Abstract
Embryogenesis in Arabidopsis thaliana is marked by a predictable sequence of oriented cell divisions, which precede cell fate determination. We show that mutation of the TORMOZ (TOZ) gene yields embryos with aberrant cell division planes and arrested embryos that appear not to have established normal patterning. The defects in toz mutants differ from previously described mutations that affect embryonic cell division patterns. Longitudinal division planes of the proembryo are frequently replaced by transverse divisions and less frequently by oblique divisions, while divisions of the suspensor cells, which divide only transversely, appear generally unaffected. Expression patterns of selected embryo patterning genes are altered in the mutant embryos, implying that the positional cues required for their proper expression are perturbed by the misoriented divisions. The TOZ gene encodes a nucleolar protein containing WD repeats. Putative TOZ orthologs exist in other eukaryotes including Saccharomyces cerevisiae, where the protein is predicted to function in 18S rRNA biogenesis. We find that disruption of the Sp TOZ gene results in cell division defects in Schizosaccharomyces pombe. Previous studies in yeast and animal cells have identified nucleolar proteins that regulate the exit from M phase and cytokinesis, including factors involved in pre-rRNA processing. Our study suggests that in plant cells, nucleolar functions might interact with the processes of regulated cell divisions and influence the selection of longitudinal division planes during embryogenesis.
INTRODUCTION
In multicellular organisms, the control of cell division and cell expansion is coordinated with the growth and development of organs and tissues. In higher plants, where the cells are enclosed in rigid cell walls, not only the timing but also the planes of cell division are subject to developmental control. The first reported studies of the angles of cell divisions in plants were recorded more than a century ago. In 1863, Hofmeister observed that the plane of cell division is usually transverse or perpendicular to the long axis of the cell (Hofmeister, 1863). Five years later, Errera proposed a rule that the plane of cell division follows the shortest distance that will halve the volume of the cell (for review, see Smith, 2001). This is generally the case with divisions within a file or layer of cells of the same cell type, whereas longitudinal or asymmetric divisions seem to be commonly associated with organ initiation or differentiation of specific cell types. It is presumed that these atypical cell divisions occur in response to specific developmental cues. For example, longitudinal cell divisions mark the earliest divisions of the Arabidopsis thaliana proembryo, and thereafter anticlinal divisions give rise to the protoderm. Due to the specificity and predictability of early divisions, the embryo in Arabidopsis is an excellent tool for the discovery of factors that may alter specific types of cell division. Furthermore, the developmental consequences of the early embryonic divisions can be followed genetically by the changes in expression pattern of specific marker genes.
Genetic screens have identified a number of Arabidopsis genes required for completion of cytokinesis, most of them being required for the formation of the cell plate (summarized in Sollner et al., 2002). By contrast, there are few characterized genes known for the correct specification/positioning of the division site. In Arabidopsis, tonneau2 (ton2)/fass and ton1 mutant cells divide in random planes, probably due to loss of preprophase bands of microtubules during the mitotic phase (Traas et al., 1995; McClinton and Sung, 1997). The TON2 protein is a putative PP2A phosphatase subunit and may play an important role in organization of the microtubule cytoskeleton during interphase and prophase (Camilleri et al., 2002). The MOR1 gene encodes a microtubule-associated protein required for the correct positioning of the cell plates and completion of cytokinesis (Kawamura et al., 2006). In maize (Zea mays), the tangled1 mutant exhibits random planes of cell division in the leaf (Smith et al., 1996). The Tangled gene encodes a basic microtubule-associated protein that is likely to be involved in the orientation and positioning of the preprophase band and the phragmoplast (Smith, 2001). These studies show the importance of cytoplasmic factors that regulate microtubule structures during the cell cycle events leading to the establishment of the division plane.
In recent years, studies in yeast and animal systems have established the remarkable fact that the nucleolus performs important functions in the regulation of cell division that are in addition to its well-established functions in the synthesis of ribosomes (reviewed in Olson et al., 2000; Dez and Tollervey, 2004; Shaw and Doonan, 2005; Matthews and Olson, 2006). Nucleolar functions discovered in animal cells include control of cell cycle progression in G1 through regulation of p53 (Tsai and McKay, 2002), entry into S phase through regulation of polo-like kinase (Zimmerman and Erikson, 2007), and control of mitosis and cytokinesis through SUMO-specific protease (Di Bacco et al., 2006). In yeast, mitotic exit requires the cell cycle regulator Cdc14p, which is sequestered in the nucleolus during interphase by the RENT complex and released when specifically required at the G2/M transition by the FEAR pathway (Shou et al., 1999; Dez and Tollervey, 2004). Specific nucleolar factors involved in ribosome biogenesis also appear to have dual functions in control of cell division. These include the Pes1 protein in mammalian cells, which functions in cell proliferation (Grimm et al., 2006), and the Nop15p protein in yeast, which functions in cytokinesis (Oeffinger and Tollervey, 2003). In plant cells, the nucleolus has been demonstrated to play a critical role in the regulation of gene silencing (Pontes et al., 2006), but there is limited information about involvement of the nucleolus in cell division in plants. Recently, it has been shown that disruption of the Arabidopsis SLOW WALKER1 (SWA1) gene, which encodes a protein involved in rRNA processing, results in slow cell cycle progression and gametophytic lethality (Shi et al., 2005).
Here, we describe a novel Arabidopsis mutant called tormoz (toz) that affects cell divisions during embryogenesis, with transverse divisions frequently replacing longitudinal cell divisions in the embryo, but not conversely. Cloning of the TOZ gene shows that it encodes a conserved nucleolar protein with homologs in animals and fungi, which in budding yeast is a component of a complex involved in rRNA biogenesis. The fission yeast homolog of TOZ is also found to be required for normal mitotic division. Our results suggest that TOZ differs from the known genes that function in the specification of planes of cell division in plants, both in phenotypic effects and in mechanism of action.
RESULTS
toz Affects Early Embryonic Development
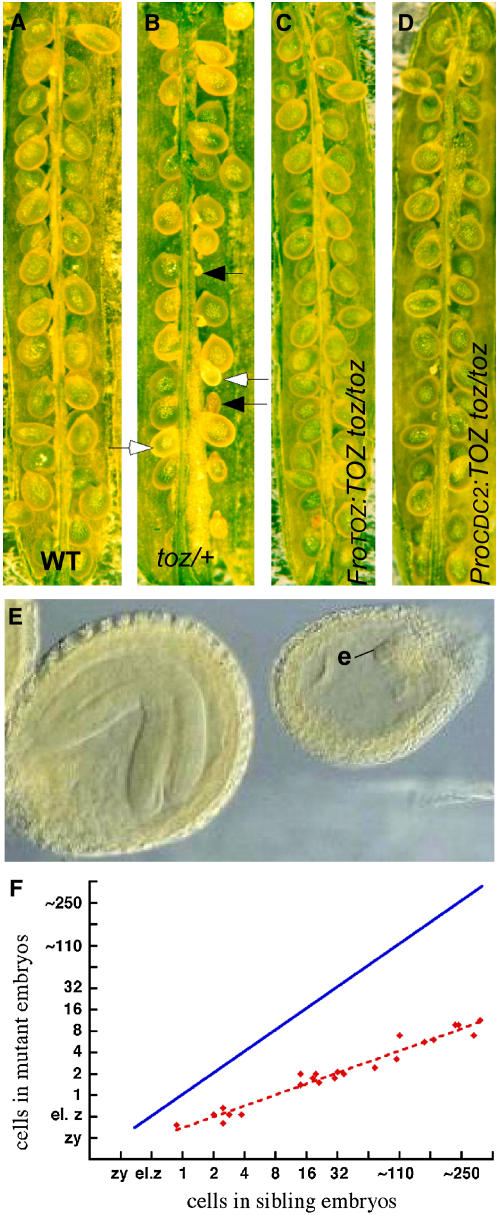
The toz mutant was isolated during a screen for embryo lethal mutants from a collection of Ds gene trap insertion lines (Sundaresan et al., 1995; Parinov et al., 1999). Self-fertilized siliques from toz/+ plants contain 25% (288/1100) aborted seed of different sizes (Figures 1A and 1B). Most mutant embryos arrest having generated a small globular mass of cells, prior to the maturation of the sibling embryos (Figure 1E). Homozygous toz/toz plants were not recovered, and toz/+ plants were otherwise indistinguishable from the wild type, indicating that the mutation is a recessive, homozygous lethal allele.
Figure 1.
Developing Seed Phenotypes.
(A) and (B) Superficial phenotypes of seeds from wild-type (A) and toz/+ (B) siliques. The closed arrows highlight aborted seeds, and the open arrows highlight living seeds with reduced rates of development.
(C) and (D) Siliques from toz/toz plants complemented with a genomic fragment (ProTOZ:TOZ) still produce a few aborted seed (C), whereas the toz/toz siliques complemented by the TOZ cDNA driven by the CDC2a promoter (ProCDC2:TOZ) show very few aborted embryos (D).
(E) Late development of embryos in a single silique, comparing the toz mutants (e) to the sibling embryo at an advanced stage.
(F) The comparison of growth rates, where each point on the graph represents the average number of cells in mutant versus sibling embryos within a single silique. The solid blue line shows the expected positions of the points if there were a 1:1 correspondence between cell divisions in the wild type versus toz mutant embryos.
As the engineered Ds element harbors a kanamycin (kan) resistance gene, seed from self-fertilized toz/+ plants was found to be segregating 1.5:1 (1171:769) kan resistant:kan sensitive plants, reduced from the expected ratio of 3:1. This is also lower than the 2:1 segregation ratio expected for an embryo lethal mutant, thus suggesting an additional loss of Ds transmission occurs through the gametes. To further investigate, reciprocal crosses of toz/+ with wild-type plants were performed. When wild-type pollen fertilized toz/+ pistils, the resultant seed segregated only 0.59:1 (270/460), whereas the kan ratio for toz/+ pollen crossed to wild-type pistils was 1:1 (538/526), indicating that transmission of Ds was not affected through the pollen, only through the ovules. To investigate whether disruption to development occurred during or after the formation of the female gametophyte, different stages of postmeiotic ovules from toz/+ plants were carefully studied. No morphological defects were found either during their development or at maturity (data not shown), suggesting that the defect may occur during or just after fertilization (i.e., before the seed abortion causes deposition of pigments). This is consistent with the different stages of development of aborted seed found in the toz/+ siliques. These data suggest that the embryo lethal phenotype is primarily due to abortion of the homozygote toz/toz seeds, yet a late megagametophytic or a very early maternal defect may also occur, as described for the MEE mutants, which display gametophytic inheritance but affect early embryogenesis (Pagnussat et al., 2005).
