Abstract
We investigated the genetic pathway in Arabidopsis thaliana targeted during infection by cucumber mosaic virus (CMV) 2b protein, known to suppress non-cell-autonomous transgene silencing and salicylic acid (SA)–mediated virus resistance. We show that 2b expressed from the CMV genome drastically reduced the accumulation of 21-, 22-, and 24-nucleotide classes of viral small interfering RNAs (siRNAs) produced by Dicer-like4 (DCL4), DCL2, and DCL3, respectively. The defect of a CMV 2b–deletion mutant (CMV-Δ2b) in plant infection was efficiently rescued in Arabidopsis mutants producing neither 21- nor 22-nucleotide viral siRNAs. Since genetic analysis further identifies a unique antiviral role for DCL3 upstream of DCL4, our data indicate that inhibition of the accumulation of distinct viral siRNAs plays a key role in 2b suppression of antiviral silencing. Strikingly, disease symptoms caused by CMV-Δ2b in Arabidopsis mutants defective in antiviral silencing were as severe as those caused by CMV, demonstrating an indirect role for the silencing suppressor activity in virus virulence. We found that production of CMV siRNAs without 2b interference depended largely on RNA-dependent RNA polymerase 1 (RDR1) inducible by SA. Given the known role of RDR6-dependent transgene siRNAs in non-cell-autonomous silencing, our results suggest a model in which 2b inhibits the production of RDR1-dependent viral siRNAs that confer SA-dependent virus resistance by directing non-cell-autonomous antiviral silencing.
INTRODUCTION
RNA silencing refers to related gene-silencing mechanisms with specificity determined by small RNAs of 21 to 30 nucleotides in length (Meister and Tuschl, 2004; Baulcombe, 2005). In Arabidopsis thaliana, RNA silencing is mediated by microRNAs (miRNAs) and three size classes of small interfering RNAs (siRNAs) of 21, 22, and 24 nucleotides long (Herr, 2005). These small RNAs are the products of four Dicer-like (DCL), double-stranded (ds) RNA-specific ribonucleases encoded by the Arabidopsis genome. miRNAs are processed by DCL1 from stem-loop structures of primary nuclear transcripts, and only partial loss-of-function dcl1 mutants are viable because of the essential roles of miRNAs in development (Chen, 2005). Both genetic and biochemical analyses have shown that the 21-, 22-, and 24-nucleotide classes of siRNAs are produced by DCL4, DCL2, and DCL3, respectively (Herr, 2005; Qi and Hannon, 2005). For example, trans-acting siRNAs mediating silencing of endogenous genes and secondary siRNAs required for cell-to-cell silencing spread are 21 nucleotides long and DCL4 dependent (Herr, 2005; Qi and Hannon, 2005; Voinnet, 2005). By contrast, the siRNAs derived from transposons, repeated sequences, and heterochromatin implicated in the methylation of DNA and chromatin are 24 nucleotides long and DCL3 dependent (Matzke and Birchler, 2005). The precursors of these endogenous siRNAs are most likely dsRNAs, because of the requirement for a distinct RNA-dependent RNA polymerase gene (RDR) for the production of each siRNA class, RDR6 and RDR2 for 21- and 24-nucleotide classes of siRNAs, respectively (Herr, 2005; Qi and Hannon, 2005).
The first biological function established for RNA silencing was as a defense mechanism against viruses in plants (Voinnet, 2005). Virus-derived siRNAs (viRNAs) detected in infected plants and insects include both positive and negative polarities and cover the entire viral genome (Hamilton and Baulcombe, 1999; Li et al., 2002). In Drosophila melanogaster, infection with (+)-strand RNA viruses results in the production of the 22-nucleotide viRNAs by Dicer2, known to process exogenous dsRNA into siRNAs but dispensable for miRNA production (Wang et al., 2006). By contrast, R2D2, which acts downstream of Dicer2 to load siRNAs into the Argonaute2 (AGO2)-containing RNA-induced silencing complex, is essential for the activity but dispensable for the production of viRNAs (Wang et al., 2006). Recent genetic studies in Arabidopsis (Xie et al., 2004; Bouche et al., 2006; Deleris et al., 2006; Fusaro et al., 2006) showed that DCL4 acts as the primary virus sensor to produce 21-nucleotide siRNAs of potent antiviral activity. In the absence of DCL4, 22- and 24-nucleotide viRNAs are produced by DCL2 and DCL3, respectively, although it appears that only the 22-nucleotide viRNAs are sufficient to mediate independent antiviral silencing (Deleris et al., 2006). Whether or not the biogenesis of viRNAs in Arabidopsis requires any additional host proteins is unknown.
