Abstract
Hormones play a central role in the coordination of internal developmental processes with environmental signals. Herein, a combination of physiological, genetic, cellular, and whole-genome expression profiling approaches has been employed to investigate the mechanisms of interaction between two key plant hormones: ethylene and auxin. Quantification of the morphological effects of ethylene and auxin in a variety of mutant backgrounds indicates that auxin biosynthesis, transport, signaling, and response are required for the ethylene-induced growth inhibition in roots but not in hypocotyls of dark-grown seedlings. Analysis of the activation of early auxin and ethylene responses at the cellular level, as well as of global changes in gene expression in the wild type versus auxin and ethylene mutants, suggests a simple mechanistic model for the interaction between these two hormones in roots, according to which ethylene and auxin can reciprocally regulate each other's biosyntheses, influence each other's response pathways, and/or act independently on the same target genes. This model not only implies existence of several levels of interaction but also provides a likely explanation for the strong ethylene response defects observed in auxin mutants.
INTRODUCTION
Plants need to adjust most of their physiological and developmental processes to constantly changing environments. Different internal and external signals often converge on a set of plant hormones that are responsible for the execution of specific responses. With only a handful of plant hormones available, the large array of responses mediated by these compounds is probably achieved by a combinatorial mechanism of interactions between the hormones and other signals (Bennett et al., 2005). Although in the literature there are plenty of examples that highlight the importance of hormonal crosstalk in the control of particular processes (Gazzarrini and McCourt, 2003), only recently light was shed on some of the molecular mechanisms that underlie these relationships (Fu and Harberd, 2003; Lorenzo et al., 2003; Li et al., 2004; Nemhauser et al., 2004; Stepanova et al., 2005).
Ethylene and auxin have a long history of reported interactions both at the physiological and molecular level. The antagonistic effects of these hormones in the control of abscission of fruits and flowers (Brown, 1997) as opposed to their synergistic effects in the regulation of root elongation, root hair formation, and growth in Arabidopsis thaliana (Pitts et al., 1998; Rahman et al., 2002; Swarup et al., 2002) illustrate some of the intricacies of the ethylene–auxin crosstalk. Mutant analysis has uncovered additional levels of complexity in the relationship between these two hormones. For example, in etiolated seedlings grown in rich media, ethylene and auxin appear to control hypocotyl elongation independently (Collett et al., 2000), whereas inhibition of root growth by ethylene under the same conditions seems to require auxin (Swarup et al., 2002; this work). Conversely, when seedlings are grown in the light in low-nutrient media, the ethylene promotion of the hypocotyl elongation also becomes dependent on auxin homeostasis (Vandenbussche et al., 2003). Together, these physiological studies indicate that ethylene and auxin are able to interact in a number of different ways contingent on the cell type, developmental stage, and environmental conditions. This complexity is likely a manifestation of the similarly elaborate net of the underlying molecular mechanisms. Interactions at the biosynthetic, signaling, and response levels have previously been proposed to explain the large variety of hormone-mediated effects in plants (Alonso and Ecker, 2001; Gazzarrini and McCourt, 2003). Intense research efforts in the past 20 years have uncovered several of the key molecular components of the individual hormonal pathways (Gray, 2004), opening the possibility of exploring the cross-pathway interactions at the molecular level.
Ethylene is synthesized from the amino acid Met by the consecutive action of three enzymatic activities: S-adenosyl-l-methionine (SAM) synthase, 1-aminocyclopropane-1-carboxylic acid (ACC) synthase, and ACC oxidase. ACC synthase catalyzes the main regulatory step in this biosynthetic pathway, the conversion of SAM to ACC (Wang et al., 2002). Auxin is known to stimulate ethylene production by activating this particular biosynthetic step (Abel et al., 1995). In fact, transcription of eight out of the nine Arabidopsis ACS genes is upregulated by auxin, and canonical auxin response elements have been found in the promoters of several of these biosynthetic genes (Tsuchisaka and Theologis, 2004). Once produced, ethylene is sensed by a family of receptors that show similarity to the bacterial two-component His kinases. Ethylene binding to the receptors causes inactivation of a Raf-like kinase CTR1 and the consequent derepression of EIN2, a protein of unknown biochemical function that is essential for the ethylene response. Downstream of EIN2, a family of transcription factors composed of EIN3 and EIN3-like proteins triggers a transcriptional cascade that results in the activation/repression of hundreds of target genes (Alonso and Stepanova, 2004; Guo and Ecker, 2004). The first step in the cascade initiated by EIN3/EILs involves other transcription factors, such as the AP2/EREBP family members ERF1 (Solano et al., 1998) and EDF1-4 (Alonso et al., 2003b). These different EIN3 targets probably represent branching points in the ethylene response that can be turned on or off not only by ethylene but also by other factors and therefore could be used to provide specificity in response to ethylene. For example, the ethylene stimulation of defense responses requires simultaneous activation of ERF1 both by ethylene and jasmonate, another plant hormone involved in stress responses (Lorenzo et al., 2003). In the case of ethylene and auxin, several common target genes have also been identified (Zhong and Burns, 2003). It is not known, however, whether or not they participate in common biological processes and what type of interaction, if any, is responsible for these coregulations.
In contrast with the ethylene biosynthetic pathway, much less is known about the genes responsible for the biosynthesis of auxins (Cohen et al., 2003). The most common active auxin, indole-3-acetic acid (IAA), can be synthesized from Trp or from Trp precursors (Bartel, 1997). Although Trp is not a limiting element in the auxin production under normal conditions (Bartel, 1997), mutations in the anthranilate synthase subunits WEI2/ASA1/TIR7 and WEI7/ASB1 that catalyze the first committed step in Trp biosynthesis result in reduction of auxin levels (Ljung et al., 2005; Stepanova et al., 2005). Moreover, both subunits are transcriptionally regulated by ethylene and play a key role in the ethylene-mediated inhibition of root elongation (Stepanova et al., 2005). Plants can use several pathways in the conversion of Trp to IAA (Bartel, 1997); however, only few of the genes coding for the predicted enzymes have been identified: a YUCCA family of flavin-containing monooxygenases responsible for the conversion of tryptamine to N-hydroxyl-tryptamine (Mikkelsen et al., 2004), two cytochrome P450s, CYP450B2 and B3, that convert Trp to indole-3-acetaldoxime (Zhao et al., 2002), and six nitrilases that can catalyze the synthesis of IAA from indole-3-acetonitrile (Normanly et al., 1997; Arabidopsis Genome Initiative, 2000). Unlike ethylene that can travel through the plant by mere diffusion, IAA is transported by a complex net of carriers, such as AUX1, PIN1, and related proteins (Swarup and Bennett, 2003). Auxin presence in the cell is sensed by the F-box TIR1 and TIR1-like proteins (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Upon binding to the hormone, the SCFTIR1 complex is activated, resulting in the ubiquitination of the IAA transcriptional corepressors and their consequent degradation by the proteosome (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). The IAA proteins interact with auxin response factors (ARFs), forming inactive complexes (Leyser, 2002). The reduction in IAA proteins levels leads to the release of ARFs and the subsequent initiation of the auxin responses. The presence in the Arabidopsis genome of 29 IAA and 23 ARF genes (Arabidopsis Genome Initiative, 2000; Liscum and Reed, 2002) provides a possible combinatorial mechanism to generate a large diversity of responses activated by the same hormone. Characterization of several ARF mutants indicates a large degree of specialization among different family members, supporting the idea that a variety of auxin responses can be achieved by the activation of specific ARFs (Hardtke et al., 2004; Nagpal et al., 2005; Okushima et al., 2005).
In summary, ethylene and auxin are known to interact in the regulation of several biological processes, such as root elongation, differential growth of hypocotyls, and root hair formation (Stepanova and Alonso, 2005). Nonetheless, despite the accumulated knowledge on each individual hormone signaling and response pathway, very little is known about the mechanisms that govern the interactions between ethylene and auxin. The ethylene-mediated regulation of auxin biosynthesis through the activation of WEI2/ASA1 and WEI7/ASB1, as well as the reciprocal effect of auxin on ethylene biosynthesis through the activation of several ACC synthases, represent two elegant, but still anecdotally scarce, examples of the molecular mechanisms controlling the ethylene–auxin crosstalk. The rudimentary state of knowledge of the ethylene–auxin interactions reflects that of most of other hormones as well, where, in the best-case scenarios, single elements of the interaction have been identified (Abel et al., 1995; Li et al., 2004; Stepanova et al., 2005), but systematic analysis has not been performed. An exception from this general trend is the recent study of the relations between auxin and brassinosteroids in the regulation of hypocotyl elongation, where a multidisciplinary approach uncovered transcriptional regulation as a major node in this crosstalk (Nemhauser et al., 2004).
To better understand the molecular mechanisms behind the ethylene–auxin interactions, a comprehensive study relying on physiological, cellular, genetic, and genomic approaches was performed. Initial characterization of the morphological effects of the two hormones in different tissues and mutant backgrounds confirmed the intricate complexity of these relations and allowed for the selection of a robust experimental system. The use of early auxin and ethylene response reporter genes not only provided a more precise map of auxin-mediated ethylene responses at the cellular level, but also pointed to the existence of auxin-dependent ethylene effects. Analysis of the ethylene and auxin transcriptional responses at the genome level further supported the role of mutual biosynthesis regulation in the interaction between these hormones and indicated the presence of an additional mechanism, presumably a sensitization of the response to one hormone by the presence of the other hormone. Finally, examination of the functional annotation of the characterized genes and independent quantitative RT-PCR experiments validated the involvement of multilevel interactions in the crosstalk between ethylene and auxin.