Aberrant Longitudinal Cell Divisions in toz Mutant Embryos
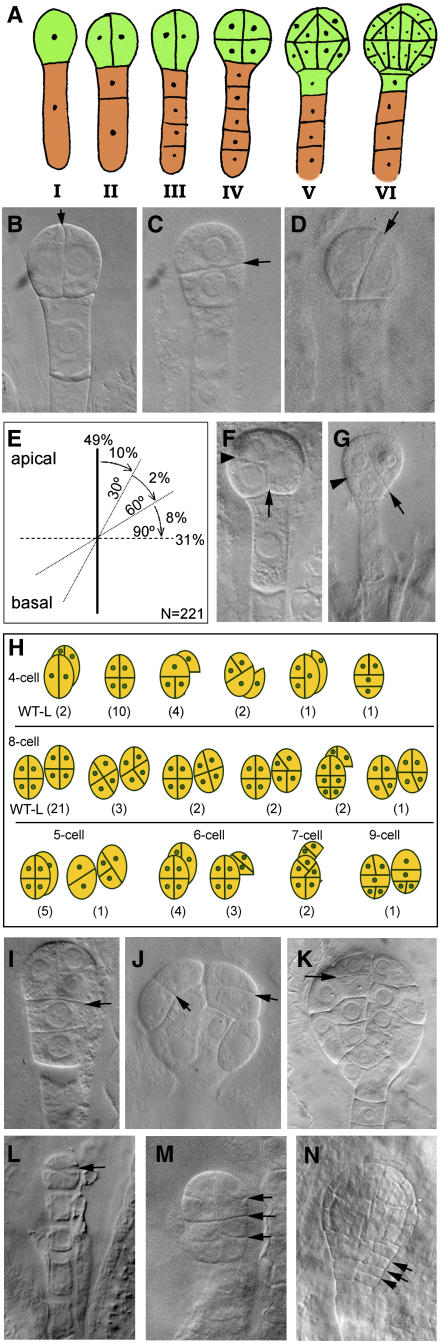
To understand the nature of the toz mutant phenotype during embryo development, heterozygous siliques were examined after whole-mount clearing. The toz mutants can be easily identified because they grow much more slowly than the sibling embryos. For example, when a sibling embryo is at the early globular stage (∼32 cells), the mutant seeds most often contain two cell proembryos (Figure 1F). The most unusual aspect of the toz mutant phenotype, however, was the cell division pattern. The strict pattern of division for wild-type embryos has been summarized in Figure 2A and has been well described elsewhere (Jürgens et al., 1994). In summary, after fertilization, the zygote elongates and then divides asymmetrically, generating a smaller apical and a larger basal cell (Figure 2A, panel I). Initially, the proembryo is exclusively derived from divisions of the apical cell. The first two divisions of the apical cell occur in the longitudinal plane, generating four cells, namely the quadrant embryo (Figure 2A, panels II and III). These cells all undergo transverse divisions to yield eight cells (Figure 2A, panel IV). Each cell then divides periclinally to give rise to the outer layer forming the protoderm that will become the L1 (Figure 2A, panel V). Globular embryos are defined from 32 cells onwards (Figure 2A, panel VI). The basal cell resulting from the first zygotic division only divides in the transverse orientation. At the 16-cell stage, however, the uppermost cell of the suspensor is defined to become the hypophysis, which is the derivative of the embryonic root, but the cells proximal to the hypophysis remain as a single layer of linearly placed cells comprising the suspensor (Figure 2A, panel V).
Figure 2.
Early Embryo Development.
(A) Wild-type embryos follow a predictable pattern of cell division. The stages represent sections of one-cell (I), two-cell (II), four-cell (III), eight-cell (IV), 16-cell (V), and 32-cell (VI) proembryos (green).
(B) to (D) Whole-mount cleared toz embryos showing the first division of the apical cell (arrows) with cell divisions that are longitudinal as in the wild type (B), transverse (C), or oblique (D).
(E) The division planes observed in two-cell proembryos with angles relative to the apical-basal axis are illustrated.
(F) A toz proembryo with three cells. The first division was longitudinal (arrow) and the second transverse (arrowhead).
(G) A four-cell proembryo. The first division was oblique (arrow); the left cell then divided transversely relative to the right cell wall (arrowhead). The right cell divided in the plane of the paper such that only one cell is visible.
(H) Illustration of the variety of embryos and associated division patterns observed in toz proembryos when sibling embryos were at the heart stage. The pairs of nearly superimposed circles, or those offset to the right of each diagram, represent daughters of a division that occurred in the plane of the page. The numbers in parentheses give the numbers observed for each pattern, with the left-most illustration representing toz embryos exhibiting the wild-type-like pattern (indicated by WT-L).
(I) to (K) Multicell toz embryos at later stages, with arrows highlighting unpredictable divisions. The embryo in (I) consists of six cells (the upper left cell overlays another cell that is outside the plane of view). toz cells can be large (J) and give rise to globular embryos with atypical cell patterning (K).
(L) to (N) Phenotypes of embryos from transgenic lines harboring the TOZ antisense cDNA driven by the At CDC2 promoter. Transverse divisions that replace longitudinal divisions were observed in two-cell proembryos (L). Transverse divisions absent in nontransformed controls were frequently observed in older embryos ([M] and [N], arrows).
In self-fertilized toz/+ plants, we found that the first division of the zygote formed an apical and basal cell indistinguishable from the wild type. However, in the next division, the apical cells often divided abnormally (Figures 2B to 2E). Approximately half (108/221) of the apical cells in the slowly growing mutant proembryos divided parallel with the apical-basal axis (longitudinally) as occurs in the wild type (Figure 2B), 31% (69/221) divided in the transverse orientation (Figure 2C), while the remaining 20% (44/221) showed oblique divisions of the apical cell (Figure 2D). Of the oblique divisions, the majority of the division angles were within 30° to either the longitudinal or transverse axis of the cell, with only 4/221 divisions being between 30° and 60° (Figure 2E). By comparison, only 2% (3/150) of wild-type embryos executed transverse divisions at the two-cell proembryo stage, and no oblique divisions were observed.
In the wild type, the two-cell proembryo undergoes synchronized longitudinal divisions resulting in a four-cell proembryo. However, in toz mutants, asynchronous division of the proembryo frequently generated three-cell proembryos, which permit unambiguous assignments of the sequence of divisions. These proembryos (n = 77) also showed abnormal division planes for the first division in similar proportion to division planes of two-cell proembryos examined in Figure 2E (i.e., longitudinal [58%], transverse [36%], or oblique [5%]) (Figure 2F; see Supplemental Figure 1 online). Notably, the sequence of divisions did not show any striking trends (i.e., longitudinal divisions could be followed by transverse divisions at frequencies comparable to the opposite sequence); in all cases, oblique divisions were infrequent (see Supplemental Figure 1 online).
The cell division patterns for four-cell proembryos were also recorded, recalling that in the wild type, the first four cells of the proembryo arise solely from longitudinal divisions of the apical cell (Figure 2A, panel III). As depicted in Figures 2G and 2H and Supplemental Figures 2A to 2D online, many different combinations of divisions were observed for 228 four-celled proembryos examined, with 77% of embryos having at least one transverse or oblique division in place of a longitudinal division observed for wild-type embryos. In addition, there appeared to be little preference given to the selection of the division plane based on the preceding division plane, although the sequence of divisions cannot be determined unambiguously for most four-cell proembryos.
When the progeny from toz/+ plants were at the heart stage of development, the toz mutant embryos were mainly at the eight-cell stage, but with a range from four to nine cells (Figure 2H). From this sample it was observed that where many longitudinal divisions were expected they were not present, yet the transverse divisions were always predictably situated. These are either transverse corresponding to the apical-basal axis of the embryo, or in several instances, they are transverse relative to the preceding division, which can apparently redefine the axis of the cell (Figures 2G and 2H). We observed no examples of longitudinal divisions where transverse divisions were expected, as would be evidenced by cells with increased longitudinal divisions compared with the wild type. Therefore, although we cannot be certain that only longitudinal division planes are affected in this mutant, it is clear that the toz mutation predominantly affects divisions in the apical-basal axis of the cell. Asynchronous cell divisions in toz mutants were further exemplified by embryos with atypical cell numbers (five, six, seven, or nine cells; Figures 2H and 2I). Mutant embryos with larger numbers of cells showed a multitude of patterns and forms with little regularity associated with wild-type apical or basal differentiation (Figures 2J and 2K; see Supplemental Figures 2E to 2J online). The suspensor divisions, on the other hand, divided more slowly than the wild-type sibling embryos but were always transverse. Apart from one observation in many hundreds, the suspensor cells were not affected in cell shape or final number.
We also observed the integrity of the nuclei in >200 cells from mutant embryos that had been stained both with 4′,6-diamidino-2-phenylindole (DAPI) and calcofluor to highlight nuclei and cells walls, respectively. Apart from slightly enlarged nuclei, we observed no abnormalities during mitosis or the integrity of nuclear material at interphase. Therefore, it seems that the toz mutation primarily affects the ability of the embryo proper to execute regulated divisions in reference to the choice of division plane and the timing of the division, particularly longitudinal divisions in the plane of the apical-basal axis.
toz Embryos Show Abnormal Expression of Patterning Genes
Growth arrest can occur at any time from the zygotic stage until ∼11 d after fertilization. The toz mutant embryos that show more advanced cell proliferation, but still fail to show any growth patterns reminiscent of cotyledon initiation, or basal cell structures usually associated with wild-type embryos. To determine whether the expression of genes associated with organogenesis was active, we performed mRNA in situ hybridization. The marker genes selected included SHOOT MERISTEMLESS (STM) (Long et al., 1996; Long and Barton, 1998; McConnell and Barton, 1998), to identify cells of the shoot meristem, and MONOPTEROS (MP) (Hardtke and Berleth, 1998; Hamann et al., 2002) that can distinguish the radial symmetry of early embryos and the root primordia of older embryos. We also tested FILAMENTOUS FLOWER (FIL) (Sawa et al., 1999; Bowman, 2000) and AINTEGUMENTA (ANT) (Elliott et al., 1996; Long and Barton, 1998) that are expressed during and at the onset of the embryonic lateral organs, the cotyledons.
The earliest of these genes to be expressed is MP. In wild-type embryos, MP signal is uniformly detected in the early proembryo stage (Hamann et al., 2002). When the protoderm forms 16 cells, the signal becomes excluded from the outer layer of cells and from the hypophysis. From the globular stages onwards, the signal is progressively restricted to mark the provascular tissue (Hamann et al., 2002). In torpedo-stage embryos, MP was found in the developing vasculature and the root tip (Figure 3A). MP expression in toz embryos was inconsistent (Figures 3B to 3D). In younger embryos, sometimes there was no expression at all (Figure 3B), whereas embryos of neighboring sibling embryos showed normal expression, constituting a positive control (Figure 3A). MP, however, was most often detected in older toz embryos, but the pattern was different for each embryo. Unlike the wild type, expression in toz embryos was found in a cell in the same location as the hypophysis, which in wild-type embryos subsequently gives rise to the quiescent center of the root (Figure 3C). Furthermore, MP was also regularly detected in the protoderm, which does not occur in the wild type (Figures 3C and 3D). Even so, it was apparent that MP expression was strongest in the cells at the center of the embryos, consistent with a semblance of radial symmetry.