Identification of potyviral helper component–proteinase (HC-Pro) and the cucumber mosaic virus (CMV) 2b protein as viral suppressors of RNA silencing/RNA interference (RNAi) (VSR) revealed a new virus counterdefensive strategy (Anandalakshmi et al., 1998; Beclin et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998). Since 1998, numerous VSRs encoded by both plant and animal viruses have been reported (Roth et al., 2004; Li and Ding, 2006). Extensive analyses in systems that assay for the suppression of transgene silencing have shown that HC-Pro and 2b, both required for long-distance virus movement and for disease symptom expression in infected plants, represent prototypes of two distinct groups of VSRs (Li and Ding, 2001; Roth et al., 2004). HC-Pro is a cytoplasmic protein that inhibits the processing of dsRNA into siRNAs without interfering with either RNA-directed DNA methylation (RdDM) or intercellular silencing spread mediated by the silencing signal (Llave et al., 2000; Mallory et al., 2001, 2002; Dunoyer et al., 2004). However, HC-Pro suppression of miRNA silencing is associated with an increased accumulation of miRNA duplexes, suggesting that HC-Pro may act downstream of the production of miRNAs (Mallory et al., 2002; Chapman et al., 2004). This is consistent with the recent observation that HC-Pro, similar to the tombusviral p19 (Silhavy et al., 2002; Lakatos et al., 2004), prevents the assembly of siRNAs into the active RNA-induced silencing complex by binding to and sequestering duplex siRNAs (Lakatos et al., 2006; Merai et al., 2006). By contrast, CMV 2b, which accumulates in the nucleus and cytoplasm of plant cells (Lucy et al., 2000; Mayers et al., 2000; Wang et al., 2004), is a potent inhibitor of both intercellular silencing spread and RdDM (Guo and Ding, 2002). Notably, CMV 2b also can block salicylic acid (SA)–mediated virus resistance (Ji and Ding, 2001). A model was proposed to account for the observed activities of 2b in which SA may induce virus resistance by potentiating antiviral silencing (Ji and Ding, 2001).
Here, we investigated the genetic basis of 2b suppression of antiviral silencing in Arabidopsis plants infected with CMV. CMV contains a (+)-strand RNA genome that encodes five proteins among three RNA molecules (Palukaitis and Garcia-Arenal, 2003). Two replicase subunits and the movement protein are translated directly from the genomic RNAs, whereas coat protein and 2b are each translated from RNAs 4 and 4A, both of which are subgenomic RNAs (Ding et al., 1994). We found that CMV infection resulted in the production of 21-, 22-, and 24-nucleotide classes of viRNAs in Arabidopsis by DCL4, DCL2, and DCL3, respectively, as reported recently (Bouche et al., 2006; Fusaro et al., 2006), and deletion of 2b from the CMV genome caused a characteristic defect in the systemic infection of Arabidopsis plants, as observed previously in other host species (Ding et al., 1995a; Ji and Ding, 2001). Examining virus infection and the accumulation of viRNAs in dcl and rdr mutants of Arabidopsis inoculated with CMV-Δ2b revealed distinct roles for the three classes of viRNAs in antiviral silencing. These analyses indicate that 2b suppression of antiviral silencing depended on inhibition of the accumulation of the three classes of viRNAs in infected Arabidopsis plants. Although the silencing suppressor activity of 2b was required to establish infection, it was dispensable in eliciting disease symptoms, indicating an indirect role for viral silencing suppressors in virus virulence. Notably, we demonstrate a specific recognition of CMV by RDR1 and a key role for RDR1 in the production of CMV siRNAs. A potential role of the RDR1-dependent secondary viRNAs in non-cell-autonomous antiviral silencing is discussed.