RESULTS
Ethylene-Mediated Root Growth Inhibition Requires Normal Auxin Levels/Activity
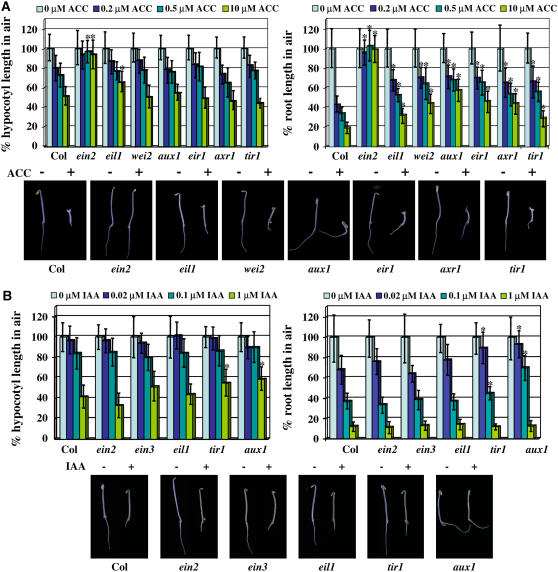
Using simple and widely used morphological assays, the role of auxin in the response to ethylene and, vice versa, the role of ethylene in the response to auxin were investigated. The effect of the two hormones on seedlings' growth in the dark has been extensively exploited previously, allowing for the identification of mutants with altered responsiveness to ethylene and/or auxin (Estelle and Somerville, 1987; Guzman and Ecker, 1990). At most concentrations, both hormones inhibit growth of hypocotyls and roots of young etiolated Arabidopsis seedlings (Stepanova et al., 2005). To investigate the reciprocal roles played by ethylene and auxin in the response to auxin and ethylene, respectively, we took advantage of the large battery of well-characterized ethylene and auxin mutants already available. Mutations that impair auxin transport (import into [aux1] or export from [eir1/pin2] the cells), perception (tir1), signaling (axr1), or biosynthesis (wei2/tir7) were found to result in reduced response to ethylene or its biosynthetic precursor ACC with respect to the effect of the hormone on root growth but not on hypocotyl elongation (Figure 1A). In aux1, where the auxin flux in roots is greatly impaired (Marchant et al., 1999), the ethylene effect on root growth was reduced to as little as 20 to 30% of the wild-type response level. Although we did not observe a significant alteration of the ethylene effect on hypocotyl length in any of the auxin mutants tested compared with the wild type (Figure 1), the aux1 mutant was found to form the characteristic exaggerated apical hook at a reduced frequency (data not shown).
Figure 1.
Auxin Mutants Display Root-Specific Ethylene Insensitivity.
(A) Relative organ size of 3-d-old etiolated seedlings grown in the presence of 0, 0.2, 0.5, and 10 μM ACC (ethylene precursor). The following genotypes were examined: Col-0 (wild type), ein2-5, ein3-1, eil1-1, wei2-1, aux1-7, eir1, axr1-12, and tir1-101. All of the mutants tested are in the Col-0 background. The response of each genotype to ethylene was expressed as the percentage of the organ length at a particular concentration of ACC with respect to the average length of that organ in the absence of the ethylene precursor. In the bottom panel, images of representative 3-d-old etiolated seedlings grown in the absence (−) or in the presence (+) of 10 μM ACC are displayed. Genotypes are as indicated.
(B) Relative organ size of 3-d-old etiolated seedlings grown in the presence of 0, 0.02, 0.1, and 1 μM IAA (auxin). The following genotypes were examined: Col-0 (wild type), ein2-5, ein3-1, eil1-1, tir1-101, and aux1-7. All of the mutants tested are in the Col-0 background. The response of each genotype to auxin was expressed as the percentage of the organ length at a particular concentration of IAA with respect to the average length of that organ in the absence of auxin. In the bottom panel, images of representative 3-d-old etiolated seedlings grown in the absence (−) or in the presence (+) of 0.1 μM IAA are displayed. Genotypes are as indicated.
An asterisk indicates a P value < 0.0001 (analysis of variance).
By contrast, mutants thought to be completely insensitive to ethylene, such as ein2-5, were found to show normal or near normal response to exogenous auxin (Figure 1B). Similarly, the auxin response of partially ethylene-resistant mutants, such as ein3-1 and eil1-1, was not significantly different from that of wild-type plants (Figure 1B). Together, these results suggest that normal levels of auxin biosynthesis, transport, and/or response are required for the growth inhibitory effect of ethylene in roots but not in hypocotyls. Conversely, ethylene is not required for the auxin-mediated inhibition of hypocotyl and root growth in etiolated seedlings.
High Levels of Auxin Activity in the Transition Zones Are Required for the Ethylene Effects on Root Growth
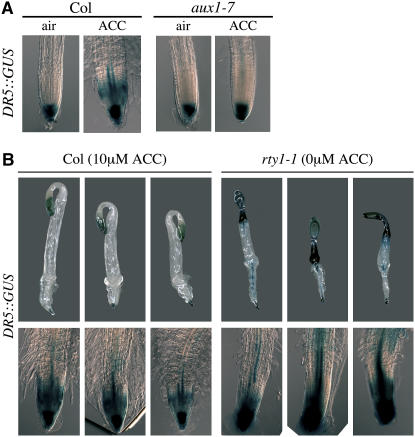
To investigate the mechanisms by which auxin mediates and/or promotes the ethylene response in roots, we examined the effects of ethylene on the expression of the auxin reporter DR5:β-glucuronidase (GUS) in wild-type plants and aux1, which is defective in an auxin influx carrier and shows a dramatic reduction in the response to ethylene (Figure 1A).
In wild-type roots, DR5:GUS is expressed in the quiescent zones and surrounding columella cells of root tips (Figure 2A), herein referred to as zone 1, according to the classification by Birnbaum et al. (2003) (Figure 7B). Treatment with exogenously supplied ethylene or its precursor ACC promotes an increase in the levels of DR5:GUS in zone 1, as indicated by the expansion of the area (or, at lower incubation times, higher intensity) of staining (Figure 2A) (Stepanova et al., 2005; data not shown). An increase in the DR5:GUS activity was also observed in the transition zone (or zone 2) of seedlings' roots, with the DR5:GUS staining never reaching into the root cells of the fast elongation zone (or zone 3) (Birnbaum et al., 2003) (Figure 2A). In the aux1 mutant, where the morphological effects of ethylene are dramatically reduced compared with the wild type (Figure 1A), the ethylene-mediated increase in the levels of DR5:GUS is strictly limited to cells of zone 1 and does not involve the transition zone (or zone 2) (Figure 2A). In fact, the cells in zone 1 of aux1 show levels of DR5:GUS activity similar to those of wild-type seedlings, both in the presence and in the absence of the ethylene precursor ACC (Figure 2A). By contrast, unlike wild-type plants, ACC-treated aux1 seedlings do not display any detectable DR5:GUS activity in the root transition zones (zone 2).
Figure 2.
Expression of DR5:GUS in Root Transition Zones Correlates with Total Root Length.
(A) Expression patterns of the DR5:GUS auxin reporter in roots of 3-d-old etiolated Col-0 and aux1-7 seedlings grown in the absence (air) or presence (ACC) of 10 μM ACC. GUS staining was performed overnight. Imaging conditions were identical for all of the plants in the experiment. The images displayed are representative of at least three independent experiments with >20 seedlings examined per experiment.
(B) Expression patterns of the DR5:GUS auxin reporter in 3-d-old etiolated Col-0 seedlings grown in the presence of 10 μM ACC and rty1-1 seedlings grown in the absence of ACC. The top panel depicts three representative seedlings per genotype/treatment, while the bottom panel shows enlarged images of corresponding root tips. The images displayed are representative of two independent experiments.
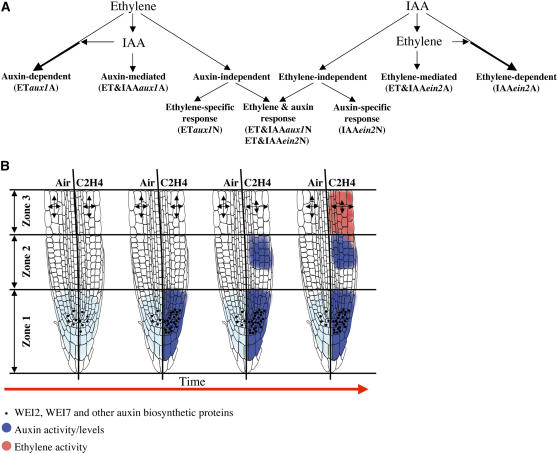
Figure 7.
Schematic Representation of the Mechanistic Model of Ethylene–Auxin Crosstalk in Roots of Etiolated Arabidopsis Seedlings.
(A) The model assumes existence of at least three different types of molecular interactions between ethylene and auxin. A subset of ethylene responses (left side of the panel) is dependent on auxin levels (ETaux1A). In this case, the role of auxin is restricted to promoting (or attenuating) the ethylene effect. By contrast, the auxin-mediated responses (ET&IAAaux1A) correspond to those changes in gene expression that are directly triggered by auxin, but in this case, by an ethylene-induced auxin activity. Finally, those ethylene effects that are not affected by the levels of auxin are classified as auxin independent, with some of these changes being independently stimulated by auxin (ET&IAAaux1N). Equivalent interactions can be defined among auxin responses (right side of the panel).
(B) The molecular interactions postulated above can be integrated with the morphological and cellular observations in a spatial/temporal model of the ethylene responses. From left to right, the time progression of the effects of ethylene on the levels of auxin activity (shown in blue), auxin biosynthetic genes (depicted as black dots), and auxin-dependent ethylene responses (marked in red) is indicated. At time 0 (before starting the ethylene treatment), the levels of auxin activity are low (shown in light blue) and are concentrated in the root zone 1. When ethylene is applied, the levels of WEI2/ASA1/TIR7, WEI7/ASB1, and potentially other biosynthetic genes (shown as black dots) increase, and the activity of auxin in zone 1 goes up. Next, the auxin activity in zone 2 becomes elevated, presumably through an AUX1-dependent transport activity from zone 1. This boost in auxin levels leads to changes in growth pattern of the cells in zone 3 from longitudinal to radial and to stimulation of ethylene responses in zone 3. Each root shown in this model is divided into two parts, with the left side of the root representing untreated roots, and the right side depicting ethylene-treated roots. Arrows in zone 3 of the roots indicate the longitudinal and radial components of root elongation. Zones 1, 2, and 3 are defined according to Birnbaum et al. (2003).