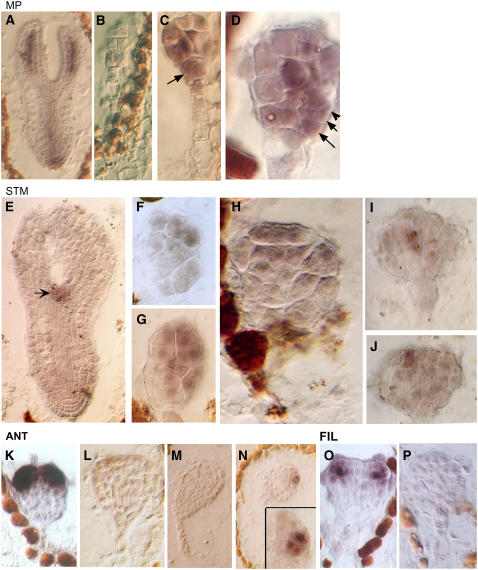
Figure 3.
Gene Expression Patterns in TOZ Embryos.
Analysis of gene expression of MP ([A] to [D]), STM ([E] to [J]), ANT ([K] to [N]), and FIL ([O] and [P]).
(A) Sibling embryo of wild-type phenotype expresses MP in vascular tissue and root tip as previously described (Hamann et al., 2002).
(B) to (D) In toz embryos, the MP signal may be absent (B) or expressed nonuniformly ([C] and [D]). The cells in the presumptive hypophysis may continue to express MP (C), and cells in the outermost layer may also express MP ([D], arrows).
(E) STM expression marks the shoot apical meristem in wild-type cells (arrow).
(F) to (H) In mutant cells, STM is widely expressed across a number of apically positioned cells.
(I) and (J) In an older embryo, STM is confined to a few cells in the subdermal or epidermal layers as shown by consecutive sections.
(K) and (L) In the wild type, ANT is first detected at the late globular stage, just prior to the outgrowth of the lateral organ primordial (K). However, in toz embryos, which consist of many more cells, no ANT expression was observed (L).
(M) and (N) It was not until a very late stage that a few cells showed ANT signal, as depicted by sections 1 and 4 of the same embryo (inset).
(O) and (P) FIL expression in the wild type (O) is absent in the mutant embryo (P).
Expression of the shoot apical meristem marker STM was also examined (Figures 3E to 3J). In wild-type embryos, STM is first expressed as a band around the central apical region of late globular-stage embryos. During the transition to heart stage, the expression becomes restricted to the central core where the shoot apical meristem is being formed (Long and Barton, 1998). In toz embryos, STM was found to be very variably expressed and often in a broad range of cells (Figures 3F to 3H). In toz mutants, the expression could be restricted to the L1 (Figure 3F) or found in both the L1 and L2 (Figure 3H), and occasionally it was also seen extending into the basal half of the embryo (Figure 3G). In one case, an older embryo showed expression in one or two central cells, yet in an adjacent section, the expression was restricted to two epidermal cells that were off center (Figures 3I and 3J). In wild-type embryos with a similar number of cells at the mid to late globular stages, STM is usually expressed in a broader range of cells (Long and Barton, 1998). Thus, it seems that STM expression is initiated broadly in the apical half of the mutant embryos but is not necessarily maintained.
To determine if expression of the genes that are required for lateral organ initiation and patterning are affected, expression of the genes ANT and FIL were examined in toz mutant embryos. ANT is correlated with the initiation of organ primordia and is first expressed at the late globular stage (Figure 3K), whereas FIL is first expressed slightly later at the onset of cotyledon development: the heart stage (Figure 3O) (Long and Barton, 1998; Siegfried et al., 1999; Bowman et al., 2002). ANT expression was generally not detected in toz mutants, even in relatively advanced embryos, such as depicted in Figure 4L. However, for one embryo, ANT signal was observed at a very late stage but only in a few cells of a section glancing the edge of the embryo (Figures 3M and 3N). On the other hand, FIL expression was not detected in any of the mutant embryos examined irrespective of their size (Figure 3P; data not shown). These results imply that the expression of the genes marking onset of primordia growth was largely absent and is consistent with the nonappearance of cotyledon development in toz mutant embryos.
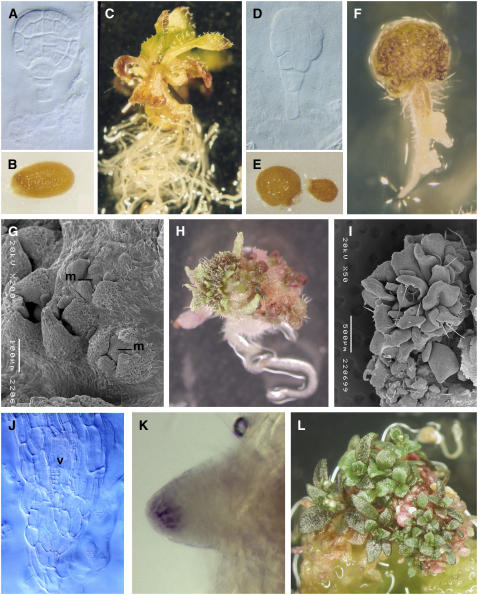
Figure 4.
Phenotypes of ex Planta–Cultured Embryos.
Wild-type ([A] to [C]) and toz ([D] to [L]) tissue were propagated by in vitro culture from early stages.
(A) to (E) Wild-type embryos were cultured from preglobular, globular (A), or older stages. Most often wild-type embryos resulted in normal seed (B) and seedlings after 3 weeks (C). Mutant embryos (D) were cultured from several stages but with similar results. Sometimes the seed failed to germinate (right seed in [E]) or was abnormally shaped (left seed in [E]) compared with the wild type (B).
(F) to (I) Approximately 2 weeks after germination, the apical region becomes a green mass of cells (F) that often gives rise to lateral organs ([H] and [L]). Developmental progression was observed by scanning electron microscopy of toz plants and revealed small centers of young lateral organ primordia developing around a central mound (m) of cells (G), which apparently induces prolific leaf production ([H] and [I]).
(J) Whole-mount cleared sections of a primary root show that the vasculature (v) forms in the center of the root as in the wild type, whereas the root apex lacks regular no cell patterning.
(K) Lugol staining detected the presence of amyloplasts in columella cells of some later toz roots.
(L) Four to six weeks after germination, toz plantlets showed prolific vegetative tissue on a mass of cells, but little root tissue was observed.
Developmental Defects of Cultured toz Mutant Seedlings
As some of the toz mutants progressed until the sibling embryos were mature, we considered whether the developmental arrest might have occurred due to maternal abortion rather than problems with the seed development per se. Some embryo lethal mutants are able to be rescued if released from the developmental and physiological restrictions of the mother plant (Vernon and Meinke, 1995). To test this, mutant seed containing embryos were removed from sterilized siliques and grown on basal tissue culture media without hormones. Whole ovules taken at various stages of wild-type development were cultured simultaneously (e.g., Figure 4A). Wild-type embryos from eight-cell to globular stages were cultured and most often (<70%) gave rise to viable seed and seedlings (Figures 4B and 4C). Seedlings cultured from embryos grown at younger stages often displayed multiple shoots and usually had extensive root development (data not shown). This effect has been also reported by previous investigators when culturing embryos (Kost et al., 1992; Sauer and Friml, 2004). The range of developmental stages assayed and the resultant phenotypes for both wild type and mutant embryos is presented in Table 1.
Table 1.
Percentage of Embryos That Progressed to Various Stages of Development for in Vitro Culture of Wild-Type and Mutant (toz) Ovules
| Genotype | No Seed Growtha | Not Germinatedb | Germinate and Diec | Germinate and Lived | Potential as Plante | Total |
|---|---|---|---|---|---|---|
| Wild type | 14.7% | 6.3 | 3.2 | 7.4 | 68.4 | 95 |
| toz | 21.6% | 19.5% | 15.1% | 43.8% | 0.0 | 185 |
Seed contained embryos between four-cell proembryo to mid-globular stages (∼65 cells; according to Jürgens and Mayer [1994]).
Seed that did not grow after transfer to medium.
Seed that enlarged significantly but seedlings failed to germinate.
Seedlings that did germinate but showed no further development.
Seedlings that continued to grow but failed to produce a viable root system.
Seedlings that generated an extensive root and shoot system that could survive if transplanted to soil.
Mutant embryos from a range of stages (e.g., late stage, Figure 4D) were also grown on minimal media without hormones. Approximately 80% grew to mature seed, but they had a different form compared with the wild type (Figure 4E, Table 1). The postgermination phenotypes showed a range of highly abnormal features but generally showed distinct apical and basal regions (Figures 4F and 4H). One to three weeks after germination, the apex usually comprised of a dense mound of small cells, and scanning electron microscopy revealed that small groups of leaf primordia appeared to form in spirals around a central bulge of cells, which could be functioning as a shoot apical meristem (Figure 4G). As the seedlings developed, some areas of the apex produced an abundance of small lateral organs that eventually covered the entire apical area (Figures 4H and 4I). Sections of the seedlings showed that there continued to be atypical cell division patterns in the shoot apex and organ primordia (data not shown). In contrast with wild-type embryos, the ex planta–cultured toz mutants showed limited root development (Figures 4C and 4H). Primary roots were always very short (<2 cm), and between 2 and 4 weeks after germination, they ceased to grow. Clearing of the roots showed an abnormal cell pattern at the tip (Figure 4J). Lugol staining of the roots was performed to determine if the roots had terminally differentiated as would be indicated by the absence of root columella cells. One month after germination, none of the primary roots tested yielded the iodine precipitate; however, some of the young lateral roots of toz mutants showed faint staining (Figure 4K). This indicates that even though primary roots terminally differentiate, lateral root development may be initiated. However, lateral roots tips often became disorganized, forming callus-like tissue before ceasing to grow (Figure 4F). The toz mutants displayed these phenotypes consistently irrespective of whether the seeds were excised early (<20 embryonic cells) or at the latest stages of development, implying that abortion within the seed was a factor in toz embryo lethality in planta and supporting the notion that the observed phenotypes are associated with the disruption of TOZ function rather than artifacts of the technique of embryo rescue.