RESULTS
Expression of CMV 2b Inhibits the Accumulation of Three Classes of viRNAs
To investigate the genetic target of CMV 2b in infected plants, we inoculated wild-type and silencing-defective mutant Arabidopsis plants with purified virions of either wild-type CMV (CMV) or its 2b-deletion mutant (CMV-Δ2b). Our analysis of CMV-derived (−)-strand siRNAs in single, double, and triple dcl mutants (Xie et al., 2005; Deleris et al., 2006; Henderson et al., 2006) revealed that CMV infection led to the accumulation of 21-, 22-, and 24-nucleotide viRNAs that required DCL4, DCL2, and DCL3, respectively (see Supplemental Figure 1 online). This result is consistent with the biogenesis of virus-derived siRNAs reported recently by several groups (Blevins et al., 2006; Bouche et al., 2006; Deleris et al., 2006; Fusaro et al., 2006).
We previously described CMV-Δ2b, which contains a point mutation converting the fourth codon (UUG) of the 2b open reading frame to UAG (Ding et al., 1995b). Whereas CMV-Δ2b is genetically stable in Nicotiana species (Ding et al., 1995a; Ji and Ding, 2001), the introduced stop codon UAG was rapidly reverted to UGG and the 2b open reading frame was restored after infection of Arabidopsis. Thus, we used a derivative of CMV-Δ2b in which most of the 2b coding sequence was deleted (Li et al., 2002). Equal amounts of CMV and CMV-Δ2b virions purified from Nicotiana glutinosa were inoculated onto the four fully extended leaves of each Arabidopsis plant at 40 ng/leaf. Five days after inoculation, total high and low molecular weight RNAs were extracted from the inoculated leaves pooled from 16 to 24 plants for the detection of virus and viRNAs. Although CMV-Δ2b was defective in systemic infection (see below), the genomic and subgenomic RNAs of both CMV and CMV-Δ2b accumulated to similar levels in the inoculated leaves of wild-type plants and the single dcl mutant Arabidopsis plants (Figure 1B, lanes 1 to 8). As expected, RNA 2 of CMV-Δ2b migrated faster than that of CMV because of the deletion.
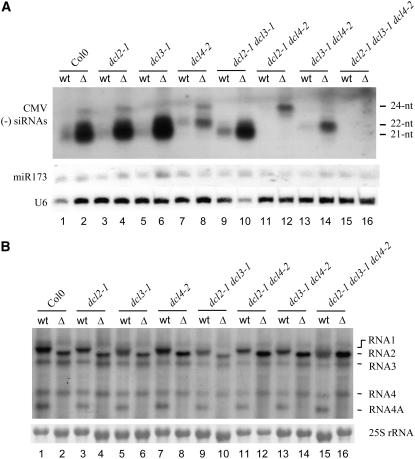
Figure 1.
Expression of 2b Inhibits the Accumulation of All Three Classes of viRNAs.
(A) Detection of (−)-strand CMV siRNAs in the inoculated leaves of wild-type Columbia (Col-0), dcl2-1, dcl3-1, dcl4-2, dcl2-1 dcl3-1, dcl2-1 dcl4-2, dcl3-1 dcl4-2, and dcl2-1 dcl3-1 dcl4-2 plants at 5 d after infection with either CMV (wt) or CMV-Δ2b (Δ). The membrane was also probed for miR173 and U6 RNAs. Positions of the 21-, 22-, and 24-nucleotide (nt) siRNAs are indicated.
(B) Detection of CMV genomic and subgenomic RNAs (indicated at right) in the inoculated leaves of the same plants used in (A). 25S rRNA served as a loading control.
In contrast with the equivalent levels of virus genomic RNAs in the leaves inoculated with either CMV or CMV-Δ2b, the three classes of viral (−)-strand siRNAs accumulated to drastically higher levels in the CMV-Δ2b–inoculated leaves than in CMV-inoculated leaves (Figure 1A). For example, a significantly reduced accumulation of viral 21-nucleotide siRNAs was observed in wild-type, dcl2, dcl3, and dcl2 dcl3 plants infected with CMV compared with those plants infected with CMV-Δ2b. The levels of the 22-nucleotide siRNAs were drastically lower in wild-type, dcl3, and dcl4 plants infected with CMV than in those plants infected with CMV-Δ2b. Similarly, whereas the 24-nucleotide class of viRNAs was detected as a major band in wild-type, dcl2, dcl4, and dcl2 dcl4 plants infected with CMV-Δ2b, it was undetectable in the same plants inoculated with the 2b-expressing wild-type CMV. However, no major difference in the accumulation of miR173 was detected following infection with either CMV or CMV-Δ2b (Figure 1A), supporting an earlier observation that CMV infection does not alter miRNA accumulation (Bouche et al., 2006). These results indicate that expression of 2b from CMV dramatically reduced the accumulation of all three classes of viRNAs in infected Arabidopsis plants.
