such as cytoplasmic catalase and superoxide dismutase isoenzymes, alkyl hydroxide reductases, peroxiredoxins, glutathione peroxidase, and cytochrome C peroxidase (Lee et al., 1999; Dumond et al., 2000; Gash et al., 2000).
Ustilago maydis is the causative agent of maize (Zea mays) smut disease. The disease cycle is initiated by fusion of compatible haploid cells. The resulting dikaryon switches to filamentous growth on the leaf surface, forms appressoria, and penetrates host cells in a process that is likely promoted by lytic enzymes. During penetration, the plasma membrane of the host invaginates and surrounds the hyphae. Plant cells stay alive, and there are no apparent defense responses triggered (Basse, 2005). After penetration, U. maydis extends into the deeper layers of the tissue. Massive fungal proliferation occurs within cells or in the apoplast, followed by hyphal fragmentation, karyogamy, and spore formation (Snetselaar and Mims, 1994; Banuett and Herskowitz, 1996). These events take place in tumor tissue that develops in response to yet unknown fungal signals. Recent insights from the genome sequence have revealed that a number of U. maydis gene clusters coding for secreted proteins of unknown function play decisive roles in shaping the biotrophic interaction with the host (Kämper et al., 2006). However, at present, it is not yet clear at which stages these proteins are required and whether they shield fungal hyphae or interfere with host defense responses. Given the situation that many plant pathogens are recognized by their hosts through conserved pathogen-associated molecular patterns (Nürnberger and Brunner, 2002) that elicit an oxidative burst, we reasoned that a H2O2 detoxification system of U. maydis might help to overcome this host response.
In this work, we characterized a Yap1-related protein of U. maydis. We show that it plays an important function for growth under oxidative stress conditions and is required for full virulence.
RESULTS
Identification and Characterization of an AP-1 Homolog in U. maydis
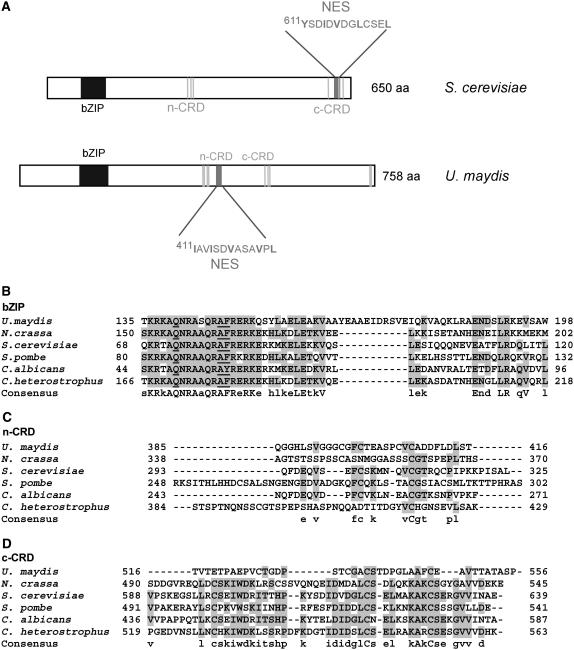
A search for AP-1–related proteins in the U. maydis genome (Munich Information Center for Protein Sequences Ustilago maydis Database, http://mips.gsf.de/genre/proj/ustilago) revealed six predicted open reading frames that showed similarity to the bZIP motif (um00567, um01513, um02191, um04916, um10256, and um11176), but of these only um02191 also displayed similarity to the CRDs (Figures 1B to 1D). Additionally, in between the two CRDs, the U. maydis protein contains a hydrophobic consensus nuclear export sequence that is characteristic for AP-1–like transcription factors and allows the binding of export substrates to Crm1p (Figure 1A) (Yan et al., 1998). In Yap1p of S. cerevisiae, such a motif colocalizes with the c-CRD domain (Figure 1A). Outside these conserved domains the AP-1-like protein from U. maydis, termed Yap1 (the respective gene is termed yap1), displays weak similarity to other Yap1 proteins (data not shown). The yap1 open reading frame is not expected to be interrupted by introns and encodes a protein of 758 amino acids that is predicted to be localized in the nucleus (http://psort.nibb.jp).
Figure 1.
Domain Organization of Yap1.
(A) Yap1p of S. cerevisiae and Yap1 of U. maydis are compared. The vertical gray lines indicate the position of Cys residues. NES (dark-gray bar), nuclear export sequence; aa, amino acids.
(B) Alignment of the bZIP domain of AP-1–like proteins from U. maydis, Neurospora crassa, S .cerevisiae, S. pombe, C. albicans, and C. heterostrophus. Accession numbers for these proteins are in Methods. Uppercase letters indicate identity in all proteins compared, and lowercase letters indicate that three or more proteins carry this residue.
(C) Alignment of the n-CRD domains of proteins listed in (A). Shading follows the scheme given in (A).
(D) Alignment of the c-CRD domains of proteins listed in (A). Numbers give amino acid positions in the respective proteins.
Under Oxidative Stress Conditions, U. maydis yap1 Complements the Growth Defect of a yap1 Mutant of S. cerevisiae
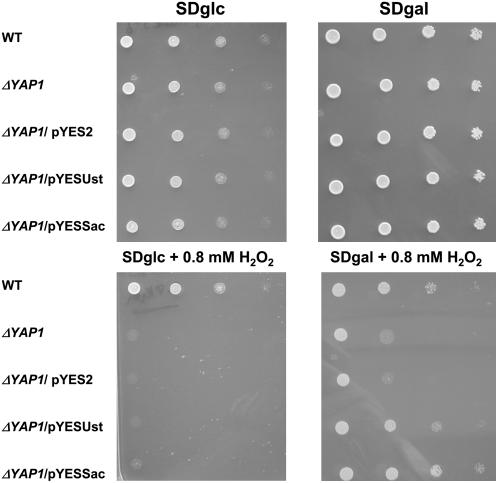
To assess the function of U. maydis yap1, we analyzed whether yap1 can replace yeast YAP1. To this end, the full-length yap1 gene from U. maydis was expressed under control of the GAL4 promoter in an S. cerevisiae Δyap1 strain. As controls, the Δyap1 strain was transformed either with empty vector or with the full-length YAP1 gene from S. cerevisiae (see Methods). When serial dilutions of all strains were spotted on glucose- or galactose-containing medium to activate the expression of GAL4, no growth differences were observed (Figure 2). On glucose-containing medium to which 0.8 mM hydrogen peroxide was added, only the wild-type strain could grow (Figure 2). On medium containing galactose and hydrogen peroxide, the wild type and the Δyap1/pYESUst (expressing U. maydis yap1) and Δyap1/pYESSac strains (expressing the S. cerevisiae YAP1 gene) formed colonies, while growth of the Δyap1 mutant and Δyap1/pYES2 (carrying the empty vector) was significantly inhibited (Figure 2). This indicates that the ability of S. cerevisiae to cope with H2O2 stress can be complemented by yap1 of U. maydis. However, as judged from the size of single colonies formed, the complementation by yap1 from U. maydis is less efficient than complementation by S cerevisiae YAP1 (Figure 2, bottom right panel).
Figure 2.
yap1 from U. maydis Complements the Hydrogen Peroxide Sensitivity of an S. cerevisiae Δyap1 Strain.
The growth of S. cerevisiae BY4742 (WT), S. cerevisiae BY4742ΔYML007W (Δyap1), and derivatives of this strain transformed with the empty vectors pYES2, pYESUst, and pYESSac was tested on SD plates with glucose (top left panel), SD plates with galactose and raffinose (top right panel), SD plates with glucose supplemented with 0.8 mM H2O2 (bottom left panel), and SD plates with glucose and raffinose supplemented with 0.8 mM H2O2 (bottom right panel). Serial 10-fold dilutions of cultures indicated on the left were spotted.
yap1 Affects Survival of U. maydis under Oxidative Stress Conditions
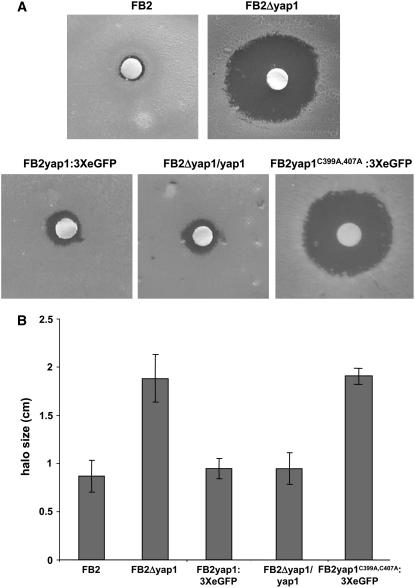
To analyze the cellular functions of yap1 in U. maydis, we constructed yap1 deletion derivatives of the compatible haploid strains FB1 and FB2. In these strains, the yap1 gene was replaced with a transcriptional fusion of Pyap1-enhanced green fluorescent protein (eGFP) and a hygromycin resistance cassette (see Methods for details). By DNA gel blot analysis, it was shown that in approximately one-third of the transformants the expected homologous recombination event had occurred (data not shown). When exposed to hydrogen peroxide in an agar diffusion test (see Methods for details), a growth inhibition halo was observed, and compared with wild-type strains, this halo was wider in all the mutants (Figure 3; data not shown). In addition, all yap1 deletion mutants produced a brown pigment with unknown composition (Figure 3A). In liquid complete medium (CM) with glucose, the growth rate of FB2 and FB2Δyap1 was indistinguishable; however, when 10 mM H2O2 was added, FB2Δyap1 was unable to grow while FB2 still could proliferate (data not shown). To verify that the described phenotypes are caused by the deletion of yap1, one copy of the yap1 gene under the control of its own promoter was inserted in the carboxin locus (cbx) of the FB2Δyap1 and FB1Δyap1 strains. These strains no longer produced the brown pigment and showed resistance to hydrogen peroxide comparable to wild-type strains (FB2Δyap1/yap1; Figure 3; data not shown), illustrating that the phenotypes associated with deletion of the yap1 gene can be complemented by introducing a single copy of yap1.