The TOZ Gene Is Preferentially Expressed in Dividing Cells
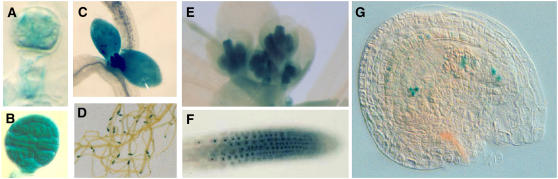
The toz allele results from an insertion of a gene trap Ds element that harbors the Escherichia coli gene uidA encoding the β-glucuronidase (GUS) enzyme. Gene trap insertions result in expression of the GUS gene under the control of the regulatory elements of the gene at the endogenous chromosomal location (Sundaresan et al., 1995). Therefore, heterozygous embryos, seedlings, and inflorescences were examined for TOZ expression by reporter GUS activity. Activity of the GUS enzyme was detected in young proliferating tissues, such as root tips, meristems, young leaves, and floral buds, and it also was observed in the mature embryo sac of the female gametophyte (Figure 5). In excised heterozygous embryos, expression was faintly detected very early in the proembryo (Figure 5A) and strongly in embryos with more than eight cells (Figure 5B). Expression in the embryo proper remained strong until cell division ceased prior to dormancy. In the seedling, expression was concentrated at the developing apex (Figure 5C) in all root tips (Figure 5D) and later in the developing flowers of the inflorescence (Figure 5E). When using high concentrations of ferricyanide inhibitors that prevent the diffusion of intermediates, the blue precipitate could be localized to the nucleus of the root tip cells and of the cells of the mature embryo sac within the ovule (Figures 5F and 5G). In summary, GUS expression was most prevalent in actively dividing cells in a variety of tissues and meristematic regions. These conclusions are consistent with data from transcriptome databases (see below).
Figure 5.
TOZ Expression Patterns.
In toz/+ heterozygous plants, GUS is expressed weakly at the four-cell proembryo stage (A) but strongly at the mid-globular stage (B). In the seedling, GUS staining is predominant in the shoot apex, young leaves (C), and root tips (D). In the inflorescence, the precipitate is strongest in the young stamens and carpels. Using a different staining solution that restricts the diffusion of intermediates, GUS can be localized to the nucleus in the roots (G) and also in each of the cells of the embryo sac in the mature ovule (H).
TOZ Encodes a Conserved WD40 Repeat Protein
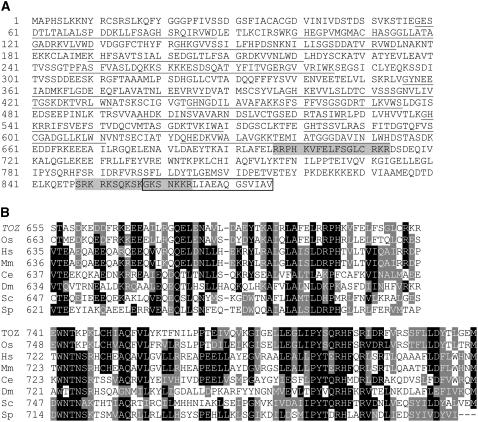
The toz mutant line was found to carry a single Ds insertion by DNA gel blots (data not shown). The genomic sequences flanking the left and right borders of the Ds insert were amplified by thermal asymmetric interlaced PCR and sequenced. BLAST searches revealed that the transposon had inserted two base pairs downstream of a stop codon of a gene encoding a WD40 repeat protein of unknown function (At5g16750). Several full-length cDNAs were isolated, sequenced, and compared with the genomic sequence. The TOZ gene was found to comprise 14 exons, which is consistent with the gene predictions reported in the relevant databases. Further analysis predicts a protein of 876 amino acids that contains 12 WD repeat domains (not eight as reported by The Arabidopsis Information Resource database; Figure 6A), but otherwise it contains no significant homology with other functional domains. Search for protein targeting signals (PSORT; http://psort.ims.u-tokyo.ac.jp/) strongly suggests that the sequence is nuclear localized (P = 0.98). The sequence predicts two nuclear localization signals at positions 696 to 713 and 848 to 865 (Figure 6A). Two independent publicly accessible transcriptome databases were examined for TOZ expression, the Bio-Array Resource database at http://bbc.botany.utoronto.ca/ and the AtGenExpress database at http://www.weigelworld.org/resources/microarray/AtGenExpress/, which is a compilation of microarray data. These databases indicated that TOZ expression is most abundant in actively dividing tissues, especially the shoot apical meristem, and lowest in nondividing tissues, such as mature leaves, confirming the conclusions from the reporter gene trap expression studies.
Figure 6.
Molecular Analysis of the TOZ Gene.
(A) The predicted translation from the cDNA sequence. The 12 WD40 repeat sequences have been underlined, the two putative nuclear localization signals are shaded, and the boxed amino acids represent those deleted in the mutant toz protein.
(B) Comparative amino acid interspecies homology for the TOZ domain at the C-terminal region. Os, Oryza sativa; Hs, Homo sapiens; Mm, Mus musculus; Ce, Caenorhabditis elegans; Sc, Saccharomyces cerevisiae; Sp, Saccharomyces pombe. A full list of TOZ homologs with accession numbers is available in Supplemental Table 1 online.
For verification of the gene identity by complementation, a genomic clone of 7.8 kb containing the TOZ transcribed region and 1 kb of 5′ promoter sequences was constructed (ProTOZ:TOZ; see Methods). After transformation of toz/+ plants, all primary transformants (T1) were tested to determine whether they also carried the Ds toz allele. Complementation of the phenotype was initially analyzed in the following generation (T2) where the kan segregation ratio served as a marker for the transmission of the toz allele. Six of eight lines independently transformed with the genomic construct showed a significant increase in kan segregation ratios from 1.5:1 (untransformed) to between 2.4:1 and 2.8:1. From one transformant, we recovered a line that was homozygous for the toz mutation and fully viable (Figure 1C), confirming that the mutation could be completely complemented using the genomic TOZ fragment.
For additional confirmation, we also used constructs containing the TOZ cDNA driven by the Arabidopsis CDC2a promoter, as this promoter can be used to direct expression specifically in proliferating tissues (Martinez et al., 1992). Only five independent lines were recovered that harbored the ProCDC2:TOZ construct, but four lines gave measures of complementation (kan segregation ratios between 2.2:1 and 2.9:1), and the fifth was homozygous for the Ds element, indicating complete complementation. The restored phenotype was also confirmed by analyzing embryo development in subsequent generations of toz/toz plants where the transforming construct was also homozygous (Figure 1D). In these cases, abnormal embryo development was ∼10% in the complemented lines, which is slightly greater than observed for wild-type plants (∼2%), but it still represents a high degree of complementation. In addition, upon outcrossing, we observed that complementation occurred only when the cDNA construct cosegregated with the toz allele in the subsequent generations.
To obtain additional plants with compromised TOZ activity, we generated nine lines harboring an antisense TOZ cDNA construct driven by the pCDC2 promoter in a wild-type background (ProCDC2:ZOT). Eight lines showed significantly reduced transmission of the antisense construct, with segregation of the selection marker (hygromycin) between 0.05:1 and 1.8:1. Analysis of the developing siliques showed embryos with typical toz mutant phenotypes (Figure 2L). Interestingly, the antisense TOZ lines sometimes generated mutant embryos that exhibited many more transverse divisions than observed for toz mutants (Figure 2M) or transverse divisions replaced other types of cell patterning (Figure 2N). We also counted the number of toz-like embryos in siliques from each line and found that those with the most mutant embryos also had the poorest transmission of the transgene, and vice versa, suggesting that the phenotypes were indeed caused by the presence of the construct (see Supplemental Figure 3 online). These observations further confirmed our conclusion that toz mutant phenotypes arose from disruption of TOZ gene activity.
TOZ Is Nuclear and Localizes to the Nucleolus
The Ds element resides in the 3′ untranslated region two bases downstream of the stop codon of the TOZ gene. Because expression of the GUS gene in a gene trap insertion normally relies on splicing from an intron or a coding exon (Sundaresan et al., 1995), we decided to investigate the basis for GUS expression. RT-PCR was performed using primers located in the 12th exon of the TOZ gene and within the GUS coding region (see Supplemental Figures 4A and 4B online, lane 5). Sequencing of the amplified fragment revealed that a TOZ-GUS transcriptional fusion resulted from a splicing event from the 14th exon of toz to the intron acceptor sequence of the GUS gene, giving rise to a deletion of the last 59 bp of the coding sequence. Significantly, a predicted intron donor site (P = 0.94, predicted by Net plant gene; http://www.cbs.dtu.dk/services/NetPGene/index.html) is present exactly where the splice occurred. Further analysis predicts that the transcript encodes a toz-uidA translational fusion, effectively generating a deletion of 19 amino acids at the C terminus of toz (Figure 6A) and addition of 604 amino acids of the GUS protein. We also determined the efficiency of the aberrant splicing by choosing primers that would amplify unspliced sequences. The RT-PCR product from the same 5′ position of the TOZ gene to the 3′ terminus of the Ds fragment was sequenced (see Supplemental Figure 4B online, lane 4), and the product was found to be of cDNA origin (no intron sequences) but contained the full-length TOZ transcript followed by the Ds 3′ flanking sequence. These results indicate that the toz Ds allele is not a null mutant for TOZ RNA transcripts. A comparison with RT-PCR using primers to the transcript wild-type TOZ allele shows that the amount of functional transcript from the mutant toz allele can be estimated to be at ∼1% of wild-type levels (see Supplemental Figure 4B online, lanes 3 and 4).
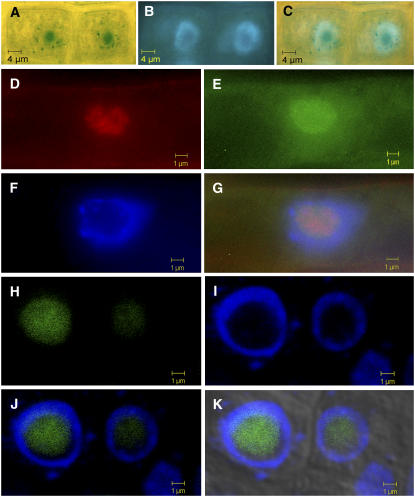
Considering that the gene trap insertion resulted in a GUS protein fused to the N-terminal 857 amino acids of the TOZ protein, it was likely that subcellular localization of the mutant toz-GUS hybrid protein could be determined. We performed GUS staining using higher concentrations of FeCN inhibitors to reduce diffusion and found that the root tissue showed a clear nuclear staining pattern (Figures 5F, 5G, and 7A). When the tissue was counterstained with DAPI, it revealed that the precipitate is localized to the nonchromatin region within the nucleus (Figures 7A to 7C). To test whether it was possibly targeted to the nucleolus, GUS polyclonal antibodies were used in whole-mount immunolocalization assays using root tissue. As a control, the roots were also double-labeled with antibodies to a mammalian nucleolar protein fibrillarin (which shows 69% sequence similarity with the Arabidopsis protein At FIB1) (Barneche et al., 2000). As shown in Figures 7D to 7G, both the TOZ-GUS fusion and fibrillarin proteins colocalize, confirming that the TOZ-GUS fusion protein is predominately restricted to the nucleolus. To further confirm the localization of the TOZ protein, we also generated fusions of TOZ to green fluorescent protein (GFP) and red fluorescent protein (RFP; DsRed). Analysis of plants carrying the TOZ-GFP fusion showed that the GFP signal was localized to the nucleolus (Figures 7H to 7K). Similar localization was obtained with the other fusions, GFP-TOZ, RFP-TOZ, and TOZ-RFP (see Supplemental Figure 5 online; data not shown). None of these fusions was able to complement the toz mutation, suggesting that the TOZ protein might not be able to tolerate significant N-terminal or C-terminal fusions. Nevertheless, the consistent nucleolar signal observed with all four types of fusions, together with the GUS fusion data, provide strong support for the nucleolar localization of the TOZ protein.