DCL4- and DCL2-Mediated Pathways Are the Main Genetic Targets of CMV 2b
The above results suggest that CMV 2b may suppress the RNA-silencing antiviral immunity by inhibiting the accumulation of viRNAs in CMV-infected plants. Thus, we next investigated whether the defect of CMV-Δ2b in systemic infection was genetically rescued in Arabidopsis plants defective in the synthesis of one or more classes of the viRNAs. The 21-nucleotide class of CMV-specific siRNAs was undetectable in the inoculated and the upper systemically infected leaves of all of the dcl mutants that contain the dcl4-2 allele, including dcl4-2, dcl2-1 dcl4-2, dcl3-1 dcl4-2, and dcl2-1 dcl3-1 dcl4-2 plants (Figures 1A and 2B). Similarly, the 22- and 24-nucleotide classes of CMV siRNAs were absent in the dcl mutants that contain dcl2-1 and dcl3-1, respectively (Figures 1A and 2B). Thus, CMV infection did not lead to the accumulation of two classes of CMV siRNAs in double dcl mutants: 21- and 22-nucleotide siRNAs in dcl2-1 dcl4-2 plants (Figures 1A and 2B, lanes 11 and 12), 21- and 24-nucleotide siRNAs in dcl3-1 dcl4-2 plants (Figures 1A and 2B, lanes 13 and 14), and 22- and 24-nucleotide siRNAs in dcl2-1 dcl3-1 plants (Figures 1A and 2A, lanes 9 and 10). Also as expected, none of the three classes of CMV siRNAs was detectable in dcl2-1 dcl3-1 dcl4-2 plants (Figures 1A and 2B, lanes 15 and 16).
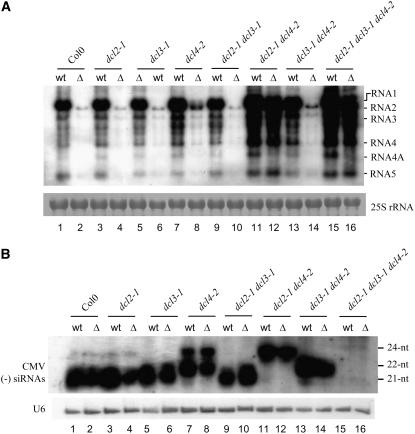
Figure 2.
CMV-Δ2b Is Rescued in dcl2-1 dcl4-2 and dcl2-1 dcl3-1 dcl4-2 Plants.
(A) Detection of CMV genomic/subgenomic RNAs from the upper systemically infected leaves of wild-type Col-0, dcl2-1, dcl3-1, and dcl4-2 single mutants, dcl2-1 dcl3-1, dcl2-1 dcl4-2, and dcl3-1 dcl4-2 double mutants, and the dcl2-1 dcl3-1 dcl4-2 triple mutant at 14 d after infection with CMV (wt) or CMV-Δ2b (Δ). 25S RNA served as a loading control. The CMV species are indicated at right.
(B) Detection of (−)-strand CMV siRNAs in the upper systemically infected leaves of the same plants used in (A). The membrane was also probed for U6 RNA. Positions of the 21-, 22-, and 24-nucleotide (nt) siRNAs are indicated.
RNA gel blot analysis showed that CMV-Δ2b accumulated to much lower levels than CMV in the upper systemically infected leaves of wild-type Arabidopsis plants at 14 d after inoculation (DAI) with equal amounts of CMV and CMV-Δ2b virions (Figure 2A, compare lanes 1 and 2). This recapitulated the previous observation made in other host species that CMV-Δ2b exhibits a defect in the systemic virus spread (Ding et al., 1995a; Ji and Ding, 2001). CMV-Δ2b remained defective in the systemic infection of dcl2-1, dcl3-1, dcl4-2, dcl2-1 dcl3-1, and dcl3-1 dcl4-2 plants because CMV-Δ2b accumulated to much lower levels than CMV did in the upper leaves (Figure 2A), similar to what occurred in wild-type plants. We noted that viRNAs accumulated to similarly high levels in the upper leaves of these plants infected with either CMV-Δ2b or CMV (Figure 2B) in spite of their different accumulation levels of the viral genomic and subgenomic RNAs. Thus, low levels of CMV RNA accumulation were associated with high-level accumulation of CMV siRNAs in the absence of 2b expression, further supporting a role for 2b in inhibiting the accumulation of viRNAs. We found that both CMV and CMV-Δ2b accumulated to similar levels in the upper leaves of dcl2-1 dcl4-2 and dcl2-1 dcl3-1 dcl4-2 plants (Figure 2A, compare lanes 11/12 and 15/16), indicating that systemic infection by CMV-Δ2b was as efficient as that by CMV in these mutant plants.