Figure 3.
Sensitivity of U. maydis Wild-Type and yap1 Mutant Strains to Oxidative Stress.
(A) Sensitivity of strains to H2O2 was assessed in an agar diffusion test in which a filter soaked with H2O2 (30% [v/v]) was placed on a CM-glucose agar plate seeded with the strains indicated on top.
(B) Halo size was quantified for the strains given in (A). Error bars indicate standard deviations derived from three independent experiments consisting of three replicas each.
The Cellular Localization of Yap1 Is Modulated by H2O2
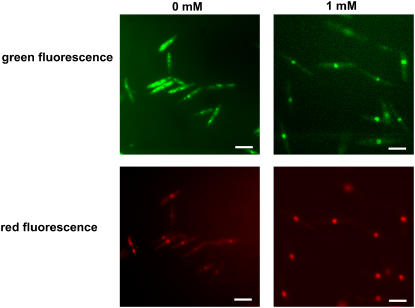
To study the regulation of yap1 under oxidative stress conditions, the yap1 deletion strain carrying the yap1 promoter-eGFP fusion was exposed to different concentrations of H2O2. Fluorimetric analysis indicated a dose-dependent transient increase in fluorescence, which, however, never exceeded the basal level by twofold (data not shown). To assess whether Yap1 protein localization changes upon H2O2 stress, as is the case in S. cerevisiae (Coleman et al., 1999), we generated a C-terminal yap1:3XeGFP fusion in the native yap1 locus in strains FB1 and FB2. With respect to sensitivity to H2O2, these strains were indistinguishable form their respective wild-type strains (Figure 3A), illustrating that the fusion to 3XeGFP did not interfere with function of the fusion protein. For localization studies, we integrated a gene encoding a fusion protein consisting of a nuclear localization signal fused to three copies of the red fluorescence protein (3XRFP) into strain FB2yap1:3XeGFP. In the absence of oxidative stress conditions, the Yap1:3XeGFP fusion protein was distributed throughout the cells showing some accumulation in vacuoles (Figure 4, top row). By contrast, after exposure to 1 mM H2O2, fluorescence was concentrated in a single spot (Figure 4, top row). This spot showed RFP fluorescence (Figure 4, bottom row), indicating nuclear localization of Yap1 after the addition of H2O2. Thus, the behavior of Yap1 protein in U. maydis is similar to what has been observed in S. cerevisiae (Kuge et al., 1997). On these grounds, we consider it likely that nuclear localization coincides with transcriptionally active Yap1 protein also in U. maydis.
Figure 4.
Subcellular Localization of Yap1 in the Presence of H2O2.
Strain FB2yap1:3XeGFPcbx:pANNE3090 was cultured in liquid CM-glucose medium, exposed to the indicated concentrations of H2O2 for 1 h, and assayed microscopically for GFP and RFP fluorescence. FB1yap1:3XeGFPcbx:pANNE3090 displayed the same behavior (data not shown). Bars = 10 μm.
The Deletion of yap1 Attenuates Virulence
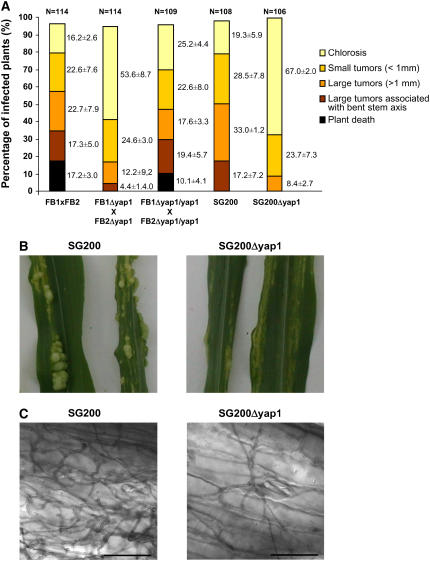
When spotted on charcoal potato dextrose (PD) medium, mixtures of compatible yap1 mutant strains produced a clear Fuz+ phenotype (Banuett and Herskowitz, 1989), indicating that yap1 is neither required during cell fusion nor for formation of the dikaryotic filament (data not shown). When the same mixture of yap1 mutant strains were used to infect maize seedlings, virulence of the yap1 mutants was severely reduced when compared with the respective wild-type strains (Figure 5A). Virulence was almost restored to the wild-type level when the complemented yap1 mutant strains were scored (Figure 5A). The most prominent difference to the wild type was the absence of dead plants in infections with the compatible yap1 mutant strains (Figure 5A). yap1 was also deleted in the solopathogenic haploid strain SG200 that produces less severe disease symptoms than a mixture of compatible haploid strains (Kämper et al., 2006; Figure 5A). Relative to SG200, the SG200Δyap1 strain caused significantly attenuated disease symptoms (Figures 5A and 5B). To elucidate at which stage of in planta development the yap1 mutants are affected, we quantified appressorium development in SG200 and SG200Δyap1 strains in 10 infected plants harvested 2 d after inoculation. The percentage of cells that formed appressoria relative to those that only formed filaments revealed no significant differences (24.2% ± 5.1% for SG200 and 21.5% ± 3.8% for SG200Δyap1). The post-penetration stage was analyzed by chlorazole black E staining after removing cells from the leaf surface (see Methods). The SG200Δyap1 deletion strain was consistently less efficient than SG200 in colonizing the plant (Figure 5C). Taken together, these results define yap1 as a virulence factor in U. maydis.
Figure 5.
Pathogenicity of yap1 Mutant Strains.
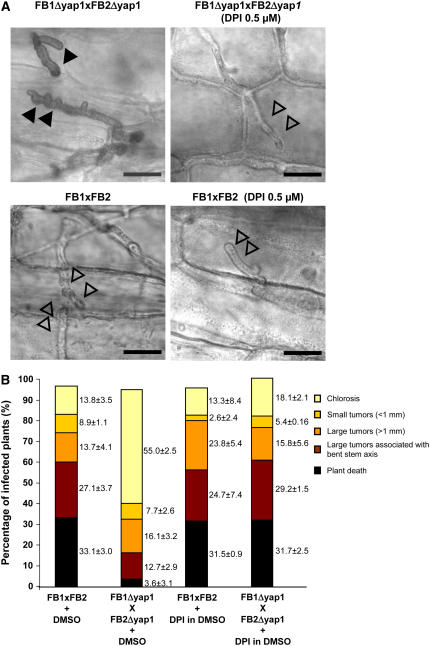
(A) Disease symptoms of compatible wild-type strains, compatible yap1 mutant strains, their complemented derivatives, and the solopathogenic strain SG200 and its Δyap1 derivative are depicted. Strains are listed under each column. For each strain or strain combination, three independent infections were performed, and the total number of infected plants is indicated above each column. Symptoms were scored 14 d after infection (see Methods for details). The color code for disease rating is given on the right. Numbers at the right of the bars indicate the average percentage in each disease category and includes the standard deviation calculated from the three independent experiments.
(B) Tumor morphology of strain SG200 and its Δyap1 derivative.
(C) Intracellular hyphae of SG200 and SG200Δyap1 strains 2 d after inoculation visualized by chlorazole black E staining. Bars = 50 μm.