Figure 7.
Subcellular Localization of TOZ.
(A) to (G) Root cells from toz/+ plants. Cells yielding blue precipitate (A) were counterstained with DAPI (B). The overlay shows that the GUS is predominantly restricted from the chromatin region of the nucleus (C). Immunofluorescence staining was used to confirm whether toz-GUS was restricted to the nucleolus. Signal from antibodies to the nucleolar-localized human fibrillarin gene is shown in red (D), in the toz-GUS protein (E), and by DAPI staining (F). The overlay (G) shows that the fibrillarin signal mostly colocalized with toz-GUS, yet both are excluded from the chromatin region of the nucleus.
(H) to (K) Localization of TOZ-GFP fusion protein in roots. GFP fluorescence (H) localizes to the nonchromatin region of the nucleus where the nucleolus is located, as revealed by DAPI staining (I), DAPI and GFP (J), and DAPI, GFP, and bright-field images (K). The slight offset of the GFP signal relative to the DAPI signal is due to displacements in the focal planes of the two lasers at higher magnifications.
TOZ is a single-copy gene in Arabidopsis and also has single-copy homologs in a diverse range of eukaryotes, including rice (Oryza sativa), frogs, mice, humans, yeast, worms, flies, fungi, and other single-celled eukaryotes (Figure 6B). The plant homolog from O. sativa, for which full-length cDNA sequence is available, shares 60% identity and 77% similarity with TOZ over the full-length protein. From the animal and yeast homologs, the identity (and similarity) is reduced, ranging from 36% (55% similarity) for Homo sapiens to 29% (46% similarity) for Saccharomyces cerevisiae. The human protein, known as a Tbl3 (transducin β-like 3), and the mouse protein are predictably highly homologous to each other, sharing 88% identity and 94% similarity. When the C-terminal region (not including the WD40 repeats) is compared across species, it reveals a number of amino acids conserved for all of the proteins, as illustrated in Figure 6B. This region was recently recognized as a domain by Prodom (PD034404 - http://protein.toulouse.inra.fr/prodom/current/html/home.php), but the significance is still to be elucidated. To distinguish TOZ and its putative orthologs from other WD40 proteins, we refer to the homologous C-terminal region as the TOZ domain. Taken together, it appears that TOZ has single-gene homologs in a diverse range of eukaryotes, suggesting that they may share a similar conserved function in all eukaryotic species.
Of the TOZ homologs in different species, only the S. cerevisiae protein UTP13 (YLR222c) has been associated with a defined biochemical function. This protein was identified as being part of a large nucleolar U3 ribonucleoprotein complex required for 18S rRNA biogenesis (Dragon et al., 2002). In the first instance, the pre-rRNA transcribes as a single molecule containing the 18S, 5.6S, and 26S rRNA species. These are initially processed by the U3 complex within the nucleolus, releasing the mature 18S rRNA species that then forms part of the small ribosomal subunit. Another group (Grandi et al., 2002) reported that YLR222C was additionally found as part of the 90S preribosomal complex. Ribosome assembly is initiated in the nucleolus and then is transported into the cytoplasm where the subunits mature. YLR222C was not found to be part of the preribosomal subunits after export from the nucleolus. However, it was determined to be one of the major components of the 90S complex during the early formation of the ribosomes, as it has increased stoichiometric proportions when compared with the other proteins, suggesting a structural role in complex formation. We examined whether the toz mutation disrupts rRNA processing. By real-time RT-PCR analysis of toz mutant embryos cultured ex planta, we could not detect any major differences in unprocessed rRNA transcripts relative to sibling wild-type or heterozygous embryos compared with the controls using primers for either the 18S or the 26S RNA, with any change being at best a slight decrease corresponding to one cycle of amplification (see Supplemental Figure 6 online). This result might be due to residual TOZ function in the toz mutant, or possible redundancy of TOZ with other factors, and suggests that the toz phenotype may not arise solely from low levels of processed rRNA.
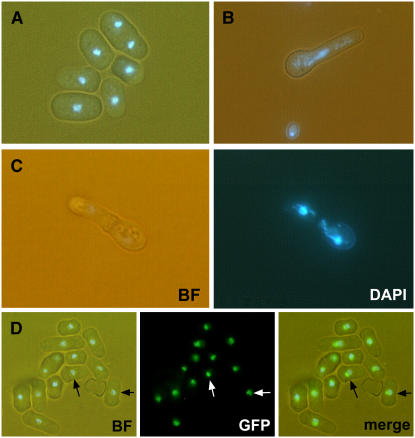
To assess the possible functions of the fungal homologs of TOZ in cell division, we generated disruptions of the TOZ homolog in the fission yeast Schizosaccharomyces pombe. As shown in Figures 8A to 8C, upon germination, the haploid spores carrying the disruption underwent one to three rounds of division and terminated with cell separation phenotypes. These included nuclei that were widely dispersed in the elongated cells (Figure 8B) or DNA in which the newly formed septum bisects the nuclear material (Figure 8C), which are similar to the CELL UNTIMELY TORN mutant phenotypes of S. pombe (Yanagida, 1998; Hirano, 2000). Even though our attempts to complement the S. pombe disruption with the Arabidopsis TOZ gene were not successful, the two proteins might yet perform related functions, as GFP localization of the Sp TOZ:GFP fusion protein also localizes to the nucleolus (Figure 8D), and in both cases, the mutations result in defects that affect cell division.
Figure 8.
Analysis of Sp TOZ in S. pombe.
(A) and (B) are superimposed bright-field and DAPI images. (C) shows bright-field and DAPI as separate panels. (D) shows superposed bright-field and DAPI images (left) as well as GFP (center) and merged images (right).
(A) Wild-type pombe have distinct small nuclei as shown by DAPI staining.
(B) and (C) By contrast, the deletion mutants (ΔSptoz) show germinated spores with dispersed nuclei. Some have undergone division, but the newly formed septum cuts the lagging DNA strands (C).
(D) GFP was engineered to produce a translational fusion with Sp TOZ and transformed into wild-type pombe for homologous recombination at the Sp TOZ locus. Sp TOZ:GFP detection shows close proximity to the nonchromatin region of the S. pombe nucleus (arrows).
DISCUSSION
The toz mutant phenotype is in contrast with other known mutations affecting orientation of division planes. In the ton/fass mutants, both longitudinal and transverse divisions are affected, including divisions of the suspensor and those of the proembryo, and are frequently replaced by oblique division planes (Torres-Ruiz and Jürgens, 1994). We find that in toz mutants, it is nearly always, if not exclusively, the longitudinal division planes that are affected, and these are generally replaced by transverse divisions rather than oblique divisions. Divisions of cells that normally divide in the transverse orientation were not affected, such as cells of the suspensor or the elongated zygote. In several instances, divisions that appear oblique to the apical-basal axis of the cell were actually perpendicular to a preceding oblique division, suggesting that they are in fact transverse divisions corresponding to a newly established axis (Figures 2G and 2H). The predominance of transverse divisions in toz mutants can be interpreted by considering such divisions as the ground state for the division choice. This implies that the division site may predominantly form perpendicular with the axis of growth, which is in general agreement with the rule proposed by Errera and states that the plane of cell division follows the shortest distance that will halve the volume of the cell (for review, see Smith, 2001). The selection of oriented division planes that occurs in the wild-type proembryo must require additional functions or signals that were disrupted in toz mutants. Staining with antibodies to tubulin and to Knolle reveals that toz mutant cells do form preprophase bands and normal cell plates, so that the defects in these mutants are unlikely to arise from an inability to form the appropriate microtubule structures for cell division (data not shown).
Effect of Misoriented Cell Divisions on Embryonic Patterning and Bilateral Symmetry
Many mutants disrupted in embryonic patterning display abnormal cell divisions as the earliest defect. In Arabidopsis, the ton1 or fass embryos have completely deregulated division planes, affecting all cells and divisions in the proembryo and the suspensor. The embryos and seedlings have morphological abnormalities, such as a compressed apical-basal axis, but still make recognizable organs and tissues according to the same developmental program as wild-type plants (Torres-Ruiz and Jürgens, 1994; Camilleri et al., 2002). Additionally, in the maize tangled mutant, the division planes in the leaves are near random, yet the overall leaf shape is not affected (Smith et al., 1996). It is therefore surprising that although in the toz mutant embryos the division planes are not as severely affected as in these other mutants, yet the toz mutation results in much stronger effects on development of the embryo. These observations can be reconciled if we consider the proposal of Meyerowitz (1996) that higher plants have mechanisms to adjust for aberrant planes of cell division through global controls that compensate for individual abnormal divisions by adjustments in the division plane of subsequent divisions. It is conceivable that the preference for transverse divisions in toz embryos, compared with the randomization of all divisions in the ton/fass mutants, results in an inability to adjust division planes to compensate for the previous wrong divisions and gives rise to more serious developmental defects. Studies with other mutants also indicate that under certain circumstances corresponding to specific types of divisions, compensatory mechanisms are unable to correct for the developmental effects of incorrect divisions. In the scarecrow mutation in Arabidopsis, the absence of periclinal divisions in a set of initials at the root apical meristem results in a failure of specification and differentiation of the cortex and endoderm cell layers (Di Laurenzio et al., 1996). Analysis of the maize Extra cell layers1 mutant (Kessler et al., 2002) shows that multiple layers of epidermal cells and abnormal thick narrow leaves arise from cells of the protoderm undergoing periclinal divisions instead of anticlinal divisions.