These results showed that, first, either the 21- or 22-nucleotide class of CMV siRNAs alone, but not the 24-nucleotide class of viRNAs, was sufficient to repress the systemic spread of CMV-Δ2b in Arabidopsis plants. Second, efficient systemic infection of CMV-Δ2b occurred only when the host plants were defective in the synthesis of both the 21- and 22-nucleotide classes of CMV siRNAs, and eliminating the production of 24-nucleotide siRNAs did not further enhance CMV-Δ2b infection when both 21- and 22-nucleotide siRNAs were absent. Third, our findings also showed that elimination of both the 21- and 22-nucleotide siRNA pathways effectively compensated for the loss of 2b expression and allowed robust systemic infection of CMV-Δ2b. Thus, we conclude that the 21- and 22-nucleotide siRNA pathways are the key genetic targets of CMV 2b, although 2b inhibited the accumulation of all three classes of CMV siRNAs. In support of this conclusion, we found that a CMV 2b transgene integrated in wild-type Arabidopsis plants suppressed the silencing of potato virus X amplicon (Li, 2001) and rescued the systemic infection of the p38-deletion turnip crinkle virus (TCV) mutant (our unpublished data), known to be targeted by the 21- and 22-nucleotide siRNA pathways in Arabidopsis (Deleris et al., 2006).
CMV 2b Is Dispensable in the Induction of Enhanced Disease Susceptibility
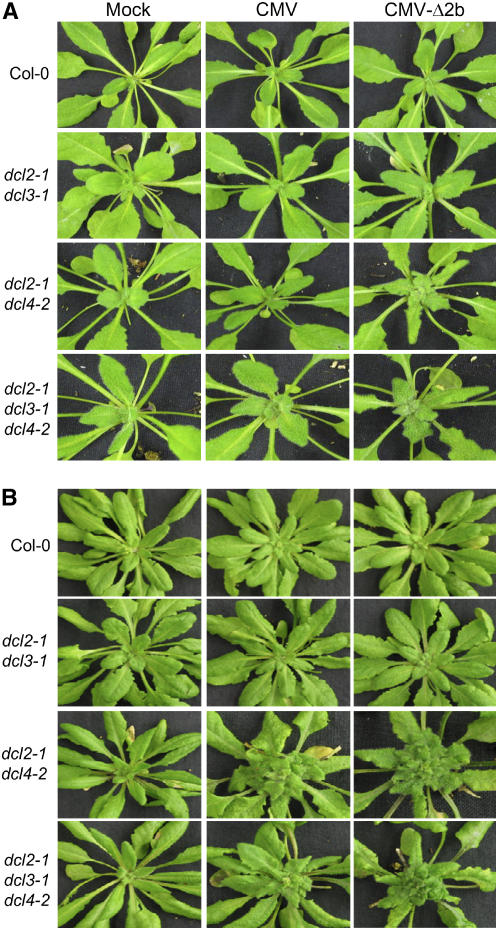
The Q strain of CMV used in this work is highly virulent in cucumber (Cucumis sativus) and N. glutinosa plants, but it causes only mild mottling in Nicotiana tabacum (Ding et al., 1995a; Ji and Ding, 2001) and very mild dwarfing in wild-type Arabidopsis (Figure 3B, top middle). Infection with CMV resulted in much higher levels of virus accumulation (Figure 2A, lanes 11 and 15) and induced the development of severe systemic disease symptoms (Figure 3B) in both dcl2-1 dcl4-2 and dcl2-1 dcl3-1 dcl4-2 plants compared with CMV infection in wild-type plants. Such enhanced disease susceptibility (EDS) phenotypes were not observed following CMV infection in dcl2-1 dcl3-1 (Figures 2A, lane 9, and 3B) or any other single and double dcl mutants (data not shown). As indicated above, robust systemic infection by CMV-Δ2b occurred only in dcl2-1 dcl4-2 and dcl2-1 dcl3-1 dcl4-2 plants. Notably, CMV-Δ2b also accumulated to higher levels and induced severe disease symptoms in these plants compared with CMV infection in wild-type plants (Figures 2A and 3B). Thus, we conclude that the expression of 2b was not required for the induction of severe disease symptoms in these immunity-defective plants.