Yap1 Is Activated during the Early Stages of Biotrophic Growth
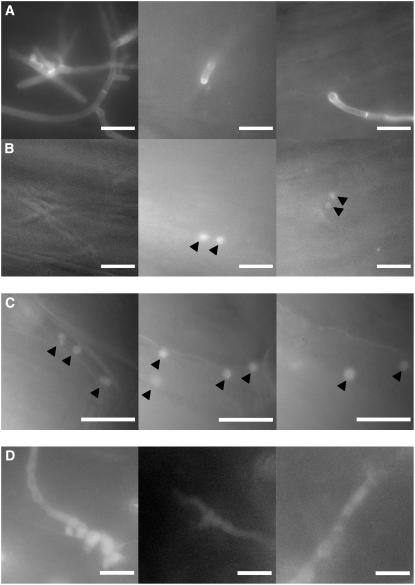
To investigate whether Yap1 is activated during fungal development on the leaf surface and/or during biotrophic growth, a mixture of FB1yap1:3XeGFP and FB2yap1:3XeGFP strains was inoculated into maize seedlings and fluorescence was monitored. Fungal cells on the leaf surface prior to penetration (visualized by calcofluor staining; Figure 6A, left panel) showed only diffuse cytoplasmic fluorescence (Figure 6B, left panel). The same was true for appressoria that had not yet penetrated (data not shown). However, in appressoria that had penetrated the plant cuticle (visualized by calcofluor staining; Figure 6A, right panel), fluorescence always localized to the two nuclei of the dikaryon (Figure 6B, right panel). Nuclear localization of the Yap1-3XeGFP fusion protein was evident until ∼2 to 3 d after infection (Figure 6C) but disappeared when massive proliferation started (Figure 6D; data not shown). Diffuse cytoplasmic fluorescence remained throughout sporogenesis (data not shown). The activation of Yap1 during the early phases of biotrophic development of U. maydis suggests that the role of yap1 during pathogenesis might be confined to these stages.
Figure 6.
Subcellular Localization of Yap1 during the Life Cycle of U. maydis.
(A) Compatible strains FB1yap1:3XeGFP and FB2yap1:3XeGFP were inoculated in maize seedlings. Hyphae and appressoria on the leaf surface were visualized by calcofluor staining 16 h after infection.
(B) GFP fluorescence of the same leaf area shown in (A). Hyphae that have not yet penetrated do not show fluorescent nuclei (left panel). Hyphae that have penetrated show fluorescent nuclei (arrowheads), and these could be visualized in a different focus plane, indicating that the appressoria shown in (A) had already penetrated.
(C) Fluorescent nuclei (arrowheads) in intracellularly growing hyphae 2 d after infection.
(D) Intracellular hyphae with diffuse fluorescence 6 d after infection.
Bars = 10 μm.
Cys-399 and Cys-407 Are Crucial for Functionality of Yap1
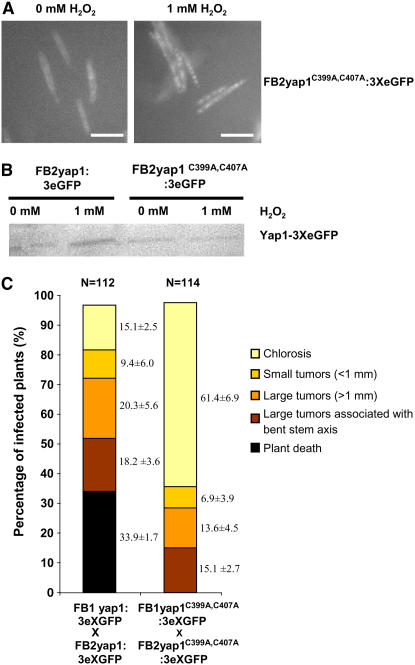
In S. cerevisiae, nuclear export of Yap1p is blocked by the formation of two disulfide bridges, and this is required for full transcriptional activity in response to H2O2 (Kuge et al., 2001). To assess whether such a mode of action also applies to U. maydis, we generated a mutant yap1 allele in strains FB1 and FB2 in which two Cys residues in the n-CRD domain were substituted by Ala, yap1C399A C407A:e3GFP. The respective Yap1C399A C407A:eGFP fusion protein showed cytoplasmic localization, both in the absence and presence of 1 mM H2O2 (Figure 7A; data not shown). Protein gel blot analyses demonstrated that the respective mutant Yap1 protein is expressed under both conditions (Figure 7B). With respect to H2O2 sensitivity and production of the brown pigment, these mutant strains behaved like Δyap1 strains (Figures 3A and 3B). The behavior of the FB1yap1 C399A C407A:e3GFP and FB2yap1 C399A C407A:e3GFP strains in planta (Figure 7C) was comparable to the Δyap1 strains, and with respect to wild-type strains, a significant reduction in virulence was observed (Figure 7C). In the mutant strains, the fusion protein showed cytoplasmic localization throughout the infection (data not shown), indicating that there are no other means to activate Yap1p under these conditions. From these results, we infer a crucial role of the disulfide bridge between Cys-399 and Cys-407 for the function of Yap1 in U. maydis.
Figure 7.
Cys-399 and Cys-407 Are Important for Functionality of Yap1.
(A) Strain FB2yap1C399A,C407:3XeGFP was cultured in liquid CM-glucose medium, exposed to 1 mM H2O2 for 1 h, and assayed microscopically for GFP fluorescence (right panel). In the left panel, cells are depicted without H2O2 treatment. Bars = 50 μm.
(B) Proteins isolated from the strains indicated on top either treated with H2O2 or left untreated were separated by SDS-PAGE. Yap1:3XeGFP and Yap1C399A,C407A:3XeGFP were detected by protein gel blot analysis using monoclonal GFP IgG mouse antibody.
(C) Disease rating of wild-type and yap1C399A,C407A mutant strains. The disease rating scheme is described in Figure 5.
U. maydis yap1 Prevents the Accumulation of ROS
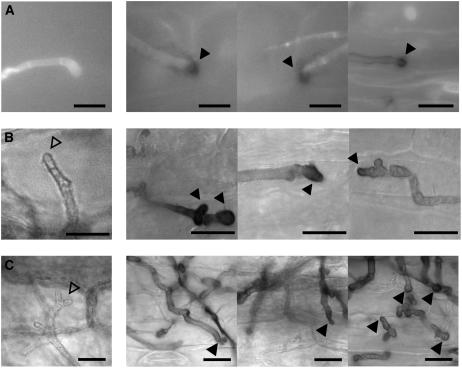
To gain insights into the mechanism that leads to the activation of Yap1 during the early plant colonization stages, we have analyzed the production of H2O2 in leaves of maize seedlings infected with U. maydis. To detect H2O2, we used the diamino benzidine (DAB) uptake technique (Thordal-Christensen et al., 1997; Fryer et al., 2002). In the presence of H2O2, DAB is converted to dark-brown polymers. In infections with wild-type strains, we have not seen an accumulation of DAB polymers, irrespective of the stage at which hyphae were analyzed (Figures 8A to 8C; data not shown). By contrast, an accumulation of H2O2 was observed in the vicinity of Δyap1 hyphal tips already during penetration (Figure 8A). H2O2 could be detected around the Δyap1 hyphae during the first 3 d after infection (Figures 8A to 8C, right panels; data not shown). During these stages, hyphal tips of Δyap1 strains often appeared swollen (Figures 8B and 8C, right panels), while hyphal tips from wild-type strains were straight (Figures 8B and 8C, left panels). DAB staining decreased at later time points and became undetectable during proliferation in tumor tissue and spore maturation (data not shown). These results suggest that in wild-type strains, yap1 is responsible for preventing the accumulation of H2O2 in the vicinity of hyphae during early stages of biotrophic growth.
Figure 8.
H2O2 Accumulation during the Early Stages of Biotrophic Growth of U. maydis Δyap1 Strains.
(A) Plant samples inoculated with either mixtures of FB1XFB2 (left panel) or FB1Δyap1XFB2Δyap1 strains (right panels) were stained with calcofluor and DAB 1 d after inoculation for visualization of hyphae and appressoria on the leaf surface by light microscopy using the 4‘,6’-diamidino-2-phenylindole filter. The closed arrowheads mark DAB precipitates.
(B) Samples were stained with DAB only, and intracellular hyphae were visualized by light microscopy 1 d after infection. DAB precipitates are marked with closed arrowheads. Intracellularly growing hyphae not surrounded by DAB precipitate are marked with open arrowheads.
(C) Samples were as in (B) 2 d after infection.
Bars = 10 μm.
The Inhibition of Plant NADPH Oxidase Restores Virulence of yap1 Mutant Strains
To determine whether the accumulation of H2O2 observed around yap1 mutant hyphae originates from the host, we applied diphenylene iodonium (DPI), an inhibitor of NADPH oxidase (Morre, 2002), together with the inoculum of U. maydis strains. Neither this compound nor its solvent DMSO affected the sensitivity of U. maydis wild-type strains or the Δyap1 mutant to H2O2 (data not shown). Plant NADPH oxidases are involved in the production of reactive oxygen intermediates in response to pathogens (Torres and Dangl, 2005). U. maydis lacks genes related to gp91phox (Aguirre et al., 2005), the transmembrane catalytic subunit of mammalian, plant, and fungal NADPH oxidases that catalyze the conversion of molecular oxygen to superoxide (Torres et al., 2002; Lambeth, 2004; Tanaka et al., 2006). Two days after maize seedling infection, H2O2 visualized by DAB staining was found in the vicinity of Δyap1 hyphal tips, while H2O2 production was no longer observed in the vicinity of Δyap1 hyphae when DPI was added to the inoculum. To exclude adverse effects of DPI, wild-type strains were also infected with DPI added (Figure 9A). In the infections with compatible wild-type strains, no H2O2 accumulation was detectable, fungal development visualized by microscopy was normal (Figure 9A), and disease symptoms were unaffected by DPI addition (Figure 9B). Interestingly, however, the presence of DPI in the inoculum of the compatible yap1 mutant strains prevented the appearance of bulbous hyphae and lead to an increase in virulence, reaching levels comparable to infections with wild-type strains (Figure 9B).