These results indicate that ethylene triggers activation of the DR5:GUS reporter expression, presumably as a reflection of an increase in auxin signaling and response in the cells of root transition zone upon exposure to ethylene, and that the DR5:GUS levels in these cells correlate with the degree of ethylene-triggered growth inhibition. By contrast, no correlation between the degree of ethylene sensitivity and the DR5:GUS activity was observed in zone 1 of the root, suggesting that the auxin effective in mediating the ethylene-stimulated growth inhibition is the auxin in the root transition zone.
This observation, together with the phenotypic measurements described above, suggests that the ethylene-mediated inhibition of root growth is dependent on the increase in the levels of auxin signaling in the cells of the root transition zone. Since auxin can inhibit root growth even in the absence of ethylene signaling, as was shown for ein2 plants treated with auxin (Figure 1B), it is possible that ethylene inhibits root growth by increasing the levels of auxin in specific regions of roots, the root transition zones, and that this increase in auxin prevents the elongation of these cells at a later developmental stage once the cells become part of the zone of fast elongation. Alternatively, it is also possible that this transient increase in auxin in the transition zone is a prerequisite for the ethylene-mediated inhibition of root growth but is not by itself sufficient to account for the totality of the ethylene effects on root growth. To test this second possibility, we compared the levels of DR5:GUS in root transition zones of ethylene-treated wild-type plants and those of the auxin-overproducing mutant rty1-1 grown in air. While the DR5:GUS activity in the roots of air-grown rty1-1 seedlings was clearly higher than that of ethylene-treated wild-type plants, the overall root length of these rty1-1 plants was greater than that of ethylene-treated wild-type seedlings (King et al., 1995; Figure 2B). These results suggest that although the elevated levels of auxin signaling and response in ethylene-treated plants are likely to be responsible for a part of the growth inhibitory effect of ethylene, they are not sufficient to account for the totality of this effect.
A Transient Increase in Auxin Levels in Root Transition Zones Is Required for Full Activation of EIN3 in Root Elongation Zones
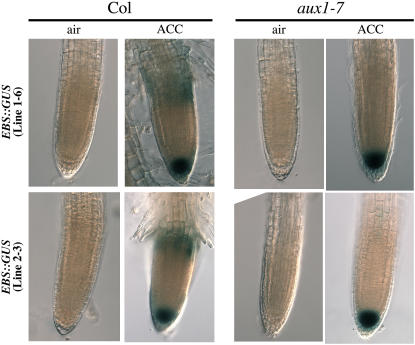
Next, we investigated whether or not the aforementioned increase in auxin (as judged by the elevated activity of DR5:GUS) in root transition zones could play additional roles in the response to ethylene. To test this possibility, the levels of expression of another reporter construct, EBS:GUS, in which the GUS reporter gene is driven by a synthetic EIN3-responsive promoter (A.N. Stepanova and J.R. Ecker, unpublished data), were examined. Several independent transgenic lines were generated in the Columbia (Col) background, and two representative lines were used in this study. To avoid potential artifacts due to positional effects of the transgenes, these two independent EBS:GUS lines were introgressed into the aux1 mutant background by crossing. In the absence of ethylene, no detectable expression of EBS:GUS in roots of wild-type or aux1 plants was observed, whereas ethylene treatment resulted in a dramatic increase in the activity of this ethylene reporter in root tips of both wild-type and aux1 seedlings (Figure 3). The lack of EBS:GUS expression in root tips of untreated plants, as well as in the transition zones of ethylene-treated wild-type plants where the levels of auxin activity are high (Figure 2A), suggests that high levels of auxin are not sufficient to activate the expression of this ethylene reporter. Furthermore, exogenous auxin treatment did not stimulate GUS activity in these plants (data not shown). Importantly, treatment of wild-type but not of aux1 plants with exogenous ethylene was able to induce activity of EBS:GUS in the cells of root elongation zones (Figure 3). A plausible interpretation of these results is that the increase in auxin signaling observed in the transition zones of ethylene-treated wild-type seedlings is required for the activation of the ethylene response in these cells later on in development, once they reach the elongation zone. The reduced auxin transport from the root apex to the transition zones in aux1 not only prevents the accumulation of DR5:GUS in response to ethylene but also that of EBS:GUS in the elongation zones. Thus, the ethylene-stimulated increase in the levels of auxin in root transition zones is not only required for the auxin-mediated inhibition of root growth but also appears necessary for the sensitization of these cells to other ethylene responses.
Figure 3.
Expression Pattern of the Ethylene Reporter EBS:GUS Is Altered in the aux1 Mutant Plants.
Activity of the EBS:GUS ethylene reporter in roots of 3-d-old etiolated seedlings grown in media not supplemented (air) or supplemented (ACC) with 10 μM ACC. Transgenic lines carrying the EBS:GUS construct were originally generated in Col-0 plants and then introgressed into aux1-7 by crossing. Two representative lines in both Col and aux1-7 backgrounds are shown. GUS staining was performed overnight. Imaging conditions were identical for all of the plants in the experiment. The images displayed are representative of at least three independent experiments with >20 seedlings examined per experiment.
In conclusion, the results described above are consistent with a mechanistic model in which an ethylene-mediated increase in the activity levels of auxin signal in the cells of root transition zones is directly responsible for part, but not all, of the growth inhibitory effect of ethylene. In addition to this direct auxin effect on cell elongation, the ethylene-mediated increase in auxin signaling is also required for sensitizing to ethylene the cells leaving the transition zones. Hence, this model predicts three types of ethylene responses: (1) auxin-mediated responses, such as part of the growth inhibition, in which the ethylene effects are an indirect result of the increase in auxin, (2) auxin-dependent responses, such as the activation of EBS:GUS in root elongation zones, in which the activation by ethylene, although direct, is modulated by the status of the auxin pathway, and (3) auxin-independent responses, such as part of the growth inhibitory effect uncovered by the rty1-1 analysis, in which the ethylene responses are not affected by the levels of auxin.
Gene Expression Analysis Supports the Existence of Several Levels of Interaction between Ethylene and Auxin
To further explore the three types of ethylene responses (mediated by, dependent on, or independent of auxin), global changes in gene expression profiles in response to ethylene were examined in roots of wild-type and aux1 mutant seedlings. In addition, because of the well-known stimulatory effect of auxin on ethylene biosynthesis (Abel et al., 1995; Woeste et al., 1999), the reciprocal possibility of ethylene-mediated and/or ethylene-dependent auxin responses was tested by comparing the auxin effects in roots of wild-type and ein2 mutant seedlings. In these studies, Affymetrix arrays covering >22,000 Arabidopsis genes were employed.
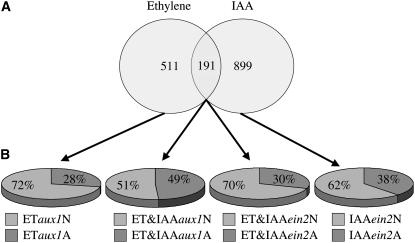
First, genes regulated by ethylene and/or auxin in wild-type plants were identified. Using a linear model approach (P < 0.05, fold change >1.5; Smyth, 2004), genes potentially regulated by these hormones were identified. To reduce the number of false negatives among genes regulated by the two hormones, a low-stringency/high-sensitivity false discovery rate cutoff of 0.15 was adopted. Using these criteria, 702 and 1090 genes were found to be regulated by ethylene and auxin, respectively, in roots of wild-type plants (Figure 4; see Supplemental Table 1 online). Comparison between these two sets of genes indicated that 191 genes were significantly affected by both hormones (27 and 18% of all the ethylene- and auxin-regulated genes, respectively). This is in agreement with the recent study by Nemhauser et al. (2006) that analyzed the effect of several different hormones on whole seedlings and also came to the conclusion that the overlap between the groups of genes regulated by different hormones is relatively small (33% of the ethylene- and 23% of the auxin-regulated genes).
Figure 4.
Ethylene and Auxin Alter Gene Expression in Both Hormone-Dependent and Hormone-Independent Ways.
(A) Venn diagram showing the number of genes regulated by ethylene and auxin in roots of wild-type Col-0 plants and the overlap between these two groups of genes.
(B) Graphic representation of the percentages of genes that had an altered response to ethylene and/or auxin in the aux1 and/or ein2 mutant backgrounds compared with that in the wild type. Pie diagrams, from left to right, display the percentages of ethylene-regulated genes that had an altered response to this hormone in the aux1 mutant plants, percentages of genes regulated by both ethylene and auxin that had an altered response to ethylene in aux1 mutant plants, percentages of genes regulated by both ethylene and auxin that had an altered response to auxin in the ein2 mutant plants, and percentage of auxin-regulated genes that had an altered response to this hormone in the ein2 mutant plants.