Figure 3.
Expression of 2b Does Not Play a Direct Role in Eliciting Disease Symptoms.
Wild-type, dcl2-1 dcl3-1, dcl2-1 dcl4-2, and dcl2-1 dcl3-1 dcl4-2 plants were photographed at 5 d (A) and 14 d (B) after infection with CMV, CMV-Δ2b, or buffer (mock).
Intriguingly, we found that EDS development was much faster in dcl2-1 dcl4-2 and dcl2-1 dcl3-1 dcl4-2 plants infected with CMV-Δ2b than in the same plants infected with CMV. The disease symptoms were clearly visible in the upper systemically infected leaves of dcl2-1 dcl4-2 and dcl2-1 dcl3-1 dcl4-2 plants at 5 DAI with CMV-Δ2b, and the growth of the infected plants was arrested from this point (Figure 3A). By contrast, disease symptoms were invisible in the same mutant plants inoculated with CMV at 5 DAI (Figure 3A), and plant growth was not significantly affected until ∼10 DAI. A 10-fold increase in CMV virion inoculum resulted in higher virus accumulation but failed to induce early disease symptoms in dcl2-1 dcl4-2 and dcl2-1 dcl3-1 dcl4-2 plants (data not shown). These data suggest that the expression of 2b delayed the development of severe disease symptoms in the early stages of infection.
DCL3 Could Act Upstream of DCL4 to Enhance Antiviral Silencing
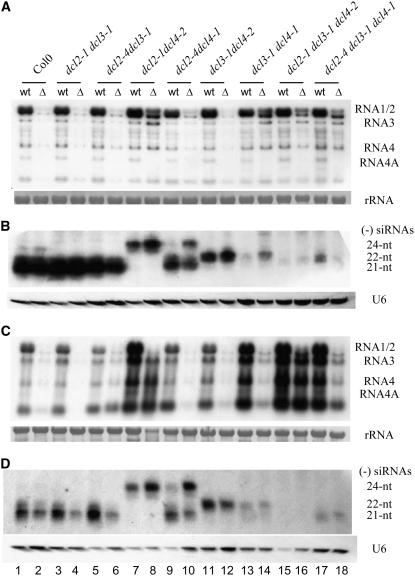
In contrast with the complete rescue of CMV-Δ2b in dcl2-1 dcl4-2 plants, we found that CMV-Δ2b remained defective in systemic infection of dcl2-4 dcl4-1 plants (Figures 4A and 4C, compare lanes 8 and 10). The accumulation of CMV-Δ2b in the upper systemically infected leaves of dcl2-4 dcl4-1 at either 5 or 14 DAI was as low as in wild-type plants (Figures 4A and 4C, compare lanes 2 and 10). dcl4-2 was isolated directly from Col-0 and contains a T-DNA inserted in an exon of the DCL4 gene, whereas T-DNA was inserted in an intron in dcl4-1, which was originally identified in the Wassilewskija ecotype and backcrossed twice to Col-0 (Gasciolli et al., 2005; Xie et al., 2005). Since the accumulation of 21-nucleotide trans-acting siRNAs was reduced in dcl4-1 plants but was undetectable in dcl4-2 (Gasciolli et al., 2005; Xie et al., 2005), we hypothesized that the resistance of dcl2-4 dcl4-1 plants to CMV-Δ2b might be mediated by the partially active dcl4-1 allele. Indeed, although not as abundant as in plants carrying wild-type DCL4 (Figures 4B and 4D, lanes 1 to 6), the 21-nucleotide CMV siRNAs reproducibly accumulated to high levels in dcl2-4 dcl4-1 plants (Figures 4B and 4D, lanes 9 and 10). By contrast, 22-nucleotide CMV siRNAs remained undetectable in dcl2-4 dcl4-1 plants, as in dcl2-1 dcl4-2 plants (Figures 4B and 4D, lanes 7 to 10). These results show that, unlike dcl4-2, dcl4-1 represented an incomplete loss-of-function allele in the production of viral 21-nucleotide siRNAs and that this allele was as effective as wild-type DCL4 in repressing the systemic infection of CMV-Δ2b in the absence of DCL2.