Figure 9.
DPI Restores Virulence of U. maydis Δyap1 Mutants.
(A) Maize seedlings were inoculated with mixtures of FB1XFB2 or FB1Δyap1XFB2Δyap1 to which DMSO or DPI dissolved in DMSO were added. Infected plant tissue samples were collected 2 d after infection and stained with DAB as indicated. DAB precipitates are marked with closed arrowheads, and hyphae without DAB precipitate in the vicinity are marked by open arrowheads. Bars = 10 μm.
(B) Symptoms produced by the indicated U. maydis strain combinations were scored following the scheme described in Figure 5.
These results indicate that the H2O2 accumulated around yap1 mutant hyphae is produced by the host. The finding that inhibiting this accumulation restores virulence implies that Yap1 in U. maydis plays a prominent role in the detoxification of these molecules during plant colonization.
Identification of Yap1 Target Genes by Microarray Analysis
To analyze in more detail the role of Yap1 in the detoxification of ROS, transcript profiles of strains FB1 and FB1Δyap1 were generated from cells grown in CM in the absence or presence of 5 mM H2O2 using the Affymetrix U. maydis DNA arrays. A set of 221 genes was downregulated in the comparison of FB1Δyap1/FB1 with a fold change of >1.5 (see Supplemental Table 1 online). The majority of these Yap1-dependent genes (203) was upregulated in FB1 when exposed to H2O2 (see fold change FB1/FB1+H2O2; see Supplemental Table 1 online). Typical Yap1 binding sites TT/GAC/GT/CA/A (Harshman et al., 1988; Fernandes et al., 1997; Toone and Jones, 1999) were found in the promoter regions of 212 of these genes (see Supplemental Table 1 online). With respect to oxidative stress, the Yap1-regulated genes fall into three categories, namely, ROS detoxifying enzymes, such as peroxidases and catalases, genes involved in the biosynthesis of low molecular mass antioxidants (ascorbic acid, glutathione, tocopherols, NADH, and NADPH), and genes encoding enzymes regenerating the reduced forms of antioxidants (reviewed in Blokhina et al., 2003). Among the genes putatively involved in the detoxification of ROS were the haeme peroxidase um10672, the cytochrome C peroxidase um01947, and the alkyl hydroperoxide reductase um02153. Among the Yap1-regulated genes involved in antioxidant function, we detected um04930 and um02592 encoding glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase, respectively, which are both involved in the generation of NADPH. Other Yap1-regulated genes are involved in the metabolism of antioxidants like glutathione (um05561, coding for a glutathione S-transferase), ascorbate (um04922, coding for a diketogulonate reductase), and folate (um02010, coding for a 2-keto-3-deoxy-D-arabino-heptulosonate 7-phosphate synthase). Among putative enzymes regenerating the reduced form of antioxidants, we detected um04578 coding for thioredoxin 2 as a Yap1-regulated gene. Besides these genes presumably directly involved in coping with oxidative stress, a large number of genes related to DNA stability and repair, protein folding and elimination of damaged proteins, transport and drug resistance, signaling, cell wall and membrane components, and lipid metabolism were upregulated after H2O2 treatment. The majority of these proved to be Yap1 dependent in their expression (see Supplemental Table 1 online).
Yap1 Regulates the Expression of Two Peroxidase Genes
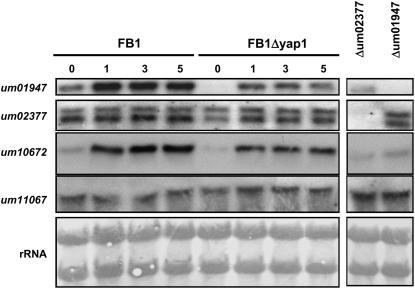
Among the Yap1-regulated genes identified were genes encoding cytochrome C peroxidases precursor (um01947) and haeme peroxidase protein (um10672). In addition, U. maydis possesses one gene (um11067) related to catalase CTT2 of S. cerevisiae and one additional gene coding for a cytochrome C peroxidase precursor (um02377), neither of which was regulated by yap1 according to the array analysis. To verify the array data, RNA gel blot analyses were performed. These showed that the expression of um01947 and um10672 was upregulated in the U. maydis wild type after addition of H2O2, while the expression of um02377 and um11067 was not altered significantly (Figure 10). The identity of the transcripts seen with probes for um01947 and um02377 was verified using RNA from respective deletion strains (Figure 10). In the Δyap1 background without the addition of H2O2, genes um01947 and um10672 were barely expressed, while after H2O2 treatment, transcript levels increased but did not reach the levels found in the respective wild-type strain (Figure 10). This indicates that these genes might be targets of Yap1. However, to explain the induction seen after H2O2 treatment, both genes must be additionally regulated by a yap1-independent mechanism. The expression levels of um02377 and um11067 were not significantly altered under these conditions (Figure 10).
Figure 10.
Yap1 Controls the Expression of Two Peroxidase Genes.
RNA gel blot of U. maydis strains FB1, FB1Δyap1, SG200Δum01947, and SG200Δum02377. Strains were grown in CM-glucose medium with H2O2 added in the concentrations (μm) indicated on top. Ten micrograms of total RNA were loaded per lane on four separate gels. Individual membranes were hybridized with DIG-labeled fragments of the genes um01947, um02377, um10672, or um11067. The two mRNAs detected with the um02377 probe is likely to reflect two different start sites or polyadenylation sites. As loading control, one of the membranes was stained with methylene blue to visualize rRNA.
A Fungal Peroxidase Gene Is Involved in the Detoxification of ROS
To assess whether the putative yap1 target genes um01947 and um10672 play an active role in the detoxification of ROS, individual deletion mutants were generated in SG200. With respect to H2O2 sensitivity, SG200Δum10672 was even more sensitive than SG200Δyap1 (data not shown), indicating that the product of this gene assumes the main responsibility for detoxification of ROS in U. maydis. SG200Δum01947 showed higher sensitivity to H2O2 than SG200 but did not reach the sensitivity level of SG200Δyap1 (data not shown), indicating a minor role of this gene in ROS degradation.
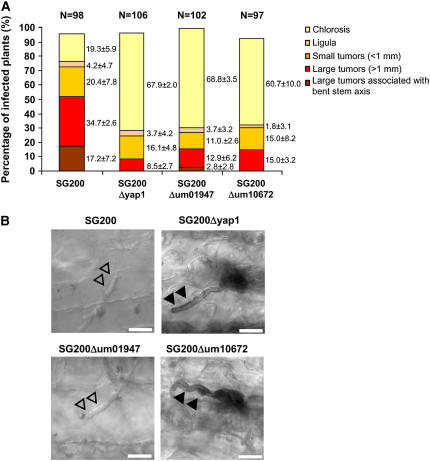
To investigate the contribution of um01947 and um10672 to virulence, SG200Δum01947 and SG200Δum10672 were inoculated in maize seedlings. Twelve days after infection, both mutant strains showed a reduction in virulence relative to SG200 comparable to SG200Δyap1 (Figure 11A). DAB staining of leaves 2 d after infection revealed brown precipitates in the infection with SG200Δum10672 comparable to SG200Δyap1 (Figure 11B), while neither SG200Δum01947 nor SG200 induced this reaction (Figure 11B). These results suggest that um10672 is the peroxidase responsible for ROS degradation during the early stages of infection.
Figure 11.
The Two Yap1-Regulated Peroxidases Affect Virulence.
(A) Plants were infected with the strains indicated below each column. Disease rating followed the scheme described in Figure 5.
(B) DAB staining of infected leaves 2 d after infection with the same strains assayed in (A). Intracellular hyphae are visualized by light microscopy and are marked by open arrowheads. DAB precipitates in the vicinity of the hyphae are marked by closed arrowheads. Bars = 10 μm.
DISCUSSION
In this work, we have characterized the U. maydis yap1 gene. Similar to Yap1p in S. cerevisiae, its product contains a bZIP domain and two CRDs. U. maydis yap1 complements the hydrogen peroxide–sensitive phenotype of a yeast yap1 mutant. The finding that U. maydis yap1 mutants are more sensitive to H2O2 than the progenitor strains, the observed nuclear localization after H2O2 exposure, and the ability to complement the yeast yap1 mutant phenotype suggests that the U. maydis Yap1 protein also functions as a redox sensor that is activated by intramolecular disulfide bridge formation. This is additionally supported by the phenotype of a yap1 mutant in which two Cys residues, Cys-399 and Cys-407, likely to be involved in disulfide bridge formation, are substituted by Ala. This mutant protein fails to be retained in the nucleus after H2O2 treatment, and in its behavior, this mutant is indistinguishable from a strain where yap1 is deleted. In the U. maydis genome, genes related to S. cerevisiae ORP1/GPX3, encoding a glutathione peroxidase-like enzyme, and TPX1, encoding an upstream activator of the redox sensor Pap1 in S. pombe (Vivancos et al., 2005), are found. Thus, it is likely that Yap1 in U. maydis is posttranslationally activated by one of these activities.