To identify genes involved in the three types of ethylene and auxin responses described above (i.e., dependent on, mediated by, and independent of the other hormone), we compared the ethylene and auxin effects in the wild type to those in the aux1 and ein2 mutants, respectively. In other words, wild-type and ein2 mutant seedlings were treated with auxin, and wild-type and aux1 seedlings were treated with ethylene. Eight different gene groups were defined (Figure 7A, Table 1). Genes that are regulated by ethylene, but not by auxin, and in which the ethylene effect is altered in the aux1 mutant background (ethylene genes altered in aux1, or ETaux1A) or in which aux1 had no effect (ethylene genes not altered in aux1, or ETaux1N) were identified. Similarly, genes that are regulated by IAA, but not by ethylene, and in which the auxin effect was (IAAein2A) or was not (IAAein2N) altered by the ein2 mutation were found. Finally, four additional groups of genes were categorized as those regulated both by ethylene and auxin (ET&IAA) and in which the ethylene effect was (ET&IAAaux1A) or was not (ET&IAAaux1N) altered in aux1, or the auxin effect was (ET&IAAein2A) or was not (ET&IAAein2N) altered in ein2. A correspondence between these gene groups and the different types of responses described above (independent of, dependent on, and mediated by the hormone) can be found (Table 1). For example, the ETaux1N and ET&IAAaux1N gene groups represent putative auxin-independent ethylene responses (i.e., genes whose expression becomes induced or repressed in response to ethylene independently of the levels of auxin in the cell). On the other hand, the ETaux1A class includes auxin-dependent ethylene-responsive genes, whose regulation by ethylene is altered in the aux1 background and therefore is likely to be modulated by auxin. Finally, the ET&IAAaux1A class corresponds to putative auxin-mediated ethylene responses, in other words, responses that are triggered by the auxin generated during the ethylene treatment. Equivalent nomenclature was established for the auxin-regulated genes. Analysis of the four ET&IAA groups suggested no clear correlation between genes affected or not by the aux1 and ein2 mutations. In other words, being categorized as aux1N (or aux1A) did not affect the probability of also being grouped as ein2A or ein2N (data not shown).
Table 1.
Description of the Different Gene Groups and the Types of Interactions They Represent
| Group Name | Criteria Used to Assign a Gene to the Group | Type of Interaction the Group Represents |
|---|---|---|
| ETaux1N | Genes are regulated by ethylene but not by IAA in Col. The ethylene effect in Col and aux1 mutants is similar. | Ethylene-regulated auxin independent. |
| ETaux1A | Genes are regulated by ethylene but not by IAA in Col. The ethylene effect in Col is different from that in aux1 mutants. | Ethylene-regulated auxin dependent. |
| IAAein2N | Genes are regulated by IAA but not by ethylene in Col. The IAA effect in Col and ein2 mutants is similar. | Auxin-regulated ethylene independent. |
| IAAein2A | Genes are regulated by IAA but not by ethylene in Col. The IAA effect in Col is different from that in ein2 mutants. | Auxin-regulated ethylene dependent. |
| ET&IAAaux1N | Genes are regulated by both ethylene and IAA in Col. The ethylene effect in Col and aux1 mutants is similar. | Ethylene- and auxin-regulated genes, but the ethylene effects are auxin independent. |
| ET&IAAaux1A | Genes are regulated by both ethylene and IAA in Col. The ethylene effect in Col is different from that in aux1 mutants. | Ethylene- and auxin-regulated genes, but the ethylene effects are likely to be auxin mediated. |
| ET&IAAein2N | Genes are regulated by both ethylene and IAA in Col. The IAA effect in Col and in ein2 mutants is similar. | Ethylene- and auxin-regulated genes, but the auxin effects are ethylene independent. |
| ET&IAAein2A | Genes are regulated by both ethylene and IAA in Col. The IAA effect in Col is different from that in ein2 mutants. | Ethylene- and auxin-regulated genes, but the auxin effects are likely to be ethylene mediated. |
It was found that 28% of the genes that were regulated by ethylene but not by auxin in our experimental conditions had an altered ethylene response in aux1 (ETaux1A) (Figure 4B; see Supplemental Table 1 online), providing additional evidence for the existence of auxin-dependent ethylene responses. Similarly, the response to auxin of 38% of the auxin-regulated genes was affected by ein2 (IAAein2A) (Figure 4B). These results not only support our previous observations involving EBS:GUS, suggesting that certain levels of auxin are required for the activation of some ethylene responses (auxin-dependent responses), but also that below certain threshold levels of ethylene signaling the response to auxin is altered.
In addition to uncovering this sensitization phenomenon, the analysis of the aux1 and ein2 effects on the expression of genes regulated by both hormones (ET&IAA) suggests an important role of the mutual regulation of the levels/activity of these two hormones in their interaction. Thus, for example, it was found that ∼50% of the genes in the ET&IAA category were differentially regulated by ethylene in the aux1 mutant background (ET&IAAaux1A) (Figure 4B; see Supplemental Table 1 online). This is much higher than the 30% of genes that were found to belong to the ET&IAAein2A category, suggesting a predominant role of ethylene-induced auxin biosynthesis/activity levels in mediating some of the ethylene responses in seedlings' roots.
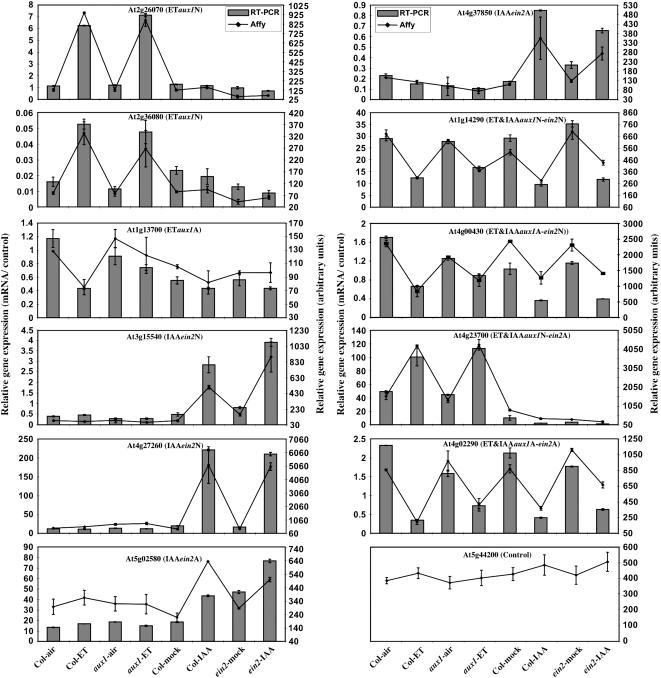
To further confirm the existence of the eight gene categories described above, 12 genes (one to two genes per category plus a housekeeping gene as a control) were selected and their expression patterns were examined by quantitative RT-PCR using RNA from a new biological replicate as a template (see Methods). Figure 5 shows that a very good correlation between the expression patterns obtained in the microarray experiments and quantitative RT-PCR was observed, although overall, the magnitude of expression level changes in the latter case appeared to be greater.
Figure 5.
The Different Levels of Interactions Observed in the Microarray Experiments Are Confirmed by Quantitative RT-PCR.
Relative expression levels of 12 selected genes (one or two per gene category) are shown. Graphs display the average and se of the normalized expression levels obtained in the microarray experiments (dots connected by lines) or in quantitative RT-PCR (solid bars). The microarray experiments and the quantitative RT-PCR were performed using RNA from independent biological replicates. The genes tested and their corresponding interaction categories (ETaux1N, ETaux1A, etc.) are as indicated. The genotypes and treatments are displayed on the horizontal axes of the bottom panels.
Taken together, these results are consistent with the experiments described above that relied on ethylene and auxin reporter genes and phenotypic measurements of the morphological effects of these two hormones. Our findings support the existence of at least three types of interactions between ethylene and auxin: one hormone sensitizing or conditioning the cells for the response to the other hormone (i.e., ethylene-dependent auxin responses and auxin-dependent ethylene responses), one hormone activating the biosynthesis (or signaling activity) of the other (i.e., ethylene-mediated auxin responses and auxin-mediated ethylene responses), and, finally, both hormones acting independently on the same subset of genes (Figure 7A) (i.e., ethylene-independent auxin responses and auxin-independent ethylene responses).
Promoter Analysis Supports the Existence of Both Direct and Ethylene-Dependent/Mediated Auxin Responses
The responses to ethylene and auxin are known to involve a very important transcriptional component; in fact, key transcription factors in the regulation of ethylene and auxin responses have been identified (Chao et al., 1997; Guilfoyle et al., 1998). In the case of ethylene, most (if not all) of ethylene responses are channeled through the plant-specific EIN3 and EIN3-Like (EIL) family of transcription factors. These transcription factors are known to regulate the expression of other transcription factors belonging to the AP2/EREBP family (Alonso et al., 2003a; Binder et al., 2004). Consensus binding sequences for both EIN3/EIL and AP2/EREBP have been identified (Solano et al., 1998). Similarly, the auxin response is known to be mediated by the degradation of the transcriptional repressors of the AUX/IAA family, releasing the ARF transcription factors from the AUX/IAA repressor complexes and allowing ARFs to bind to specific DNA elements in the promoters of early auxin-regulated genes (Guilfoyle et al., 1998). Like in the ethylene case, the DNA sequence recognized by the ARFs has been identified (Ulmasov et al., 1997). Since both ARFs and EIN3/EILs and AP2/EREBPs are considered to regulate early events in the auxin and ethylene responses, respectively, the presence of the elements recognized by these transcription factors in the promoters of a group of genes could be used as an additional diagnostic tool in the classification of the ethylene and auxin response genes.
No significant enrichment was found for the EIN3/EIL or the AP2/EREBP target DNA elements in any of the gene groups defined above. This could be explained by the fact that the ethylene response involves a transcriptional cascade (Solano et al., 1998) and that most of the genes that are regulated by this hormone are not directly regulated by EIN3/EIL. Moreover, it also suggests that in addition to AP2/EREBPs, other types of transcription factors may also be activated by EIN3 or AP2/EREBPs themselves. One could easily imagine how this transcriptional cascade could be used to achieve specificity in the ethylene response by activating or repressing some of these downstream branches under different conditions.