In most cases, it is thought that compensatory mechanisms exist that facilitate recovery of mutants that affect cell division planes and cell fate determination such that the later-stage embryos have a recognizable morphology (Traas et al., 1995; Meyerowitz, 1996). The toz mutant embryos differ in this respect, in that analysis of cell arrangements and gene expression patterns showed that toz embryos do not readily establish a radial symmetry and fail to initiate a bilateral symmetry. It is only under cultured conditions that the embryos clearly differentiate these stereotypical axes, despite the continuation of irregular cell division planes. One of the most striking defects was the inability of the large globular toz embryos to initiate lateral organs, despite that the number of cells in the mutant embryos far exceed that observed in the globular stages of wild-type embryos and are more similar to the late-heart-stage wild-type embryos (see Supplemental Figures 2G, 2I, and 2J). This was supported by analysis of the organ primordia marker genes ANT and FIL. In wild-type embryos, these genes are expressed at the onset of lateral organ development at the globular and transition stages, respectively. We only found one instance of ANT expression in a very large mutant embryo (Figure 3N) and no evidence of FIL expression (Figure 3P). This implies that it is likely that the failure to initiate the bilateral symmetry in toz embryos is not simply due to fewer cells but a defect in initiation of organogenesis. The radial and apical-basal axes were also disrupted, for example, in globular embryos of the wild type, MP is excluded from the protoderm (L1 layer) yet is expressed in the hypophysis. Our observation of the misexpression of MP in the L1 of toz mutants and its exclusion from the presumptive hypophysis suggests abnormalities associated with both the radial and apical-basal symmetries that may be caused by disruptions to the global controls over cell differentiation.
Possible Functions of the TOZ Protein
The TOZ gene product corresponds to a predicted WD40 repeat protein. Such proteins function in a variety of cellular processes, including cell cycle progression, signal transduction, gene regulation, and RNA processing. Putative single gene orthologs of TOZ that show significant conservation outside of the WD40 repeat region exist in widely divergent eukaryotic species (Figure 6B). The similar localization of these proteins in Arabidopsis and S. pombe (this study), humans (Andersen et al., 2002), and S. cerevisiae (Dragon et al., 2002) imply a conserved function. The only homolog with a putative assigned function is the budding yeast protein UTP13p. This protein has been identified as part of a conserved large ribonuclear complex, with a primary activity in pre-rRNA processing (Dragon et al., 2002) and pre-ribosome assembly (Grandi et al., 2002). In eukaryotes, three of the rRNA molecules are transcribed as a single pre-rRNA molecule that undergoes cleavage of internal and external spacer sequences and covalent modification prior to incorporation into 90S pre-ribosomal particles (Grandi et al., 2002). Another Arabidopsis gene that is likely to be involved in processing pre-rRNA was recently reported, SWA1 (Shi et al., 2005). The homolog of SWA1 from S. cerevisiae, UTP15, is predicted to form part of the same complex as UTP13 (Dragon et al., 2002), and thus it is likely to share in the same pre-rRNA processing function as TOZ. However, the swa1 mutant showed a different phenotype from toz; it disrupted the normal rate of progression of megagametophyte development, and very few embryo sacs containing the mutant allele were viable. Downregulation of SWA1 by RNA interference (RNAi) resulted in a significant reduction of rRNA processing and slower growth of the plants but without marked developmental abnormalities (Shi et al., 2005). By contrast, embryo sacs containing the toz-Ds allele appeared normal and can give rise to homozygous mutant embryos, but the embryos exhibit severe developmental defects. Furthermore, unlike the SWA1 RNAi plants, the toz mutant plants perpetuated in tissue culture exhibited only slight effects on rRNA processing (see Supplemental Figure 6 online). It is possible that these differences with SWA1 could be due to residual TOZ function in the toz mutant, which as described in Results is not a null allele and might provide significant activity for processing of the pre-rRNA and gametophyte viability. We note that our antisense toz alleles also generated the same phenotypes as the Ds insertion allele, but it is likely that these too are not null alleles. A search during this study for other insertions into the TOZ gene in publicly available collections that might correspond to null alleles revealed only a single insertion line, GABI 029G04, in which there is a T-DNA insertion in a TOZ exon. However, detailed molecular characterization of this insertion line revealed a rearrangement associated with the T-DNA so that it could not be used for determining the effects of a complete loss of TOZ function (A. Surendrarao and V. Sundaresan, unpublished results). Despite the relatively small effects on rRNA processing in the toz mutant, based on the localization studies in Arabidopsis (Figure 7; see Supplemental Figure 5 online) and S. pombe (Figure 8D) and homology to S. cerevisiae UTP13, it is likely that the TOZ protein has functions in rRNA processing. As discussed below, the effects of the toz mutation on cell division and development might arise from interactions between the nucleolus and the cell division machinery.
The Nucleolus and Regulation of Cell Division
As described in the Introduction, studies in animal and yeast cells have shown that the nucleolus has important functions in regulation of cell division. The slow growth of toz mutant cells and the asynchrony of cell division timing (as observed by the presence of three-, five-, or seven-celled embryos) could arise from defects in rRNA processing. In murine cell lines, inhibition of the nucleolar protein Bop1 results in a specific block in the maturation of the 28S and 5.8S rRNAs leading to a cell cycle arrest in G1. Significantly, they found that this was associated with downregulation of Cdk2 and Cdk4 kinase activities and hypophosphorylation of pRb, triggering the G1/S checkpoint (Pestov et al., 2001). Furthermore, the G1 arrest occurred prior to a notable disruption to the global translation rate that might occur due to depletion of the cytoplasmic ribosome pool. These results have led to the proposal that ribosome biosynthesis, mediated by rRNA processing, serves as a cell cycle checkpoint (Strezoska et al., 2002).
It is necessary to consider the possibility that the observed effects on division planes in toz mutants are a consequence of slower growth arising from defects in ribosome synthesis. Several lines of evidence from previous mutant studies in Arabidopsis argue against a simple connection between general effects on cell growth and metabolism and the selection of division planes. Many mutations in genes controlling general cellular processes (e.g., housekeeping genes) leading to embryo arrest have been described, but in general, the planes of cell division are maintained as in the wild type at least in the early divisions of the proembryo where the patterns are more predictable (Apuya et al., 2001; Baud et al., 2004; Brukhin et al., 2005). Furthermore, several mutants in ribosomal genes have been described, but none of them lead to aberrant cell division patterns in the embryo (Ito et al., 2000; Weijers et al., 2001; Nishimura et al., 2005; Kojima et al., 2007). For example, disruption of the RPS5 gene leads to insufficiency of ribosomes and embryo arrest at the globular stage, yet the patterns of cell division are similar to those in wild-type embryos (Weijers et al., 2001). Recently, it has been shown that disruption of the Arabidopsis nucleolin gene At NucL1 significantly affects rRNA processing, but homozygous mutant plants show relatively minor developmental defects despite very slow growth (Kojima et al., 2007). As mentioned above, similar results have been obtained in mutants generated by RNAi of the SWA1 gene, which is also a component of the rRNA processing complex (Shi et al., 2005). Nonetheless, we cannot presently rule out that delayed synthesis of rRNA in the toz mutant cells and consequently slower cell divisions through a checkpoint mechanism (see above), indirectly affect division site choice resulting in aberrant planes of cell division in toz embryos.
Another possible explanation for the unusual effects of the toz mutation is that in addition to rRNA processing, the TOZ protein participates in specialized nucleolar functions that interact with the cellular control of cytokinesis. In budding yeast, the Nop15p protein, which is required for pre-rRNA processing, has been shown to be essential for cytokinesis (Oeffinger and Tollervey, 2003; Dez and Tollervey, 2004). The Nop15p protein is delocalized from the nucleolus during mitosis and appears to form a transient bridge between the daughter nuclei at telophase (Oeffinger and Tollervey, 2003). BLAST searches show that Arabidopsis contains a putative Nop15p ortholog encoded by the At5g04600 gene, which could also have a function in cell division, though unfortunately this possibility cannot be easily tested because no insertion lines for this gene are currently available. Additional functions for the TOZ orthologs have not been described in the literature. A search of the yeast genome databases (http://db.yeastgenome.org) reveals only that disruption of the S. cerevisae TOZ gene UTP13 results in lethality. In view of the finding that disruption of S. pombe TOZ results in mitotic cell division defects (Figure 8; this study), the possibility of an additional function for TOZ in aspects of cell division control should be considered.
It has been shown that plant nucleoli have some unique features compared with nucleoli from other eukaryotes. In a recently conducted proteomic analysis of Arabidopsis nucleoli, components of the exon junction complex, which links transcription and processing to mRNA export and stability, were identified, suggesting the existence of novel functions related to these processes for nucleoli in plants (Pendle et al., 2005). If there are specific factors in plant cells required for establishment of the division plane at G2/M, these may also be affected by nucleolar functions. The TOZ protein could be involved in such processes directly through interactions with other proteins or indirectly by affecting nucleolar structure and organization. Further studies on the dynamics of localization of TOZ during mitosis and cytokinesis could help to distinguish between these possibilities. In either case, the unique effects of the loss of TOZ function on cell division planes and on the expression of embryo patterning genes are suggestive of an involvement of the nucleolus in the spatial and temporal control of plant cell division.
METHODS
Plant Material
The toz-1 allele was isolated as line SGT9551 during a visual screen for embryo lethal mutants from F3 populations of Ds-transposed insertion lines in Arabidopsis thaliana as described by Sundaresan et al. (1995) and Parinov et al. (1999). Plant materials were plated on growth media (4.6 g/L Murashige and Skoog salts [Sigma-Aldrich] and 2% sucrose, pH 6.8) containing kan (50 μg/mL) to further select for toz-1/+ heterozygotes. All plants were transferred to soil and grown at 22°C under long daylight conditions. Plant transformant lines containing the GFP or RFP constructs were grown vertically on Murashige and Skoog plates containing 3% sucrose. All constructs made during this study were transformed into toz/+ heterozygous or Landsberg erecta plants using Agrobacterium tumefaciens (strain AGL1) by the floral dip method (Clough and Bent, 1998) and were selected by resistance to hygromycin B (20 μg/mL).
In Vitro Cultures of Developing Seed and Seedlings
Siliques including ∼2 mm of pedicle were collected at several stages of development from wild-type (Landsberg) and toz-1/+ plants, surface sterilized (20% bleach and 0.1% Tween 20) for up to 10 min, and rinsed well in sterile water. Siliques were dissected under the microscope in sterile conditions. Developing seed were excised form the placenta and transferred to plates containing growth media (half-strength B5 salts [Sigma-Aldrich], 2% sucrose, and 0.2% phytagel [Sigma-Aldrich], pH 6.0, supplemented with 400 mg/L glutamine) based on Kost et al. (1992). To determine the developmental stage taken for in vitro growth, a few seed from each silique were transferred to a slide with clearing solution (chloralhydrate:distilled water:glycerol [8:3:0.5]) and viewed after 1 h. Culture plates were kept in the dark at 22°C until germination, when they were transferred to long-day conditions at the same temperature and transferred to fresh media every 2 to 4 weeks. Callus tissue was induced by transferring tissue to growth media containing 1 mg/L 2,4-D and 0.25 mg/L kinetin.