Compatible Δyap1 strains were indistinguishable from wild-type strains in their ability to mate, to develop dikaryotic filaments, and to form appressoria. However, they showed increased sensitivity to H2O2 and were severely attenuated in virulence (i.e., they formed fewer and smaller tumors). This most likely reflects problems during the early post-penetration stages, since 2 d after infection, a significant reduction of fungal biomass was observed in plants infected with the Δyap1 mutant compared with infections with wild-type strains. Additionally, the swollen tips of yap1 mutant hyphae after penetration are likely to indicate some stress response, which could be related to the observed reduction in fungal biomass. A Yap1:3XeGFP fusion protein localized to the nucleus immediately after penetration, and this activation was maintained for 2 to 3 d and then declined and became undetectable at late stages when massive fungal proliferation takes place and spore development is initiated. This suggests that Yap1 is needed primarily during the early stages of biotrophic growth where its presence allows more efficient colonization. In line with this reasoning, H2O2 accumulation was detected around yap1 mutant hyphae only at those time points when Yap1 protein localized to the nucleus in wild-type strains.
The generation of ROS is a hallmark of successful recognition of plant pathogens by the host (Nürnberger et al., 2004). This plant response can be elicited by fungal, bacterial, and viral pathogens and serves to activate plant defense programs (Torres and Dangl, 2005). The absence of H2O2 accumulation in infections with wild-type U. maydis strains and the strong accumulation of H2O2 around hyphae of yap1 deletion mutants indicate that Yap1 participates effectively in the detoxification of ROS. Since U. maydis does not code for genes related to NADPH oxidases (Aguirre et al., 2005), the enzymes responsible for ROS generation are likely to be of plant origin. The absence of NADPH oxidase genes in U. maydis is unusual, as most filamentous fungi have two or three NADPH oxidase isoforms (Lalucque and Silar, 2003). These proteins all have the structural core domains found in the animal gp91phox (Aguirre et al., 2005). noxA in Podospora anserina and Aspergillus nidulans is specifically induced and required during differentiation of sexual fruiting bodies (Lara-Ortíz et al., 2003; Malagnac et al., 2004). In the mutualistic Epichloë festuca/ryegrass (Lolium perenne) interaction, the deletion of noxA in the fungus changes the interaction of this biotrophic endophyte with its host from mutualistic to antagonistic (Tanaka et al., 2006). The noxA mutant accumulates significantly more biomass in infected hosts, and it has been speculated that ROS produced by NoxA during plant colonization negatively regulates hyphal tip growth, thereby preventing excessive colonization due to restricted growth of the fungus (Tanaka et al., 2006). The situation in the biotrophic interaction of U. maydis with its host is fundamentally different. U. maydis does not possess an NADPH oxidase but is likely to use its redox sensor Yap1 to detoxify H2O2 produced by the plant in response to being recognized. In the absence of yap1, H2O2 accumulates in the vicinity of fungal hyphae, and since the mutant is unable to detoxify it, this could negatively affect fungal proliferation in the infected tissue. We consider the finding that the inhibition of NADPH oxidase by DPI restores virulence to yap1 mutants as additional support for the assertion that it is the production of ROS by the plant that attenuates virulence of U. maydis yap1 mutants, although we cannot formally exclude that DPI modulates the activity of other enzymes. However, these are then not likely to be of fungal origin as the treatment of the Δyap1 mutant with DPI did not affect its sensitivity to H2O2. Thus, the Yap1-controlled ROS detoxification system serves an important function during the early U. maydis infection phase. In the Claviceps purpurea/rye interaction it has been demonstrated that the deletion of a CREB-like transcription factor, CPTF1, which positively controls catalase production, also induces an oxidative burst (Nathues et al., 2004) similar to what we have observed in yap1 mutants of U. maydis. However, in this case, it is speculated that CPTF1 negatively controls the activity of a fungal NADPH oxidase (i.e., when CPTF1 is deleted, the fungus would produce elevated levels of H2O2, which then in turn would trigger the oxidative burst) (Nathues et al., 2004).
The identification of Yap1-regulated genes by array analysis has revealed the same functional categories of genes that were found to be regulated through Yap1p in S. cerevisiae (Dumond et al., 2000). Among these genes were two Yap1-regulated peroxidase genes, um01947 and um10672, which we considered likely to be involved ROS detoxification. Interestingly, single mutations in either gene showed reduced virulence, comparable to the yap1 deletion strain. For the Δum10672 mutant, we could show that ROS accumulate around invading hyphae, while this was not observed for Δum01947 mutants. Since U. maydis lacks catalases like the YAP1-regulated CTT1 in S. cerevisiae (He and Fassler, 2005), we consider the Yap1-regulated heme peroxidase encoded by um10672 as prime activity for ROS detoxification in the U. maydis/maize system. The protein encoded by um01947 is predicted to have a mitochondrial targeting sequence; therefore, its function is likely to be confined to mitochondria. Mitochondrial integrity has already been show to be crucial for pathogenicity (Bortfeld et al., 2004).
The nuclear localization of Yap1 during the early stages of plant colonization in wild-type strains suggests that U. maydis may trigger a transient oxidative burst, which is sufficient to allow activation of Yap1 but may not be strong enough to allow its visualization by DAB staining. It is likely that at later time points there is no oxidative burst, as this should have resulted in continued nuclear localization of Yap1. Recent experiments indicate that secreted proteins of U. maydis are actively involved in suppressing plant defense responses (K. Schipper, T. Brefort, G. Doehlemann, K. Münch, and R. Kahmann, unpublished data). This situation in U. maydis contrasts with findings in the necrotrophic plant pathogen C. heterostrophus where a YAP1-related gene, CHAP1, has been studied (Lev et al., 2005). CHAP1 deletion mutants were also more sensitive to hydrogen peroxide, but their virulence was unaffected. CHAP1 showed nuclear localization already in conidial germ tubes on the leaf surface, and this localization persisted throughout the infection. Since nuclear localization of CHAP1 was only observed at high H2O2 concentrations that were not detected in plant extracts and since plant extracts triggered nuclear localization even when H2O2 was eliminated, it has been speculated that CHAP1 is induced by an as yet unidentified plant compound and serves to adapt the redox state of the cell to the plant environment (Lev et al., 2005). Our finding that strains carrying the mutant allele yap1C399A C407A:e3GFP do not show nuclear localization of the fusion protein and behave like the yap1 deletion mutant with respect to reduced virulence suggests that activation of Yap1 in U. maydis requires H2O2 and this cannot be bypassed by plant compounds. For C. heterostrophus, one could hypothesize that the levels of ROS encountered by this necrotrophic pathogen during growth may be insufficient to damage the fungus, and this would explain why CHAP1 is not needed for virulence. In this respect, it is of interest that induction of the hypersensitive response by necrotrophic fungi like Botrytis cinerea and Sclerotionia sclerotiorum actually facilitates infection (Govrin and Levine, 2000). It is thus an attractive possibility that such a situation also exists in C. heterostrophus (i.e., in this scenario, it would not be in the interest of the pathogen to detoxify ROS, as the fungus would require dead plant tissue for proliferation). Conversely, biotrophic pathogens, like U. maydis, might be more sensitive to the detrimental effects of ROS during infection and depend on the yap1 system at least during the early infection stages. In the future it will be very interesting to analyze how ROS production and detoxification systems in different fungi that live in close association with plants determine the outcome of the respective interactions and whether specific strategies are used depending on the type of interaction.
METHODS
Strains and Growth Conditions
The Escherichia coli K12 derivatives DH5α (Bethesda Research Laboratories) and Top10 (Invitrogen) were used for cloning purposes. Saccharomyces cerevisiae strains were grown in SD medium supplemented with the necessary amino acids (Yeast Protocols Handbook; Clontech) and glucose (3% [w/v]) or galactose and raffinose (2% [w/v]). The wild-type Ustilago maydis strains FB1 (a1 b1) and FB2 (a2 b2) (Banuett and Herskowitz, 1989), the solopathogenic strain SG200 (a1:mfa2 bE1 bW2) (Kämper et al., 2006), and its derivatives were grown as indicated at 28°C in liquid CM (Holliday, 1974), YEPSL (0.4% yeast extract, 0.4% peptone, and 2% sucrose), or PD (2.4% PD broth [Difco]) medium or solid PD agar. In Yap1 activation assays, the strains were inoculated in CM medium and cultured overnight, diluted 1:10 into fresh CM medium, and grown to an OD600 of 0.6. H2O2 was added in the concentrations indicated in each experiment. To assay H2O2 sensitivity, U. maydis strains were plated on PD or PD medium supplemented with 0.5 μM DPI. Filter disks of Whatman paper (5 mm) were soaked with 1 or 2 μL of H2O2 (30% [v/v]) and placed on the plates. The halo sizes were measured in four duplicates after 48 h of incubation.