Conversely, a significant enrichment for the presence of the consensus ARF biding site (TGTCTC) was observed among genes that were similarly regulated by auxin in both Col and ein2 mutants (IAAein2N) (P < 0.05) (Table 2) but not among the IAAein2A or any other of the gene groups described above. These results are coherent with our expectations that the IAAein2N genes represent direct auxin responses and therefore should be enriched for the presence of ARF binding site, whereas, for example, genes representing ethylene-mediated auxin responses (ET&IAAein2A) should not. The lack of enrichment for the ARF binding site among genes representing ethylene-dependent auxin responses (IAAein2A) indicates that the majority of these genes do not correspond to early auxin responses and, therefore, that the interaction between ethylene and auxin in the regulation of these ethylene-dependent auxin responses happens downstream of the Aux/IAAs and ARFs. The common theme emerging from the results described above is that ethylene and auxin regulate mostly independent processes or, in other words, largely nonoverlapping sets of genes. However, detailed mutant analyses also found clear evidence in support of one hormone signaling status affecting some of the other hormone-regulated processes (sets of genes) and in support of one hormone mediating response to the other hormone, most likely due to the mutual regulation of each other's biosynthetic pathways.
Table 2.
Relative Abundance of ARF Binding Sites in the Promoters of Different Gene Groups
| Group Name | P Valuea | Number of Observed Genes | Number of Expected Genes | Percentage of Genes in the Group That Contains an ARF Binding Site |
|---|---|---|---|---|
| ET | 1.00 | 77 | 62.63 | 15.30% |
| ETaux1A | 1.00 | 25 | 17.71 | 17.60% |
| ETaux1N | 1.00 | 52 | 45.04 | 14.40% |
| IAA | 0.33 | 149 | 110.40 | 16.70% |
| IAAein2A | 1.00 | 43 | 42.74 | 12.60% |
| IAAein2N | 0.01 | 106 | 68.07 | 19.30% |
| ET&IAA | 1.00 | 22 | 23.63 | 11.10% |
| ET&IAAaux1A | 1.00 | 12 | 12.50 | 12.90% |
| ET&IAAaux1N | 1.00 | 10 | 12.50 | 10.42% |
| ET&IAAein2A | 1.00 | 6 | 7.13 | 10.50% |
| ET&IAAein2N | 1.00 | 16 | 16.50 | 12.10% |
Multiple sampling-corrected probability.
Gene Functional Analysis Supports the Predominant Role of Auxin-Mediated Ethylene Responses in the Control of Root Growth
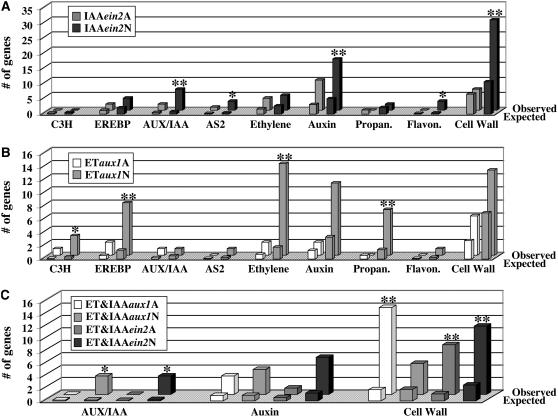
To investigate whether or not specific functions could be assigned to the gene groups defined in this study, we used the publicly available MapMan software to examine their putative functional classification. We found a significant enrichment for four functional categories (transcription factors, hormone metabolism, secondary metabolism, and cell wall modification) in one or more of the gene groups.
The AUX/IAA family of transcriptional corepressors was significantly enriched among the group of IAAein2N genes (representing auxin responses that are ethylene independent), but no enrichment was found in the IAAein2A group (representing auxin responses that are ethylene dependent) (Figure 6A). Again, these results suggest that the IAAein2N group of genes represents early steps in the auxin response. This finding is also consistent with the aforementioned observation of the IAAein2N group being significantly enriched for genes with ARF binding elements in their promoters.
Figure 6.
Gene Functional Analysis Supports the Existence of Hormone-Specific Effects and the Effects Mediated by the Interaction between Ethylene and Auxin.
Several gene function categories were found to be significantly enriched in one of the ethylene- and/or auxin-regulated gene groups. The MapMan software and the corresponding gene function databases were used to determine the significance of the enrichment and the number of observed and expected genes in each functional group (see Methods for more details). ** and * indicate a P value < 0.0001 (with the * marking functional categories containing ≤3 genes).
(A) Comparison between the number of genes (observed versus expected) in the following functional categories of auxin-regulated genes: C3H (C3H zing finger family of transcription factors), EREBP (AP2/EREBP family of transcription factors), IAA (AUX/IAA gene family), AS2 (family of transcription factors related to AS2), ethylene (genes annotated as ethylene related or ethylene metabolism), auxin (genes annotated as auxin related or auxin metabolism), propan. (genes annotated as secondary metabolism, phenylpropanoids), flavon. (genes annotated as secondary metabolism, flavonols), and cell wall (cell wall metabolism genes).
(B) Comparison between the numbers of observed and expected genes in the same functional categories as in (A) but among ethylene-regulated genes.
(C) Comparison between the number of observed and expected genes in the following functional categories of ethylene- and auxin-regulated genes: IAA (AUX/IAA gene family), auxin (auxin-related or auxin metabolism-related genes), and cell wall (cell wall metabolism genes).
Remarkably, in addition to the auxin-related AUX/IAA genes, the ASYMMETRIC LEAVES2 (AS2) family of transcription factors (Semiarti et al., 2001) was also enriched in the IAAein2N gene class (Figure 6A). Although without knowing the exact function of the auxin-regulated AS2-related genes it is not possible to evaluate the physiological significance of this finding, it suggests a tight and previously unknown connection between auxin response in roots and this family of transcription factors.
Enrichment was also found for the AP2/EREBP transcription factor family among the genes in the ETaux1N group (representing ethylene responses that are auxin independent) (Figure 6B). Several members of this gene family are known to participate in early ethylene responses (Solano et al., 1998; Alonso et al., 2003b); therefore, these results are consistent with the proposed idea that this class of genes represents responses that are directly regulated by ethylene and are not mediated by auxin. In addition to the AP2/EREBP gene family that has been previously implicated in the ethylene response, we also found a C3H zinc finger family of transcription factors being significantly enriched among the ETaux1N genes (Figure 6B), implying that it may play a role in the regulation of some yet uncharacterized downstream branches of the ethylene response.
Another functional group in which enrichment was observed was, not surprisingly, the hormone metabolism group. As expected, enrichment for the ethylene-related genes was only observed among the ETaux1N genes (Figures 6B), while auxin-related genes were significantly enriched in the IAAein2N and, to a smaller degree, in the ET&IAAein2N group (Figures 6A and 6C). As elaborated above, this suggests that the ETaux1N and IAAein2N groups represent genes directly and specifically involved in the ethylene and auxin responses, respectively.
Connection between ethylene, auxin, and secondary metabolites has been reported previously. For example, the regulation of auxin transport by flavonols (Peer et al., 2004) and the role of phenylpropanoids in ethylene-mediated responses against pathogens (Ecker and Davis, 1987; La Camera et al., 2004) are well documented. Analysis of our microarray data suggests that auxin and ethylene regulate the production of flavonols and phenylpropanoids, respectively, via changes in the transcription of potentially key biosynthetic genes (Figures 6A and 6B). Since enrichment for flavonol-related genes was observed only among IAAein2N and the enrichment for phenylpropanoid genes was seen only among ETaux1N, we suggest that ethylene and auxin act independently in the regulation of these pathways.
The last functional category in which enrichment was observed was that of cell wall–related genes (Figure 6C). As discussed above, the ethylene effect on root growth is largely mediated by an increase in the levels of auxin signaling and response in roots, presumably due to an ethylene-mediated increase in auxin biosynthesis and transport (Ljung et al., 2005; Stepanova et al., 2005; Růžička et al., 2007; Swarup et al., 2007). Based on this, the expectation would be that the genes involved in cell expansion would be regulated by both ethylene and auxin, but the ethylene effect on these genes would be mediated by auxin. This prediction was indeed reflected in the distribution of cell wall genes in the different ET, IAA, and ET and IAA gene groups. As shown in Figure 6, cell wall genes are enriched among genes that are regulated only by auxin and are not affected by ein2 (IAAein2N) and among genes that are both ethylene and auxin regulated (the ET&IAA class; Figures 6A and 6C). Moreover, the most dramatic enrichment for cell wall–related genes was observed in the ET&IAAaux1A gene category, in other words, among genes whose regulation by ethylene is likely to be mediated by auxin (Table 1). Although not as strong, some enrichment for the cell wall category was observed in the ET&IAAein2N and ET&IAAein2A groups (Figure 6C), consistent with the idea that part of the ethylene effect on root growth is auxin independent. The existence of this auxin-independent ethylene effect was previously suggested by the high levels of DR5:GUS expression and the long root phenotype of the rty1-1 mutant compared with ethylene-treated wild-type seedlings (Figure 2B).
DISCUSSION
Hormones participate in the regulation of almost every aspect of the plant life cycle and do so with an unusual functional plasticity. For example, it is not atypical to find that the same hormone controls a variety of completely different biological processes, yet at the same time, a particular biological process is similarly affected by several hormones (Gazzarrini and McCourt, 2003). Therefore, it is not possible to generally define a hormone function without specifying the tissue type, developmental stage, and environmental conditions. In other words, understanding the function of a hormone requires comprehension of a complex network of interactions between multiple signals. To gain insights into the molecular mechanisms involved in the signal crosstalk in plants, we have chosen two hormones, ethylene and auxin, and explored their role in the control of root growth in Arabidopsis.