Molecular Analysis and Plasmid Strains
Isolation of flanking DNA probes was performed by thermal asymmetric interlaced PCR using the Ds-specific primers and conditions described by Parinov et al. (1999). The flanking DNA was used to identify the corresponding gene At5g16750 by BLAST-N search of the Arabidopsis database. To test for complementation of the mutant, we generated the genomic construct pCOM2.2. The DNA flanking the 3′ end of the Ds insert was used to probe an Arabidopsis genomic DNA library (Stratagene). A phagemid containing a 6.2-kb fragment, encompassing all the TOZ gene plus 398 bp upstream and 1083 bp 3′ to the translation stop codon, was subcloned into the pCAMBIA1300 vector using KpnI-EcoRI restriction sites, generating the construct pCOM1. As the distance between the start of the TOZ coding sequence and the ATG of the closest neighboring gene was only 1 kb, an additional 520 bp of the promoter upstream from the KpnI site was isolated by PCR (primers G29R5′ [5′-GCAGCTCGCTTTTATACCTTGTC-3′] and pBam1F [5′-GCGGATCCTGGATCCGAATCTGACATCT-3′) and subcloned into pCOM1 using BamHI-KpnI restriction sites.
The TOZ cDNA clones were isolated from a size-selected (3 to 6 kb) cDNA library, CD4-16 (prepared by J. Ecker; Library:2271245), obtained from the ABRC, with use of a DNA probe corresponding responding to the 5′ end of the predicted gene (primers pC58F [5′-GTCGTGATGGCTCCTCATTC-3′] and Ex2.1R [5′-TGACACAACCCCTTTATGGC-3′]). A construct based on the pCAMBIA1300 vector containing the 35S promoter of Cauliflower mosaic virus and Nos plant terminator (gift of Weicai Yang, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) was used as a base for subcloning of the full-length TOZ cDNA using restriction sites XbaI-SalI (sense) or BamHI-KpnI (antisense). The ProCDC2:TOZ and ProCDC2:ZOT constructs were prepared in the original cDNA vector of the lambda zap library by digestion of either sense or antisense oriented clones with HindIII (filled in)-SalI and subcloning of the CDC2 promoter from PSY91 (gift from Benedikt Kost, Heidelberg Institute of Plant Sciences) using the 1.7-kb SalI-SmaI fragment. This construct was subcloned into the pCAMBIA1300 vector containing the Nos terminator sequence (cloned at EcoRI-SacI; gift from Weicai Yang, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing) using XbaI-KpnI digests for both the ProCDC2:TOZ sense and antisense inserts and the pCAMBIA1300-NOS vector. For the construction of fluorescent protein TOZ fusions, rsmGFP (Davis and Vierstra, 1998) or DsRed2 (Clontech) were PCR amplified and cloned either 3′ (TOZ-GFP and TOZ-RFP) or 5′ (GFP-TOZ and RFP-TOZ) to the TOZ full-length cDNA sequence of 876 amino acids using (Ala)8 linkers. For fluorescence studies, expression of the fusion was driven by the CDC2a promoter used successfully for complementation with the TOZ cDNA (see above) or with the 35S promoter; similar localization results were obtained with either promoter.
For RT-PCR, isolated RNA was treated with DNase (Promega). cDNA was synthesized from 2 μg of total RNA using an oligo(dT) primer and MLV reverse transcriptase. Primers used for amplification for semiquantitave PCR are as follows: 9551F forward primer (5′-TGAGCTTCGTAGGCCTCACA-3′); 9551R reverse primer in the coding region (5′-TTACGCTTCCCTGTGCTTCG -3′); GUSR reverse primer at the 5′ end of the GUS coding region (5′-TGAATGCCCACAGGCCGTCGAG-3′); and Ds3-2, the Ds-specific flanking primer (5′-CCGGTATATCCCGTTTTCG-3′). The RT-PCR products were confirmed by sequencing following gel purification.
In situ hybridizations were performed as described previously (Hamann et al., 2002).
Microscopy
Whole-mount embryos were prepared by clearing in Herr's solution (85% lactic acid, chloral hydrate, phenol, clove oil, and xylene [2:2:2:2:1]) from 2 h to overnight. Slides were viewed using a Leica DMR19 microscope using differential interference contrast optics. For scanning electron microscopy, fresh material was collected, fixed, coated, and photographed as previously described (Elliott et al., 1996). Samples were viewed using a JEOL JSM-5310 LV microscope and images captured using JEOL SEMafore software.
GUS Staining Assays
Histochemical staining for GUS activity was performed using the standard 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) solution (100 mM sodium phosphate buffer, pH 7.0, 0.1% Triton X-100, and 1 mg/mL X-Gluc [Biosynth], with the addition of either 2 or 10 mM potassium ferricyanide and potassium ferrocyanide). The samples were vacuum-infiltrated in X-Gluc solution for 10 min and kept at 37°C overnight. Samples were then cleared in 70% ethanol. Counterstaining of nuclei was performed by incubation in DAPI (1 μg/mL) for 20 min and rinsing in water for 30 min.
Antibody Staining and Confocal Laser Scanning Microscopy
Embryos were fixed and removed from seed for immunolocalization as previously described (Lauber et al., 1997). Antibodies and dilutions not reported by Lauber et al. (1997) were as follows: goat anti-fibrillarin antiserum (1:250, A-16: Sc-11335; Santa Cruz Biotechnology), rabbit anti-GUS antiserum (1:400; Molecular Probes); 4,6-dichlorotriazinyl amino fluorescein–conjugated secondary donkey anti-rabbit antibody (1:200; Dianova); and Cy3-conjugated secondary donkey anti-goat antibody (1:500; Dianova). DAPI staining was performed as described (Lauber et al., 1997). Calcoflour (1 μg/mL) and DAPI (1 μg/mL) were used to stain slides with fixed embryos for 20 min at 37°C and rinsed several times in water. Microscopy was performed with a Leica confocal laser scanning microscope with Leica TCS-NT software. The standard objective was 63 (water immersion), and scanning was performed with electronic magnification (Volker et al., 2001). Fresh tissue for confocal analysis was examined using a Zeiss confocal microscope (LSM-510), with either ×10 or ×40 standard water objectives, and with the manufacturer-supplied software.
The images in Figures 7H to 7K and Supplemental Figure 5 online were obtained with a Zeiss 510 Meta Multiphoton laser scanning microscope. The GFP was excited using the 488-nm line from a Losos argon laser. The DsRed was excited using the 543-nm line from a Losos HeNe laser. Multiphoton excitation for DAPI was performed using a Coherent Mira-90 pump laser and a Verdi-V10 laser that provides wavelengths from 700 to 1000 nm.
Yeast Methodology
Media for vegetative growth were as described by Moreno et al. (1991) and Keeney and Boeke (1994). Chemical transformations of Schizosaccharomyces pombe were performed using a lithium acetate–mediated method (Moreno et al., 1991; Keeney and Boeke, 1994).
The deletion strain for the pombe-TOZ gene (ΔSpTOZ:Ura4+) was generated by subcloning the 5′ and 3′ flanking sequences of gene SPCC16A11.02 and introducing it into the pBluescript vector containing the pombe Ura4+ gene (gift of Mohan Balasubramanian, Tamesk Lifescience Laboratories). PCR-generated fragments from the 5′ genomic DNA flank (primers pom5.1F [5′-GACGGATCCCTTTGGGACCAT-3′] and pom5.1R [5′-CAGAATTCTCCCCAATTGGCGC-3′]) were digested with BamHI and EcoRI, respectively, and cloned upstream of the Ura4+ gene. Genomic DNA sequences 3′ to the SPCC16A11.02 stop codon were cloned using primers pom3.1F′ (5′-GACCTCGAGTCAACGATCTCATTGA-3′) and pom3.1R (5′-GACGGATCCATCACTCGTAATTCAT-3′). Fragments were digested with XhoI and KpnI, respectively, to facilitate cloning. Transformation was performed with KpnI, linearized construct into chemically competent diploid cells (ΔUra−, ΔLeu−, ΔMet−).
The GFP fusion with the SpToz gene (Sp TOZ:GFP:Ura4+) was generated in pJK210, containing the GFP coding sequence (gift of Kelvin Wang, Genome Institute of Singapore), and was digested with KpnI and SmaI and ligated with a 2-kb genomic DNA fragment containing the N-terminal region of Sp TOZ (PCR product using primers pomG1F [5′-TCAGGTACCGGTAATTACGATGACAATAG-3′] and pomG1R [5′-CTCACCCGGGAATAACATAATC-3′] digested with KpnI or SmaI, respectively). Transformation was performed with PstI-linearized construct into chemically prepared haploid competent cells.
To determine if the Arabidopsis TOZ gene could complement the deletion mutant, Arabidopsis TOZ cDNA was cloned into vectors containing the thymine-repressible nmt1+ promoter (Rep1) or a similar vector containing a highly attenuated version of nmt1, denoted as Pnmt1-81 (Rep81) (Basi et al., 1993; Keeney and Boeke, 1994). Both vectors were transformed into haploid S. pombe strain (ΔUra−, ΔLeu−, ΔMet−), and transformants were selected on media containing thiamine (2 μM) to repress the expression of the construct. Triploid crosses were performed using At TOZ-1:Leu+ or At TOZ-81:Leu+ with ΔSpTOZ:Ura4+ diploid cells according to Keeney and Boeke (1994) and spores selected using random spore analysis selection on minimal media without thiamine (http://pingu.salk.edu/∼forsburg/diploids.html).
DAPI staining of cells was performed after fixation for 30 min in 3.7% paraformaldehyde in PBS. Cells were washed thrice with PBS, then DAPI was added at 1 μg/mL for 15 min and cells were mounted in antifade solution. Cells were viewed using a Leica DMLB microscope and images captured using an Optronics DEI-750T CCD camera. Leica Qwin software was used to acquire the images.
Accession Number
The Arabidopsis Genome Initiative locus identifier for TOZ is At5g16750.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Distribution of the Angles of First Division Planes in toz Proembryos.
Supplemental Figure 2. Cellular Patterning in toz Embryos.
Supplemental Figure 3. Phenotypes of the ProCdc2:TOZ Antisense Plants.
Supplemental Figure 4. Molecular Analysis of TOZ Transcripts.
Supplemental Figure 5. Localization of TOZ with GFP-TOZ and TOZ-RFP Chimeric Proteins.
Supplemental Figure 6. Real-Time RT-PCR Analysis of 18S rRNA Processing in Cultured toz Homozygous and Sibling Normal Embryos.