Mating assays were performed by cospotting compatible strains onto PD plates containing 1% charcoal. Subsequent incubation was done at 21°C (Holliday, 1974).
Pathogenicity assays were performed as described (Kämper et al., 2006). For plant infections, cultures of the U. maydis strains were grown to an OD600 of 0.6 to 0.8 in YEPSL, centrifuged, and resuspended to an OD600 of 1 and injected into young maize (Zea mays) seedlings (Early Golden Bantam; Olds Seeds). Plants were examined for tumor formation 7 to 14 d after infection. Categories for disease rating were as follows: (0) no symptoms; (1) chlorosis; (2) ligular swelling; (3) small tumors (<1 mm in diameter); (4) tumors >1 mm in diameter, not associated with bending of stem; (5) large tumors associated with bending of infected stems; and (6) dead plants (Kämper et al., 2006).
Hygromycin B was from Roche. All other chemicals were of analytical grade and were obtained from Sigma-Aldrich or Merck.
Yeast YAP1 Complementation
S. cerevisiae BY4742ΔYML007w (yap1) and the strain from which it was derived, BY4742 (MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), were obtained from Euroscarf. The full-length U. maydis yap1 (2.2 kb) and S. cerevisiae YAP1 genes (1.7 kb) were amplified using the primers pairs 5′-CTCTAAGCTTATGTCATCTCCAGCTATGGC/TCTCTCTAGATTAAGGTGTCCGCCTTTCGC-3′ and 5′-CTCTAAGCTTATGAGTGTGTCTCACGCCAG/TCTCTCTAGATTAGTTCATATGCTTATTCAAAG-3′, respectively. The obtained PCR products were digested with HinDIII and XbaI, cloned into the yeast expression vector pYES2 (Invitrogen), and transformed into BY4742ΔYML007w. Colonies were selected on synthetic CM lacking uracil. As a control, these strains were also transformed with empty pYES2 vector. Transformed yeast cells were grown on synthetic CM without uracil containing either glucose 3% (w/v) or 2% galactose and 2% raffinose, respectively. Five-microliter drops from serial dilutions from cultures with an OD600 of 0.5 were spotted on plates with and without 0.8 mM H2O2 and grown for 2 d at 30°C. This experiment was repeated three times.
Plasmids and Strain Construction
Plasmid pCR2.1TOPO (Invitrogen) was used for cloning and sequencing of fragments generated by PCR. Sequence analysis of genomic fragments and fragments generated by PCR was performed with an automated sequencer (ABI 377; Applied Biosystems) and standard bioinformatic tools.
Deletion and fusion constructs were generated according to Kämper (2004). To generate a transcriptional fusion of the yap1 promoter to egfp, a 1.0-kb fragment comprising the 5′ flank and a 1.0-kb fragment comprising the 3′ flank of the yap1 open reading frame were generated by PCR on U. maydis FB1 genomic DNA with primer combinations 5′-GCAGAGGTGCTGGAAAACTCAAG-3′/5′-GCGGGCCGCGTTGGCCGCCGCTGAAGAAGATACCAG-3′ and 5′-GCGGGCCTGAGTGGCCCGCAAATCAAAACGGAAGATG-3′/5′-CAACTCGGAACTGGCAGAAGAG-3′, respectively. These fragments were then digested with SfiI and ligated to the 3.7-kb SfiI eGFP-hygromycin resistance cassette from pUMa229 (Brachmann et al., 2004). The resulting ligation product was cloned into pCR2.1-TOPO. The flanks of plasmid, pyap1-eGFP-hyg, were sequenced and shown to match the U. maydis genomic sequence. pyap1-eGFP-hyg was subsequently used as a template to amplify the yap1 deletion construct with primers 5′-GCAGAGGTGCTGGAAAACTCAAG-3′ and 5′-CAACTCGGAACTGGCAGAAGAG-3′. This fragment was then used in transformation to generate the yap1 deletion mutants of strains FB1, FB2, and SG200. Deletion derivatives were identified by DNA gel blot analysis. To generate the transcriptional fusion between yap1 and three copies of the egfp gene, a 1.0-kb fragment comprising from 1285 to 2274 bp of the yap1 open reading frame and a 1.0-kb fragment comprising the 3′ flank of yap1 were generated by PCR on U. maydis FB1 DNA using primer combinations 5′-GTTGCATCTGCAGTTCCACTC-3′/5′-CGCGGCCGCGTTGGCCGGTGTCCGCCTTTCGCCA-3′ and primer combinations 5′-CGCGGCCTGAGTGGCCCACCACAAATCTCGTATCAC-3′/5′-GACCTTAGCAGATGGATGGTC-3′, respectively. These fragments were then digested with SfiI and ligated to the 5.2-kb SfiI fragment from the plasmid pUMa647 (K. Zarnack, unpublished data) containing 3XeGFP and the hygromycin resistance cassette. The resulting ligation product was cloned into pCR2.1-TOPO to yield pyap1-3XeGFP. Border regions of this plasmid were sequenced and shown to match the wild type. This plasmid was subsequently used as a template to amplify the yap1-3eGFP construct with primers 5′-GTTGCATCTGCAGTTCCACTC-3′ and 5′-GACCTTAGCAGATGGATGGTC-3′. This fragment was transformed in strains FB1, FB2, and SG200. Homologous integration was verified by DNA gel blot analysis.
To generate the transcriptional fusion between yap1 and three copies of the egfp gene carrying the mutation C399A and C407A, a 0.8-kb fragment of the yap1 open reading frame was generated by PCR on U. maydis FB1 DNA using primer combinations 5′-AAGGTGGCCATCTTTCTGTCGGAGGAGGCTGTGGCTTTGCCACCGAGGCTTCTCCCTGTGTTGCTGCA-3′(mutations are underlined)/5′-ATGCTTCGCGAGGCCGTTTGCTGGAGAGAAG-3′. The obtained PCR fragment was digested with MluI and NruI and was inserted into pyap1-3XeGFP, where it replaced the respective fragment of the wild-type yap1 gene. This plasmid pyap1C399-407A-3XeGFP was sequenced to verify the presence of the two mutations and subsequently used as a template to amplify the yap1C399AC407A-3eGFP construct with primers 5′-GTTGCATCTGCAGTTCCACTC-3′ and 5′-GACCTTAGCAGATGGATGGTC-3′. The resulting fragment was transformed into strains FB1 and FB2. Homologous integration was verified by DNA gel blot analysis.
To generate the deletion mutant of um01947, a 1.2-kb fragment comprising the 5′ flank and a 1.3-kb fragment comprising the 3′ flank of the um01947 open reading frame were generated by PCR on U. maydis FB1 genomic DNA with primer combinations 5′-ATGAAAGAGGTGAAGGCTGC-3′/5′-GATCGGCCATCTAGGCCTAACAAAAGAGTGCGCGTGC-3′ and 5′-GATCGGCCTGAGTGGCCTTGTCGTCCCTCTTGACAGG-3′/5′-TTAGGACAACGCGCCTTTGC-3′, respectively. These fragments were then digested with SfiI and ligated to the 2.7-kb SfiI hygromycin resistance cassette from pBS-Hyg(-) (Keon et al., 1991). The ligation products were transformed into strain SG200. Homologous integration was verified by DNA gel blot analysis.
To generate the deletion mutant of um10672, a 1.2-kb fragment comprising the 5′ flank and a 1.4-kb fragment comprising the 3′ flank of the um10672 open reading frame were generated by PCR on U. maydis FB1 genomic DNA with primer combinations 5′-AAGTTGTCGGCACTGCAAGC-3′/5′- GATCGGCCATCTAGGCCAACGCTGATCTCATAAACGC-3′ and 5′- GATCGGCCTGAGTGGCCTTCATTTGCACGATGGAAGC-3′/5′- ATGTGGGCGAGTACTACTCC-3′, respectively. These fragments were then digested with SfiI and ligated to the 2.7-kb SfiI hygromycin resistance cassette from pBS-Hyg(-) (Keon et al., 1991). The ligation products were transformed into strain SG200. Homologous integration was verified by DNA gel blot analysis.
For amino acid comparisons with Yap1, the following proteins were used: Neurospora crassa (GenBank accession number NP_013707), S. cerevisiae (GenBank accession number NP_593662), Schizosaccharomyces pombe (GenBank accession number CAB91681), Candida albicans (GenBank accession number EAL02784), and Cochliobolus heterostrophus (GenBank accession number AY486156).