Physiological studies have previously shown an interesting interaction between ethylene and auxin in the control of hypocotyl elongation in light-grown plants (Smalle et al., 1997). Under the conditions used in these assays, ethylene does not inhibit but rather promotes hypocotyl growth, and this effect is dependent on auxin (Vandenbussche et al., 2003). On the other hand, the interaction between ethylene and auxin in the control of hypocotyl elongation under dark conditions is less clear. Early studies clearly showed that in the hypocotyls of dark-grown seedlings, the inhibitory effects of auxin were not mediated by ethylene, but the reciprocal question of the role of auxin in the ethylene-mediated effect was not investigated (Collett et al., 2000). With respect to the interaction between ethylene and auxin signals in roots, several independent studies have reported a dramatically altered responsiveness of auxin mutants to ethylene (Swarup et al., 2002). However, in spite of the general awareness in the plant hormone community of this phenomenon, it has not been clearly established whether these examples were representative of a particular subclass of auxin mutants or, vice versa, reflected a common phenomenon. Even more importantly, it remained unclear what the underlying molecular mechanisms of this ethylene insensitivity of the auxin mutants were. Herein, by quantifying the ethylene and auxin effects in both hypocotyls and roots of an array of different auxin and ethylene mutants, we conclude that ethylene is not required for the auxin-induced growth inhibition in either roots or hypocotyls of dark-grown seedlings. Conversely, auxin appears to be necessary for the ethylene-induced growth inhibition in roots but not in hypocotyls. However, it should be noted that the latter inference is based on the phenotypic analysis of auxin mutants that only partially block auxin production, transport, perception, signaling, and/or response. Therefore, a possibility remains that due to, for example, potentially higher genetic redundancy in the hypocotyls than in the roots, there might be adequate residual levels of auxin activity maintained in the hypocotyls of these auxin mutants sufficient to enable their normal ethylene responsiveness. Nevertheless, we find that defects in auxin biosynthesis, transport, or response can all lead to altered ethylene responsiveness in root tissues. Interestingly, while most of the auxin mutants tested (those shown in Figure 1, as well as tir3 and axr2) show reduced ethylene sensitivity in roots, other auxin mutants, such as pin1 and ett, are not affected (data not shown). We speculate that the latter auxin mutants that show wild-type response levels to ethylene do not alter the levels/response to auxin in the transition zone of roots.
Although it is clear that auxin is required for the normal ethylene response in roots, and ethylene is able to stimulate auxin levels/signal in these tissues (Stepanova et al., 2005), this does not necessarily imply that auxin acts downstream of ethylene in the control of root elongation. The restoration of the ethylene response of aux1 and eir1 by very low levels of exogenous auxin observed by Rahman et al. (2001) made the authors speculate that auxin does not act as the executer of the ethylene response functioning downstream of ethylene but rather as a positive regulator of the ethylene response. One potential problem with such interpretation is that the levels of auxin in the roots after the different treatments were not examined. Therefore, it is possible that the low levels of the exogenous auxin applied in this study, on top of the residual ethylene-mediated increase in endogenous auxins, were sufficient to inhibit root growth of the aux1 or eir1 plants treated with both ethylene and low auxin. We have reexamined this question by monitoring activity of auxin (DR5:GUS) and ethylene (EBS:GUS) reporters in several auxin mutants. The correlation between DR5:GUS levels and the ethylene effects on root growth suggests a direct role for auxin in mediating the ethylene-induced growth inhibition. However, the analysis of the DR5:GUS expression in the auxin overproducer rty1-1 leaves the possibility of a parallel non-auxin-mediated ethylene effect open.
While the focus of the physiological studies described here was placed on a single morphological trait, the inhibition of root growth by auxin and ethylene, it is obvious that these two hormones play additional roles in root biology, and it is more than likely that the relationships between ethylene and auxin would be different when a different response is considered. As the first step to examining these other putative relationships, the expression levels of the ethylene reporter EBS:GUS in wild-type and aux1 mutant backgrounds were examined. This not only allowed us to look at early ethylene responses in a mutant with altered auxin distribution but also to do so in a spatial context. The root growth dynamics suggests that the spatial separation of the maximal auxin (transition zone) and ethylene (elongation zone) activity levels is likely the result of the underlying temporal pattern of interactions between these two hormones. A plausible mechanistic interpretation of the findings described in Figures 2 and 3, in light of our current understanding of the ethylene and auxin biology in roots, would be as follows (Figure 7B). Ethylene treatment stimulates ethylene responses in root tips and, as we have shown in a previous study (Stepanova et al., 2005), one of these responses is the transcriptional induction of two Trp biosynthetic genes, WEI2/ASA1/TIR7 and WEI7/ASB1, leading to a presumed increase in the levels of auxin in the root tips. In fact, recent studies by Růžička et al. (2007) and Swarup et al. (2007) support the role of ethylene in boosting auxin biosynthetic rates in roots, although different growth conditions were used in these two studies. This auxin is then transported to the transition zones by an AUX1- and PIN2-dependent mechanism (Růžička et al., 2007; Swarup et al., 2007). Such transient increase in auxin would lead to two outcomes: on the one hand, it would inhibit cell growth (a direct auxin-mediated ethylene effect) and, on the other, it would sensitize these cells for full response to ethylene once they leave the root transition zone (an auxin-dependent ethylene effect). This model is sufficient to explain the distribution of DR5:GUS and EBS:GUS in the wild-type plants treated with ethylene as well as the alterations observed in the expression of these reporters in the aux1 mutant background (Figures 2 and 3). It also suggests that in addition to the auxin-mediated and auxin-dependent ethylene responses, there may be a third type of response that would be dependent on the levels of auxin but would not be directly mediated by this hormone. Existence of these three types of interactions between ethylene and auxin is also supported by our microarray experiments (see below).
Whole-Genome Expression Profiling of the Ethylene–Auxin Interactions
The genomic component of the ethylene and auxin responses is critical, as indicated by the dramatic ethylene and auxin phenotypes of the ein3 eil1 and arf7 arf19 transcription factor mutants, respectively (Alonso et al., 2003a; Okushima et al., 2005). The availability of mutants affected in well-defined steps of ethylene or auxin pathways makes global gene expression studies an excellent tool for dissecting the interactions between these two hormones. We have chosen the ein2 mutant to study the role of ethylene in the auxin response because this is the only loss-of-function mutant known with a complete blockage of all ethylene responses examined to date. Similarly, we selected the aux1 mutant to investigate the role of auxin in the ethylene response due to its extreme resistance to ethylene in roots, as well as relatively minor developmental alterations, compared with other auxin mutants, such as axr2, which shows similar levels of ethylene insensitivity. One possible caveat of using aux1 in the gene expression studies, however, is that the root cells of this mutant are not completely depleted of auxin as shown by DR5:GUS expression (Sabatini et al., 1999) and not all of the root cells are affected to the same degree by the mutation. In fact, this problem would also be encountered with any other auxin mutant or pharmacological treatment since auxin is essential for plant survival. Regardless, the consequence of using aux1 in this study would be the underestimation (as opposed to the overestimation) of auxin-dependent ethylene responses; therefore, the conclusions drawn would remain valid.
The first interesting but not surprising result from the whole-genome expression analysis is that the proportion of genes coregulated by the two hormones is relatively small (27 and 18% of the ethylene- and auxin-regulated genes, respectively). In fact, these numbers are very similar to those recently found by Nemhauser et al. (2006) (33 and 22%, respectively). The small differences between the outcomes of these two studies could be explained by the differences in the experimental systems (roots versus whole seedlings, the duration of the treatment, etc.) and/or in the analysis criteria used to select the ethylene- and auxin-regulated genes.
In addition to uncovering the small fraction of genes coregulated by both hormones, we found evidence of other modes of interaction between ethylene and auxin. Twenty-eight percent of the remaining ethylene-regulated genes showed altered ethylene responses in the aux1 background. Similarly, 46% of the remaining auxin-regulated genes was affected in their response to auxin in the ein2 mutant. Presence of these different types of regulation suggests existence of auxin-independent, auxin-mediated, and auxin-dependent ethylene responses and, likewise, of ethylene-independent, ethylene-mediated, and ethylene-dependent auxin responses.
One possible problem with the interpretation of the microarray results is that the criteria used to define what an ethylene-regulated gene is or to determine whether a gene is differentially regulated by a hormone in a specific mutant background are arbitrary. Thus, for example, if a high-stringency selection (an approach that reduces the number of false positives) is employed to define what ethylene- and auxin-regulated genes are, one may end up with a large number of false negatives. That is, if a gene that was defined as regulated only by ethylene (based on the high-stringency selection) turns out to also be auxin regulated, then an ethylene response that was categorized as auxin dependent would instead be auxin mediated. To avoid this possible problem, we used high-sensitivity/low-stringency criteria to select the ethylene- and auxin-regulated genes. Three pieces of evidence support the reliability of the classification criteria used: (1) the overlap between the gene groups defined in this work and the functional categories previously associated with these hormones (such as the AP2/EREBP and AUX/IAA transcriptional regulators or hormone metabolism, both for ethylene and auxin); (2) the promoter analysis that found significant enrichment for the ARF binding sites among auxin-regulated genes, specifically, among the group of ethylene-independent genes; (3) the reproducibility of expression patterns observed for the 12 selected genes reexamined using quantitative RT-PCR.
Therefore, the existence of these different groups of genes strongly supports not only the existence of auxin-dependent and auxin-mediated ethylene effects, as was suggested by the physiological and reporter gene studies, but also the presence of ethylene-dependent and ethylene-mediated auxin responses in roots. These latter types of interactions may not be an obvious expectation, judging from the normal response of ethylene-insensitive mutants to auxin at the morphological level. On the other hand, it is well known that auxin can stimulate ethylene production (Woeste et al., 1999); therefore, some degree of at least ethylene-mediated auxin responses was expected. In fact, we observed that four ACS genes coding for the key enzyme in ethylene biosynthesis ACC synthase were regulated by auxin. Three of these genes contain ARF binding sites in their promoters (in the 500-bp region upstream of ATG), suggesting that they might be direct auxin targets.