Supplementary Material
Acknowledgments
We thank Benedikt Kost (Heidelberg Institute of Plant Sciences, Germany) for help with the in vitro culture of embryos, Imran Siddiqi (Centre for Cellular and Molecular Biology, Hyderabad, India) for investigating the possible gametophytic toz phenotype, and Weicai Yang for advice and assistance. We also thank Claire Ellis, Sergey Parinov, Kelvin Wong, Regina Zahn, and Mohan Balasubramanian (Temasek Life Science Laboratories, Singapore) for guidance and assistance with yeast genetics. R.M. thanks James H. Resau (Van Andel Research Institute, Grand Rapids, MI) for support. This research was funded by the Agency for Science and Technological Research, Singapore, and by Award IBN 0235548 to V.S. and R.M. from the National Science Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Venkatesan Sundaresan (sundar@ucdavis.edu).
Online version contains Web-only data.
References
- Andersen, J.S., Lyon, C.E., Fox, A.H., Leung, A.K., Lam, Y.W., Steen, H., Mann, M., and Lamond, A.I. (2002). Directed proteomic analysis of the human nucleolus. Curr. Biol. 12 1–11. [DOI] [PubMed] [Google Scholar]
- Apuya, N.R., Yadegari, R., Fischer, R.L., Harada, J.J., Zimmerman, L., and Goldberg, R.B. (2001). The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60α gene. Plant Physiol. 126 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barneche, F., Steinmetz, F., and Echeverria, M. (2000). Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275 27212–27220. [DOI] [PubMed] [Google Scholar]
- Basi, G., Schmid, E., and Maundrell, K. (1993). TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123 131–136. [DOI] [PubMed] [Google Scholar]
- Baud, S., Bellec, Y., Miquel, M., Bellini, C., Caboche, M., Lepiniec, L., Faure, J.-D., and Rochat, C. (2004). gurke and pasticcino3 mutants affected in embryo development are impaired in acetyl-CoA carboxylase. EMBO Rep. 5 515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L. (2000). Axial patterning in leaves and other lateral organs. Curr. Opin. Genet. Dev. 10 399–404. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Eshed, Y., and Baum, S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18 134–141. [DOI] [PubMed] [Google Scholar]
- Brukhin, V., Gheyselinck, J., Gagliardini, V., Genschik, P., and Grossniklaus, U. (2005). The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis. Plant Cell 17 2723–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri, C., Azimzadeh, J., Pastuglia, M., Bellini, C., Grandjean, O., and Bouchez, D. (2002). The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP). for use in higher plants. Plant Mol. Biol. 36 521–528. [DOI] [PubMed] [Google Scholar]
- Dez, C., and Tollervey, D. (2004). Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 7 631–637. [DOI] [PubMed] [Google Scholar]
- Di Bacco, A., Ouyang, J., Lee, H.-Y., Catic, A., Ploegh, H., and Gill, G. (2006). The SUMO-specific protease SENP5 is required for cell division. Mol. Cell. Biol. 26 4489–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio, L., Wysocka-Diller, J., Malamy, J.E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M.G., Feldmann, K.A., and Benfey, P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86 423–433. [DOI] [PubMed] [Google Scholar]
- Dragon, F., et al. (2002). A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, R.C., Betzner, A.S., Huttner, E., Oakes, M.P., Tucker, W.Q., Gerentes, D., Perez, P., and Smyth, D.R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi, P., Rybin, V., Bassler, J., Petfalski, E., Strauss, D., Marzioch, M., Schafer, T., Kuster, B., Tschochner, H., Tollervey, D., Gavin, A.C., and Hurt, E. (2002). 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10 105–115. [DOI] [PubMed] [Google Scholar]
- Grimm, T., Holzel, M., Rohrmoser, M., Harasim, T., Malamoussi, A., Gruber-Eber, A., Kremmer, E., and Eick, D. (2006). Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 34 3030–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, T., Benkova, E., Baurle, I., Kientz, M., and Jürgens, G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, T. (2000). Chromosome cohesion, condensation, and separation. Annu. Rev. Biochem. 69 115–144. [DOI] [PubMed] [Google Scholar]
- Hofmeister, W. (1863). Zusatze und Berichtigungen zu den 1851 veröffenlichen Untersuchungengen der Entwicklung höherer Krptogamen. Jb. Wiss. Bot. 3 259–293. [Google Scholar]
- Ito, T., Kim, G.T., and Shinozaki, K. (2000). Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon mutagenesis causes aberrant growth and development. Plant J. 22 257–264. [DOI] [PubMed] [Google Scholar]
- Jürgens, G., and Mayer, U. (1994). Arabidopsis. In Embryos: Colour Atlas of Development. J.B.L. Bard, ed (London: Wolfe Publishing), pp. 7–21.
- Jürgens, G., Torres Ruiz, R.A., and Berleth, T. (1994). Embryonic pattern formation in flowering plants. Annu. Rev. Genet. 28 351–371. [DOI] [PubMed] [Google Scholar]
- Kawamura, E., Himmelspach, R., Rashbrooke, M.C., Whittington, A.T., Gale, K.R., Collings, D.A., and Wasteneys, G.O. (2006). MICROTUBULE ORGANIZATION 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiol. 140 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, J.B., and Boeke, J.D. (1994). Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, S., Seiki, S., and Sinha, N. (2002). Xcl1 causes delayed oblique periclinal cell divisions in developing maize leaves, leading to cellular differentiation by lineage instead of position. Development 129 1859–1869. [DOI] [PubMed] [Google Scholar]
- Kojima, H., Suzuki, T., Kato, T., Enomoto, K., Sato, S., Kato, T., Tabata, S., Sáez-Vasquez, J., Echeverría, M., Nakagawa, T., Ishiguro, S., and Nakamura, K. (2007). Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 49 1053–1063. [DOI] [PubMed] [Google Scholar]
- Kost, B., Potrykus, I., and Neuhaus, G. (1992). Regeneration of fertile plants from excised immature zygotic embryos of Arabidopsis thaliana. Plant Cell Rep. 12 50–54. [DOI] [PubMed] [Google Scholar]
- Lauber, M.H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W., and Jürgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125 3027–3035. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Martinez, M.C., Jorgensen, J.E., Lawton, M.A., Lamb, C.J., and Doerner, P.W. (1992). Spatial pattern of cdc2 expression in relation to meristem activity and cell proliferation during plant development. Proc. Natl. Acad. Sci. USA 89 7360–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, D.A., and Olson, M. (2006). What is new in the nucleolus? EMBO Rep. 7 870–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClinton, R.S., and Sung, Z.R. (1997). Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta 201 252–260. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125 2935–2942. [DOI] [PubMed] [Google Scholar]
- Meyerowitz, E.M. (1996). Plant development: Local control, global patterning. Curr. Opin. Genet. Dev. 6 475–479. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194 795–823. [DOI] [PubMed] [Google Scholar]
- Nishimura, T., Wada, T., Yamamoto, K.T., and Okada, K. (2005). The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger, M., and Tollervey, D. (2003). Yeast Nop15p is an RNA-binding protein required for pre-rRNA processing and cytokinesis. EMBO J. 22 6573–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, M.O., Dundr, M., and Szebeni, A. (2000). The nucleolus: An old factory with unexpected capabilities. Trends Cell Biol. 10 189–196. [DOI] [PubMed] [Google Scholar]
- Pagnussat, G.C., Yu, H.J., Ngo, Q.A., Rajani, S., Mayalagu, S., Johnson, C.S., Capron, A., Xie, L.F., Ye, D., and Sundaresan, V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132 603–614. [DOI] [PubMed] [Google Scholar]
- Parinov, S., Sevugan, M., Ye, D., Yang, W.C., Kumaran, M., and Sundaresan, V. (1999). Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle, A.F., Clark, G.P., Boon, R., Lewandowska, D., Lam, Y.W., Andersen, J., Mann, M., Lamond, A.I., Brown, J.W., and Shaw, P.J. (2005). Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 16 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov, D.G., Strezoska, Z., and Lau, L.F. (2001). Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: Effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell. Biol. 21 4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes, O., Li, C.F., Nunes, P.C., Haag, J., Ream, T., Vitins, A., Jacobsen, S.E., and Pikaard, C.S. (2006). The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126 79–92. [DOI] [PubMed] [Google Scholar]
- Sauer, M., and Friml, J. (2004). In vitro culture of Arabidopsis embryos within their ovules. Plant J. 40 835–843. [DOI] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Liu, Y.G., Shibata, D., Kanaya, E., Morita, E.H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, P., and Doonan, J. (2005). The nucleolus: Playing by different rules? Cell Cycle 4 102–105. [DOI] [PubMed] [Google Scholar]
- Shi, D.Q., Liu, J., Xiang, Y.H., Ye, D., Sundaresan, V., and Yang, W.C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, W., Seol, J.H., Shevchenko, A., Baskerville, C., Moazed, D., Chen, Z.W., Jang, J., Shevchenko, A., Charbonneau, H., and Deshaies, R.J. (1999). Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97 233–244. [DOI] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128. [DOI] [PubMed] [Google Scholar]
- Smith, L.G. (2001). Plant cell division: Building walls in the right places. Nat. Rev. Mol. Cell Biol. 2 33–39. [DOI] [PubMed] [Google Scholar]
- Smith, L.G., Hake, S., and Sylvester, A.W. (1996). The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development 122 481–489. [DOI] [PubMed] [Google Scholar]
- Sollner, R., Glasser, G., Wanner, G., Somerville, C.R., Jürgens, G., and Assaad, F.F. (2002). Cytokinesis-defective mutants of Arabidopsis. Plant Physiol. 129 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strezoska, Z., Pestov, D.G., and Lau, L.F. (2002). Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J. Biol. Chem. 277 29617–29625. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9 1797–1810. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz, R.A., and Jürgens, G. (1994). Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120 2967–2978. [DOI] [PubMed] [Google Scholar]
- Tsai, R.Y.L., and McKay, R.D.G. (2002). A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16 2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas, J., Bellini, C., Nacry, P., Kronenberger, J., Bouchez, D., and Caboche, M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375 676–677. [Google Scholar]
- Vernon, D.M., and Meinke, D.W. (1995). Late embryo-defective mutants of Arabidopsis. Dev. Genet. 16 311–320. [Google Scholar]
- Volker, A., Stierhof, Y.D., and Jürgens, G. (2001). Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J. Cell Sci. 114 3001–3012. [DOI] [PubMed] [Google Scholar]
- Yanagida, M. (1998). Fission yeast cut mutations revisited: control of anaphase. Trends Cell Biol. 8 144–149. [DOI] [PubMed] [Google Scholar]
- Weijers, D., Franke-van Dijk, M., Vencken, R.J., Quint, A., Hooykaas, P., and Offringa, R. (2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128 4289–4299. [DOI] [PubMed] [Google Scholar]
- Zimmerman, W.C., and Erikson, R.L. (2007). Polo-like kinase 3 is required for entry into S phase. Proc. Natl. Acad. Sci. USA 104 1847–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.