U. maydis yap1 Complementation
A fragment of 3.7 kb, containing the yap1 gene from U. maydis and 1 kb of its promoter region, was amplified by PCR using FB1 genomic DNA as template and the primer combination 5′-GCAGAGGTGCTGGAAAACTCAAG-3′/5′-TCTCTCTAGATTAAGGTGTCCGCCTTTCGC-3′. This fragment was subcloned into pCR2.1-TOPO giving the plasmid pYAPUst. The DNA fragment containing the yap1 gene and its promoter region were sequenced and subcloned as a KpnI-EcoRV fragment into plasmid pBS-Cbx(+) (Keon et al., 1991), yielding plasmid pCbx-yap1. This plasmid was transformed into U. maydis strains FB1Δyap and FB2Δyap. Carboxin-resistant clones were recovered, and single integration of the yap1 gene in the carboxin locus was confirmed by DNA gel blot analysis.
Tagging of Nuclei with Triple RFP
To visualize nuclei, the strains FB1yap1-3XeGFP and FB2yap1-3XeGFP were transformed with the integrative plasmid pANNE3090 (kindly provided by A. Straube and G. Steinberg) carrying a fusion between the strong Potef promoter, a nuclear localization sequence, a triple RFP gene (Potef-NLS-3RFP), and a carboxin resistance marker. These strains are designated FB1yap1-3XeGFPcbx:pANNE3090 and FB2yap1-3XeGFPcbx:pANNE3090. Single integrations in the carboxin locus were verified by DNA gel blot analysis.
DNA and RNA Procedures
Standard molecular techniques were used (Sambrook et al., 1989). Transformation of U. maydis was performed as published previously (Schulz et al., 1990). U. maydis DNA was isolated as described (Hoffman and Winston, 1987). RNA was isolated following the TRIZOL reagent protocol (Invitrogen). Probes for detecting transcripts from the peroxidase genes were generated by PCR and verified by diagnostic digestions. To generate the probe for the um01947 gene, a 0.9-kb fragment was generated using the primer combination 5′-TCTTGCTGTGCTTAAGCAGC-3′ and 5′-AAGAATGCTGCGAGAGAACG-3′. To generate the probe for the um03277 gene, a 0.9-kb fragment was generated using the primer combination 5′-AAGTTTTCAGCTGGGACACC-3′ and 5′-ATGGCGCCAATAAGTTCCAG-3′. To generate the probe for um10672 gene, a 1.0-kb fragment was generated using the primer combination 5′-TGTGTGCCTTGTTGCACTCG-3′ and 5′-TTCTGTCTTCTTGCACAAGC-3′. To generate the probe for the um11067 gene, a 0.8-kb fragment was generated using the primer combination 5′-TTCTCAAGCGCTGCGTTACC-3′ and 5′-AAGCTGGCGATGCGATTACC-3′. Probes were labeled by the PCR DIG labeling kit following the manufacturer's instructions (Roche).
Immunodetection
Immunoblot analysis was performed as described (Basse et al., 2000) using a monoclonal GFP IgG mouse antibody (Roche).
Sample Preparation and Microarray Analysis
Two independent overnight cultures of U. mayidis FB1 and FB1Δyap1 grown in CM-glucose (OD600 of 0.8) were diluted in 100 mL of the same medium (OD600 of 0.2) and grown at 28°C until an OD600 of 0.6. The cultures were divided, and one half was supplemented with 5 mM H2O2. After 1 h of exposition to H2O2, cells were harvested by centrifugation and frozen in liquid nitrogen. RNA extraction, purification, cDNA generation, purification, and labeling were performed according to standard protocols (Affymetrix). DNA array analysis was performed on two biological replicates each, using custom-designed Affymetrix chips (MPIUstilagoA). Data were analyzed using a GeneArray Scanner (Agilent/Affymetrix) and GeneChip Expression Analysis software (GCOS) Microarray Suite 5.0 (Affymetrix) as described (Eichhorn et al., 2006). Data analysis was performed using the Biconductor R package (http://www.bioconductor.org/) as described (Eichhorn et al., 2006). The P values for the coefficients/contrasts of interest were adjusted for multiple testing by the false discovery rate method (Benjamini and Hochberg, 1995). For the data set of Yap1-regulated genes, genes were filtered by applying the following criteria: genes should be at least 1.5-fold downregulated in the comparison of FB1Δyap1+ 5 mM H2O2 with FB1 + 5 mM H2O2 and have a corrected P value of <0.05 in the biological duplicates analyzed.
Calcofluor and Chlorazole Black E Staining
Calcofluor staining using Fluorescent Brightener 28 (Sigma-Aldrich) for the microscopy of prepenetration stages of U. maydis and Chlorazole Black E staining for microscopy of post-penetration stages were performed as described (Brachmann et al., 2003).
Histochemical Detection of H2O2
H2O2 production was visually detected in infected plants using DAB as substrate (Orozco-Cárdenas and Ryan, 1999). Briefly, plants were decapitated at the base of the stem with a razor blade and placed in a 1-mg/mL solution of DAB for 16 h under darkness at room temperature. Leaves were decolorized by immersion in ethanol (96%) for 48 h. Brown polymerization products that result from the reaction of DAB with H2O2 were microscopically identified in regions at least 1 cm above the immersed plant parts.
DPI Treatment
DPI solved in DMSO was added at final concentrations of 0.5 μM directly to the mixtures of compatible U. maydis strains prior to inoculation. As control, DMSO alone was added to the mixtures of compatible U. maydis strains.
Microscopy Observation
For microscopy observation, a Zeiss Axiophot microscope with differential interference contrast optics was used. Calcofluor fluorescence was observed with a standard 4‘,6’-diamidino-2-phenylindole filter set. GFP fluorescence was detected with a specific filter set (band-pass 470/20, beam splitter 493, band-pass 505 to 530 nm; Zeiss). RFP fluorescence was detected with standard filters for rhodamine. DAB precipitates were visualized by differential interference contrast optics. Pictures were taken with a CCD camera (Hamamatsu). Image processing was done with Image Pro (Media Cybernetics), Adobe Photoshop 6.0, and Canvas 6.0 (Deneba Systems).
Accession Numbers
Sequence data for yap1 can be found in the GenBank/EMBL data libraries under accession number BN000987. The GenBank accession numbers of the studied peroxidases are XP_758094 (um01947), XP_758524 (um02377), XP_760856 (um10672), and XP_759546 (um11067). The Gene Expression Omnibus number for array data included in this article is GSE7518.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Table 1. List of Genes Regulated by Yap1.
Supplementary Material
Acknowledgments
We thank S. Gut for assistance with respect to the S. cerevisiae complementation assay, K. Münch for technical assistance, and M. Vranes and J. Kämper for help with the array analysis. We also thank G. Döhlemann and A. Mendoza-Mendoza for critical reading of the manuscript and the members of our network for their constructive comments. We thank G. Steinberg for providing plasmid pANNE3090. This work was supported by the European Community Project (HPRN-CT-2002-00249).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Regine Kahmann (kahmann@mpi-marburg.mpg.de).
Online version contains Web-only data.