In addition to confirming coinvolvement of several modes of interaction between ethylene and auxin in roots, the analysis of the functional annotation of the different gene groups provides support for the mechanistic model proposed above. For example, according to our model, the growth inhibition induced by ethylene would be in large part mediated by an increase in auxin, whereas the auxin effect on growth would be ethylene independent. Interestingly, when the functional categories of those genes that are regulated both by ethylene and auxin were examined, a strong enrichment for genes involved in the modification of the cell wall was found. A more detailed analysis showed that the enrichment was more significant among genes in which the ethylene regulation was aux1 dependent and their auxin regulation was ein2 independent (Figure 6). This type of regulation would be consistent with that of genes regulated by ethylene through an auxin-mediated mechanism. In the context of our model, these results imply that one of the effects of the ethylene-induced increase in auxin levels in root transition zones is to modify the cell walls of these cells. These structural changes are probably required for the establishment of the new growth pattern in which the cells expand radially rather than longitudinally.
In summary, these results suggest a simple mechanistic model to explain some of the interactions between ethylene and auxin in roots of etiolated seedlings, including the dramatic morphological ethylene defects of the auxin mutants. However, this model does not exclude the possibility for other elements, such as regulation of auxin transport (as shown in the companion manuscripts), also playing critical roles in the interaction between these two hormones.
METHODS
Strains, Growth Conditions, and Phenotypic Analysis
All of the Arabidopsis thaliana seed lines used in this study are in the Col background. The rty1-1 allele was obtained from the ABRC (CS8156). The DR5:GUS and EBS:GUS reporters were generously provided by T. Guilfoyle and J. Ecker, respectively. The reporters were introduced into mutant backgrounds by crossing to avoid possible positional chromosomal effects. For phenotypic tests, freshly propagated seeds were surface-sterilized with 50% bleach plus 0.005% Triton, washed three times with sterile water, resuspended in melted precooled 0.7% low-melting-point agarose in water and spread on the surface of AT plates (1× Murashige and Skoog salts [Caisson], pH 6.0, 1% sucrose, and 0.6% agar) supplemented with the indicated concentrations of ACC or IAA. Plates with seeds were stratified for 3 d at 4°C in the dark, exposed to light for 2 h at room temperature to improve germination, wrapped with aluminum foil, placed horizontally, and incubated in the dark at 22°C for 3 d. Plates were then unwrapped and opened, and 30 to 40 seedlings per treatment per genotype were pulled out of agar and placed horizontally side by side on the surface of fresh plates containing 0.6% agar in water. Plates were then scanned, and the images obtained were used for quantifying root and hypocotyl lengths as described (Stepanova et al., 2005). For GUS staining, 3-d-old dark-grown seedlings were pulled out of agar, fixed in ice-cold 90% acetone, and stained overnight as described by Stepanova et al. (2005).
Microarray Studies
Wild-type Col-0, aux1-7, and ein2-5 seeds were surface-sterilized and deposited onto a Nylon membrane (Sefar filtration, 03-100/47) that had been previously autoclave sterilized and laid on the surface of sterile AT plates. Seeds were stratified for 3 d at 4°C in the dark and then exposed to light for 1 h at room temperature. Plates with seeds were placed in a vertical orientation and incubated for 3 d in the dark at 22°C. For the ethylene experiments, plates were exposed to hydrocarbon-free air or air containing 10 ppm of ethylene for the last 4 h of the treatment. For the auxin experiments, after 3 d in the dark at 22°C, plates were opened under safe green light (Kodak green filter KOFSLl3810/ 1521632) and sprayed with 1 μM IAA in a 0.01% ethanol solution or with 0.01% ethanol alone. Seedlings were then incubated for an additional 4 h at 22°C in the dark.
Immediately after the 4-h treatments, 50 mL of RNAlater solution (a saturated solution of ammonium sulfate containing 25 mM sodium citrate and 10 mM EDTA, pH 5.2; US patent 6204375) was poured into the plates, and the membranes with the seedlings were transferred to new plates containing 50 mL of fresh RNAlater solution. Using a razor blade, the roots were dissected from the hypocotyls while still submerged into the RNAlater solution, transferred to a microfuge tube, and frozen at −80°C. Two biological replicates were prepared per condition (per genotype treatment combination), with each biological replicate consisting of a pool of three independent experiments (∼150 roots per treatment). Total RNA was extracted using TRIzol reagent (Invitrogen Life Technologies) and then further purified using the RNeasy mini kit (Qiagen).
cRNA synthesis, labeling, and hybridization to Arabidopsis ATH1 genome arrays from Affymetrix were performed according to manufacturer's recommendations, except that the labeling reactions were scaled down to 50%. After hybridization, arrays were scanned and Cel files were used for further analysis.
All normalization and quality controls were performed using the packages GCRMA, SIMPLEAFFY, and AFFY from BioConductor. After normalization, present, marginal, and absent flags, together with the intensity values converted from logarithmic to linear scales, were exported to GeneSpring GX. Ethylene- and auxin-regulated genes were selected using a linear model approach (Smyth, 2004) implemented in the limma package from BioConductor. This analysis was done using the Remote Analysis Computation for Gene Expression (Psarros et al., 2005). Genes that had a P value of <0.05 and a fold change between control and treated Col experiments greater than 1.5 were selected. To account for the multiple testing, a highly sensitive low-stringency false discovery rate (q-value) of 0.15 was used. Finally, only genes that were present or marginal in both replicates in the treated (when selecting upregulated genes) or in the untreated (when selecting for downregulated genes) samples were further considered.
To select ethylene- or auxin-regulated genes that had an altered response to the hormone in the aux1 or ein2 mutant backgrounds, respectively, two different criteria were applied. Starting, for example, with the ethylene-regulated genes found in Col, the change in expression between all Col untreated and treated samples was calculated as well as between untreated and treated aux1 samples. Genes that showed a significant difference (analysis of variance P < 0.05) in the response to the hormone in Col and aux1 were selected. A similar approach was used to select genes with altered response to auxin in the ein2 background. Finally, genes were selected based on the fold difference between the average change in Col and in the mutants. Only genes that show a fold change >1.3-fold were selected. To account for multiple testing, the q-value was calculated using the QVALUE package from BioConductor. Only genes that pass a cutoff q-value of 0.05 were considered to have altered expression in the mutant background compared with the wild type.
The sequence and latest gene annotation of the Arabidopsis genome (TIGR6) were loaded into GeneSpring GX. The “find potential regulatory sequences” function was used to identify specific sequences in the promoters of selected genes. No sequence ambiguities were allowed. The promoters were defined as the sequence from –25 to –425 upstream of the start codon. To determine significance of the findings, the frequency of the DNA element was compared between the selected groups of genes and the promoters of the rest of the genes in the genome.
Gene Function Analysis
To determine whether or not some functional categories were significantly overrepresented in any of the gene groups identified in this work, the MapMan v5 software was used. For each gene category (ETaux1A, ETaux1N, etc.), a MapMan experiment was generated. In these experiments, an expression value of 10 was assigned to genes of a particular category (for example, ETaux1N) and a value of 1 to the rest of the genes that were flagged as present in at least 8 out of 16 experiments. To determine whether or not a gene group was enriched for a particular functional category, each individual Arabidopsis Affy pathway in MapMan v5 was examined using the Wilcoxon rank sum test option and the Benjamini Hochberg multiple testing correction. A group of genes was considered to be enriched for a particular functional category if the corrected P value was ≤0.0001 and at least three genes from the group were found in that particular functional category.
Real-Time RT-PCR
For the RT-PCR analysis, eight RNA samples (Col air, Col ethylene, aux1 air, aux1 ethylene, Col control, Col IAA, ein2 control, and ein2 IAA) corresponding to a new biological replicate (i.e., different than those used in the microarray experiments above) were extracted using the same procedures as described above in the Microarray Studies section. Four hundred nanograms of each RNA sample were reverse transcribed in a 20 μL volume using TagMan reverse transcription reagents (Applied Biosystems) according to the manufacturer's recommendations. The samples were diluted to 100 μL with water and 2 μL of each sample (∼8 ng RNA equivalent) were PCR amplified using Power SYBR Green (Applied Biosystems) in a 10-μL reaction, containing 2 μL diluted cDNA, 5 μL Power SYBR Master Mix, 1 μL of 10 μM forward primer, 1 μL of 10 μM reverse primer, and 1 μL of water. The ABI7900 machine was used to run quantitative RT-PCR with the following 11 primer pair combinations corresponding to 11 differentially expressed genes from all eight gene categories (at least one per category) on the eight cDNA samples in triplicates in a 384-well plate: At2g26070F, 5′-TATCTCCAACTCGATAGAACCAAGTG-3′; At2g26070R, 5′-GTTTTATGCTCAAAGCTACGTGTGC-3′; At2g36080F, 5′-CGGAGTGAACATGGAGTGCCAGC-3′; At2g36080R, 5′-ATCTCCTGTGAAACTTATGTCC-3′; At1g13700F, 5′-CTTACGACAAGATTGTAGATTGG-3′; At1g13700R, 5′-GTCTCGGAAACACATTTACCTTGG-3′; At3g15540F, 5′-GGTTAGGGTATGTGAAAGTGAGC-3′; At3g15540R, 5′-GTCACCATCTTTCAAGGCCACACC-3′; At4g27260F, 5′-TCTCTGAGTTCCTCACAAGCTCT-3′; At4g27260R, 5′-CGAGACCAGGAACGAACTGGCTC-3′; At5g02580F, 5′-GCTTCTTACATCCATTTGGTGC-3′; At5g02580R, 5′-CTTTCGCTAGCTCTTTCCACACTG-3′; At4g37850F, 5′-CTAGTTCCTGGCCTTAAAAAGATGG-3′; At4g37850R, 5′-GAGAATGATTGATTATTATCGTCC-3′; At1g14290F, 5′-GATTGCATTCGAAGATAGATGAGG-3′; At1g14290R, 5′-CATCTGCATCACTTCCTGTTATCTTG-3′; At4g00430F, 5′-TGCAGTGTTCTTGGTACACTTGGC-3′; At4g00430R, 5′-GGTCCAACCCAGAAAATCCAATGGTC-3′ At4g23700F, 5′-TTGGTAAAGACAGAGGGGTTC-3′; At4g23700R, 5′-CGGTTCGTCTCCTCCACCGTTCG-3′; At4g02290F, 5′-CTCTTACTCTGGTTATCAGGATG-3′; At4g02290R, 5′-TGGCACCAGCGTGCTTGTTATCC-3′.