References
- Aguirre, J., Rios-Momberg, M., Hewitt, D., and Hansberg, W. (2005). Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13 111–118. [DOI] [PubMed] [Google Scholar]
- Alarco, A.M., and Raymond, M. (1999). The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel, K., and Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. [DOI] [PubMed] [Google Scholar]
- Apostol, I., Heinstein, P.F., and Low, P.S. (1989). Rapid stimulation of an oxidative burst during elicitation of cultured plant cells. Plant Physiol. 90 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada, K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. 50 601–639. [DOI] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1989). Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1996). Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development 122 2965–2976. [DOI] [PubMed] [Google Scholar]
- Basse, C.W. (2005). Dissecting defense-related and developmental transcriptional responses of maize during Ustilago maydis infection and subsequent tumor formation. Plant Physiol. 138 1774–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basse, C.W., Stumpferl, S., and Kahmann, R. (2000). Characterization of a Ustilago maydis gene specifically induced during the biotrophic phase: Evidence for negative as well as positive regulation. Mol. Cell. Biol. 20 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300. [Google Scholar]
- Billard, P., Dumond, H., and Bolotin-Fukuhara, M. (1997). Characterization of an AP-1-like transcription factor that mediates an oxidative stress response in Kluyveromyces lactis. Mol. Gen. Genet. 257 62–70. [DOI] [PubMed] [Google Scholar]
- Blokhina, O., Virolainen, E., and Fagerstedt, K.V. (2003). Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann. Bot. (Lond.) 91 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortfeld, M., Auffarth, K., Kahmann, R., and Basse, C.W. (2004). The Ustilago maydis a2 mating-type locus genes lga2 and rga2 compromise pathogenicity in the absence of the mitochondrial p32 family protein Mrb1. Plant Cell 16 2233–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, A., Konig, J., Julius, C., and Feldbrugge, M. (2004). A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272 216–226. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., Schirawski, J., Müller, P., and Kahmann, R. (2003). An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 22 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, E.G., Jesuino, R.S.A., Dantas, A.S., Brígido, M.M., and Felipe, M.S.S. (2005). Oxidative stress response in Paracoccidioides brasiliensis. Genet. Mol. Res. 4 409–429. [PubMed] [Google Scholar]
- Chen, S.X., and Schopfer, P. (1999). Hydroxyl-radical production in physiological reactions. A novel function of peroxidase. Eur. J. Biochem. 260 726–735. [DOI] [PubMed] [Google Scholar]
- Coleman, S.T., Epping, E.A., Steggerda, S.M., and Moye-Rowley, W.S. (1999). Yap1p activates gene transcription in an oxidant-specific fashion. Mol. Cell. Biol. 19 8302–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay, A., Isnard, A.D., and Toledano, M.B. (2000). H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19 5157–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke, N., Miura, Y., Sanchez, L.M., Park, H.J., Noritake, T., Yoshioka, H., and Kawakita, K. (1996). The oxidative burst protects plants against pathogen attack: Mechanism and role as an emergency signal for plant bio-defence. Gene 179 45–51. [DOI] [PubMed] [Google Scholar]
- Dumond, H., Danielou, N., Pinto, M., and Bolotin-Fukuhara, M. (2000). A large-scale study of Yap1p-dependent genes in normal aerobic and H2O2-stress conditions: the role of Yap1p in cell proliferation control in yeast. Mol. Microbiol. 36 830–845. [DOI] [PubMed] [Google Scholar]
- Eichhorn, H., Lessing, F., Winterberg, B., Schirawski, J., Kamper, J., Muller, P., and Kahmann, R. (2006). A ferroxidation/permeation iron uptake system is required for virulence in Ustilago maydis. Plant Cell 18 3332–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, L., Rodrigues-Pousada, C., and Struhl, K. (1997). Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 17 6982–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer, M.J., Oxborough, K., Mullineaux, P.M., and Baker, N.R. (2002). Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 53 1249–1254. [PubMed] [Google Scholar]
- Gash, A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounalaki, N., and Thireos, G. (1994). Yap1p, a yeast transcriptional activator that mediates multidrug resistance, regulates the metabolic stress response. EMBO J. 13 4036–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govrin, E.M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10 751–757. [DOI] [PubMed] [Google Scholar]
- Harshman, K.D., Moye-Rowley, W.S., and Parker, C.S. (1988). Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell 53 321–330. [DOI] [PubMed] [Google Scholar]
- He, X.J., and Fassler, J.S. (2005). Identification of novel Yap1p and Skn7p binding sites involved in the oxidative stress response of Saccharomyces cerevisiae. Mol. Microbiol. 58 1454–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata, D., Yano, K., and Miyakawa, T. (1994). Stress-induced transcriptional activation mediated by YAP1 and YAP2 genes that encode the Jun family of transcriptional activators in Saccharomyces cerevisiae. Mol. Gen. Genet. 242 250–256. [DOI] [PubMed] [Google Scholar]
- Hoffman, C.S., and Winston, F. (1987). A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57 267–272. [DOI] [PubMed] [Google Scholar]
- Holliday, R. (1974). Ustilago maydis. In Handbook of Genetics, Vol. 1, R.C. King, ed (New York: Plenum Press) pp. 575–595.
- Kämper, J. (2004). A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271 103–110. [DOI] [PubMed] [Google Scholar]
- Kämper, J., et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444 97–101. [DOI] [PubMed] [Google Scholar]
- Keon, J.P., White, G.A., and Hargreaves, J.A. (1991). Isolation, characterization and sequence of a gene conferring resistance to the systemic fungicide carboxin from the maize smut pathogen, Ustilago maydis. Curr. Genet. 19 475–481. [DOI] [PubMed] [Google Scholar]
- Kuge, S., Arita, M., Murayama, A., Maeta, K., Izawa, S., Inoue, Y., and Nomoto, A. (2001). Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21 6139–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge, S., and Jones, N. (1994). YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge, S., Jones, N., and Nomoto, A. (1997). Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 16 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalucque, H., and Silar, P. (2003). NADPH oxidase: An enzyme for multicellularity? Trends Microbiol. 11 9–12. [DOI] [PubMed] [Google Scholar]
- Lambeth, J.D. (2004). NOX enzymes and the biology of reactive oxygen. Nature Rev. Immunol. 4 181–189. [DOI] [PubMed] [Google Scholar]
- Lara-Ortíz, T., Riveros-Rosas, H., and Aguirre, J. (2003). Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50 1241–1255. [DOI] [PubMed] [Google Scholar]
- Lee, J., Godon, C., Lagniel, G., Spector, D., Garin, J., Labarre, J., and Toledano, M.B. (1999). Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274 16040–16046. [DOI] [PubMed] [Google Scholar]
- Lev, S., Hadar, R., Amedeo, P., Baker, S.E., Yoder, O.C., and Horwitz, B.A. (2005). Activation of an AP1-Like transcription factor of the maize pathogen Cochliobolus heterostrophus in response to oxidative stress and plant signals. Eukaryot. Cell 4 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac, F., Lalucque, H., Lepere, G., and Silar, P. (2004). Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41 982–997. [DOI] [PubMed] [Google Scholar]
- Morre, D.J. (2002). Preferential inhibition of the plasma membrane NADH oxidase (NOX) activity by diphenyleneiodonium chloride with NADPH as donor. Antioxid. Redox. Signal 4 207–212. [DOI] [PubMed] [Google Scholar]
- Moye-Rowley, W.S., Harshman, K.D., and Parker, C.S. (1989). Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 3 283–292. [DOI] [PubMed] [Google Scholar]
- Nathues, E., Joshi, S., Tenberge, K.B., von den Driesch, M., Oeser, B., Baumer, N., Mihlan, M., and Tudzynski, P. (2004). CPTF1, a CREB-like transcription factor, is involved in the oxidative stress response in the phytopathogen Claviceps purpurea and modulates ROS level in its host Secale cereale. Mol. Plant Microbe Interact. 17 383–393. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T., and Brunner, F. (2002). Innate immunity in plants and animals: Emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5 318–324. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 198 249–266. [DOI] [PubMed] [Google Scholar]
- Orozco-Cárdenas, M., and Ryan, C.A. (1999). Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 96 6553–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schulz, B., Banuett, F., Dahl, M., Schlesinger, R., Schafer, W., Martin, T., Herskowitz, I., and Kahmann, R. (1990). The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60 295–306. [DOI] [PubMed] [Google Scholar]
- Snetselaar, K.M., and Mims, C.W. (1994). Light and electron microscopy of Ustilago maydis hyphae in maize. Mycol. Res. 98 347–355. [Google Scholar]
- Tanaka, A., Christensen, M.J., Takemoto, D., Park, P., and Scott, B. (2006). Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell 18 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D.B. (1997). Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11 1187–1194. [Google Scholar]
- Torres, M.A., and Dangl, J.L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8 397–403. [DOI] [PubMed] [Google Scholar]
- Torres, M.A., Dangl, J.L., and Jones, J.D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone, W.M., and Jones, N. (1999). AP-1 transcription factors in yeast. Curr. Opin. Genet. Dev. 9 55–61. [DOI] [PubMed] [Google Scholar]
- Toone, W.M., Kuge, S., Samuels, M., Morgan, B.A., Toda, T., and Jones, N. (1998). Regulation of the fission yeast transcription factor Pap1 by oxidative stress: Requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 12 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone, W.M., Morgan, B.A., and Jones, N. (2001). Redox control of AP-1-like factors in yeast and beyond. Oncogene 20 2336–2346. [DOI] [PubMed] [Google Scholar]
- Vivancos, A.P., Castillo, E.A., Biteau, B., Nicot, C., Ayte, J., Toledano, M.B., and Hidalgo, E. (2005). A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc. Natl. Acad. Sci. USA 102 8875–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, M.J., Andrade, E.C., and Storz, G. (2003). The redox domain of the Yap1p transcription factor contains two disulfide bonds. Biochemistry 42 11982–11991. [DOI] [PubMed] [Google Scholar]
- Wu, A., Wemmie, J.A., Edgington, N.P., Goebl, M., Guevara, J.L., and Moye-Rowley, W.S. (1993). Yeast bZip proteins mediate pleiotropic drug and metal resistance. J. Biol. Chem. 268 18850–18858. [PubMed] [Google Scholar]
- Wu, A.L., and Moye-Rowley, W.S. (1994). GSH1, which encodes gamma-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol. 14 5832–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki, R., Fortier, P.K., Maciaszczyk, E., Thorsen, M., Leduc, A., Odhagen, A., Owsianik, G., Ulaszewski, S., Ramotar, D., and Tamas, M.J. (2004). Transcriptional activation of metalloid tolerance genes in Saccharomyces cerevisiae requires the AP-1-like proteins Yap1p and Yap8p. Mol. Biol. Cell 15 2049–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C., Lee, L.H., and Davis, L.I. (1998). Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 17 7416–7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.