In addition, a housekeeping gene, At5g44200, encoding nuclear cap binding protein, CBP20, was employed as a control: At5g44200F, 5′-AATCGCCATGGAAGAGGAGAC-3′; At5g44200R, 5′-GAATCGTGGGTTCTTCTCCGGTC-3′.
Primers were designed across exon-exon junctions of cDNA to avoid potential problems due to contaminating genomic DNA. Amplification efficiency for each primer pair was calculated using serial cDNA dilutions. For this purpose, equal volumes of all eight cDNA samples were pooled and 1.5×, 1×, 1/2×, 1/4×, and 1/8× dilutions were run in triplicates for each of the 12 primer combinations. After correcting the cycle threshold values according to the amplification efficiencies, the expression values of the 11 genes were normalized to those of the control.
Accession Number
Expression profiling data can be found in the Gene Expression Omnibus database under accession number GSE7432.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Table 1. Normalized Expression Values of Ethylene- and Auxin-Regulated Genes.
Supplementary Material
Acknowledgments
We thank Joe Ecker for providing the EBS:GUS lines prior to publication and the ABRC for the seeds of various auxin mutants. We are also grateful to Robert Franks, Malcom Bennett, and Eva Benkova for helpful comments on the manuscript. This work was supported by National Science Foundation Grants MCB 0519869 and MCB 0315992 and funds from North Carolina State University to J.M.A.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jose M. Alonso (jmalonso@unity.ncsu.edu).
Online version contains Web-only data.
References
- Abel, S., Nguyen, M.D., Chow, W., and Theologis, A. (1995). ACS4, a primary indole acetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. Structural characterization, expression in Escherichia coli, and expression characteristics in response to auxin. J. Biol. Chem. 270 19093–19099. Erratum. J. Biol. Chem. 270: 26020. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., and Ecker, J.R. (2001). The ethylene pathway: A paradigm for plant hormone signaling and interaction. Sci. STKE 2001 RE1. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., and Stepanova, A.N. (2004). The ethylene signaling pathway. Science 306 1513–1515. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003. b). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., Stepanova, A.N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F.M., and Ecker, J.R. (2003. a). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 100 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Bartel, B. (1997). Auxin biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 51–66. [DOI] [PubMed] [Google Scholar]
- Bennett, M., Bellini, C., and Van Der Straeten, D. (2005). Integrative biology: Dissecting cross-talk between plant signaling pathways. Physiol. Plant 123 109. [Google Scholar]
- Binder, B.M., Mortimore, L.A., Stepanova, A.N., Ecker, J.R., and Bleecker, A.B. (2004). Short-term growth responses to ethylene in Arabidopsis seedlings are EIN3/EIL1 independent. Plant Physiol. 136 2921–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302 1956–1960. [DOI] [PubMed] [Google Scholar]
- Brown, K.M. (1997). Ethylene and abscission. Physiol. Plant 100 567–576. [Google Scholar]
- Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., and Ecker, J. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89 1133–1144. [DOI] [PubMed] [Google Scholar]
- Cohen, J.D., Slovin, J.P., and Hendrickson, A.M. (2003). Two genetically discrete pathways convert tryptophan to auxin: More redundancy in auxin biosyntehsis. Trends Plant Sci. 8 197–199. [DOI] [PubMed] [Google Scholar]
- Collett, C.E., Harberd, N.P., and Leyser, O. (2000). Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 124 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. [DOI] [PubMed] [Google Scholar]
- Ecker, J.R., and Davis, R.W. (1987). Plant defense genes are regulated by ethylene. Proc. Natl. Acad. Sci. USA 84 5202–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle, M.A., and Somerville, C.R. (1987). Auxin resistant mutants of Arabidopsis with an altered morphology. Mol. Gen. Genet. 206 200–206. [Google Scholar]
- Fu, X., and Harberd, N.P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 740–743. [DOI] [PubMed] [Google Scholar]
- Gazzarrini, S., and McCourt, P. (2003). Cross-talk in plant hormone signalling: What Arabidopsis mutants are telling us. Ann. Bot. (Lond.) 91 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M. (2004). Hormonal regulation of plant growth and development. PLoS Biol. 2 E311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T., Hagen, G., Ulmasov, T., and Murfett, J. (1998). How does auxin turn on genes? Plant Physiol. 118 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2004). The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 7 40–49. [DOI] [PubMed] [Google Scholar]
- Guzman, P., and Ecker, J. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., Ckurshumova, W., Vidaurre, D.P., Singh, S.A., Stamatiou, G., Tiwari, S.B., Hagen, G., Guilfoyle, T.J., and Berleth, T. (2004). Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131 1089–1100. [DOI] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. [DOI] [PubMed] [Google Scholar]
- King, J., Stimart, D., Fisher, R., and Bleecker, A. (1995). A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Camera, S., Gouzerh, G., Dhondt, S., Hoffmann, L., Fritig, B., Legrand, M., and Heitz, T. (2004). Metabolic reprogramming in plant innate immunity: The contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 198 267–284. [DOI] [PubMed] [Google Scholar]
- Leyser, O. (2002). Molecular genetics of auxin signaling. Annu. Rev. Plant Biol. 53 377–398. [DOI] [PubMed] [Google Scholar]
- Li, H., Johnson, P., Stepanova, A., Alonso, J.M., and Ecker, J.R. (2004). Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7 193–204. [DOI] [PubMed] [Google Scholar]
- Liscum, E., and Reed, J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49 387–400. [PubMed] [Google Scholar]
- Ljung, K., Hull, A.K., Celenza, J., Yamada, M., Estelle, M., Normanly, J., and Sandberg, G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O., Piqueras, R., Sanchez-Serrano, J.J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant, A., Kargul, J., May, S.T., Muller, P., Delbarre, A., Perrot-Rechenmann, C., and Bennett, M.J. (1999). AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18 2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Naur, P., and Halkier, B.A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37 770–777. [DOI] [PubMed] [Google Scholar]
- Nagpal, P., Ellis, C.M., Weber, H., Ploense, S.E., Barkawi, L.S., Guilfoyle, T.J., Hagen, G., Alonso, J.M., Cohen, J.D., Farmer, E.E., Ecker, J.R., and Reed, J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132 4107–4118. [DOI] [PubMed] [Google Scholar]
- Nemhauser, J.L., Hong, F., and Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475. [DOI] [PubMed] [Google Scholar]
- Nemhauser, J.L., Mockler, T.C., and Chory, J. (2004). Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2 E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly, J., Grisafi, P., Fink, G.R., and Bartel, B. (1997). Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, W.A., Bandyopadhyay, A., Blakeslee, J.J., Makam, S.N., Chen, R.J., Masson, P.H., and Murphy, A.S. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts, R.J., Cernac, A., and Estelle, M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16 553–560. [DOI] [PubMed] [Google Scholar]
- Psarros, M., Heber, S., Sick, M., Thoppae, G., Harshman, K., and Sick, B. (2005). RACE: Remote Analysis Computation for gene Expression data. Nucleic Acids Res. 33 W638–W643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, A., Amakawa, T., Goto, N., and Tsurumi, S. (2001). Auxin is a positive regulator for ethylene-mediated response in the growth of Arabidopsis roots. Plant Cell Physiol. 42 301–307. [DOI] [PubMed] [Google Scholar]
- Rahman, A., Hosokawa, S., Oono, Y., Amakawa, T., Goto, N., and Tsurumi, S. (2002). Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol. 130 1908–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička, K., Ljung, K., Vanneste, S., Podhorská, R., Beeckman, T., Friml, J., and Benková, E. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19 2197–2212. [DOI] [PMC free article] [PubMed]
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Smalle, J., Haegman, M., Kurepa, J., Van Montagu, M., and Van Der Straeten, D. (1997). Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc. Natl. Acad. Sci. USA 94 2756–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 Article3. [DOI] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova, A.N., and Alonso, J.M. (2005). Ethylene signaling and response pathway: A unique signaling cascade with a multitude of inputs and outputs. Physiol. Plant 123 195–206. [Google Scholar]
- Stepanova, A.N., Hoyt, J.M., Hamilton, A.A., and Alonso, J.M. (2005). A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup, R., and Bennett, M. (2003). Auxin transport: The fountain of life in plants? Dev. Cell 5 824–826. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Parry, G., Graham, N., Allen, T., and Bennett, M. (2002). Auxin cross-talk: Integration of signalling pathways to control plant development. Plant Mol. Biol. 49 411–426. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Perry, P., Hagenbeek, D., Van Der Straeten, D., Beemster, G.T.S., Sandberg, G., Bhalerao, R., Ljung, K., and Bennett, M.J. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19 2186–2196. [DOI] [PMC free article] [PubMed]
- Tsuchisaka, A., and Theologis, A. (2004). Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 136 2982–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche, F., et al. (2003). The Arabidopsis mutant alh1 illustrates a cross talk between ethylene and auxin. Plant Physiol. 131 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K.L., Li, H., and Ecker, J.R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14 (suppl.): S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste, K.E., Vogel, J.P., and Kieber, J.J. (1999). Factors regulating ethylene biosynthesis in etiolated Arabidopsis thaliana seedlings. Physiol. Plant 105 478–484. [Google Scholar]
- Zhao, Y., Hull, A.K., Gupta, N.R., Goss, K.A., Alonso, J., Ecker, J.R., Normanly, J., Chory, J., and Celenza, J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, G.V., and Burns, J.K. (2003). Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol. Biol. 53 117–131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.