Figure 4.
DCL3 Enhances Antiviral Silencing by dcl4-1.
Detection of CMV genomic/subgenomic RNAs ([A] and [C]) and (−)-strand CMV siRNAs ([B] and [D]) from the upper systemically infected leaves of Arabidopsis seedlings of various genotypes as indicated at 5 d ([A] and [B]) and 14 d ([C] and [D]) after inoculation with CMV (wt) or CMV-Δ2b (Δ). 25S rRNA served as a loading control. The membrane was also probed for U6 RNA. Positions of the 21-, 22-, and 24-nucleotide (nt) siRNAs are indicated.
Notably, we found that the dcl4-1 allele failed to effectively repress CMV-Δ2b infection in dcl3-1 dcl4-1 and dcl2-4 dcl3-1 dcl4-1 plants in which DCL3 was inactivated. Accumulation of CMV-Δ2b in the upper leaves of both mutant plants (Figure 4A, lanes 14 and 8) was as high as in dcl2-1 dcl4-2 plants at 5 DAI (Figure 4A, lane 8), indicating that dcl3-1 dcl4-1 and dcl2-4 dcl3-1 dcl4-1 mutants supported the efficient systemic infection of CMV-Δ2b. By 14 DAI, the accumulation of CMV-Δ2b in dcl3-1 dcl4-1 and dcl2-4 dcl3-1 dcl4-1 plants (Figure 4A, lanes 13 and 18) was higher than that in dcl2-4 dcl4-1 plants but lower than that in dcl2-1 dcl4-2 plants (Figure 4C, lane 8), suggesting more effective antiviral silencing in dcl3-1 dcl4-1 and dcl2-4 dcl3-1 dcl4-1 plants at 14 DAI (Figure 4C, lanes 14 and 18). These results showed that the antiviral activity of the dcl4-1 allele depended on the function of DCL3, indicating a unique antiviral role of DCL3 upstream of DCL4.
In contrast with the abundant accumulation of 21-nucleotide CMV siRNAs in dcl2-4 dcl4-1 plants, the level of 21-nucleotide CMV siRNAs was extremely low in both dcl3-1 dcl4-1 and dcl2-4 dcl3-1 dcl4-1 plants, which contained the same dcl4-1 allele (Figures 4B and 4D). This difference in the accumulation of 21-nucleotide CMV siRNAs explains, first, why dcl3-1 dcl4-1 and dcl2-4 dcl3-1 dcl4-1 plants, but not dcl2-4 dcl4-1 plants, supported the efficient systemic infection of CMV-Δ2b. Second, it also indicates that in dcl3-1 dcl4-1 and dcl2-4 dcl3-1 dcl4-1 plants, the dcl3-1 allele not only eliminated production of the 24-nucleotide CMV siRNAs, as expected, but also potently inhibited production of the 21-nucleotide CMV siRNAs by the partially active dcl4-1 allele. However, production of the 21-nucleotide viRNAs by wild-type DCL4 was independent of DCL3, because the 21-nucleotide CMV siRNAs were as abundant in dcl3-1 and dcl2-4 dcl3-1 plants as in wild-type plants (Figure 1B, compare lanes 2 and 6; Figures 4B and 4D, compare lanes 1 and 5). Thus, these results indicate that DCL3 acts to amplify production of the 21-nucleotide viRNAs by the weak dcl4-1 allele.
We noted that the accumulation levels of the 22-nucleotide CMV siRNAs were also very low in dcl3-1 dcl4-1 plants (Figures 4B and 4D, lanes 13 and 14). However, the reduced accumulation of the 22-nucleotide viRNAs observed in dcl3-1 dcl4-1 plants was associated with either wild-type or partially active DCL4 but not with the presence of the dcl3-1 allele. This suggests that dcl4-1 remained active in the observed hierarchical antiviral activities of DCL4 and DCL2, in which DCL4 is dominant and inhibitory to DCL2 (Deleris et al., 2006).
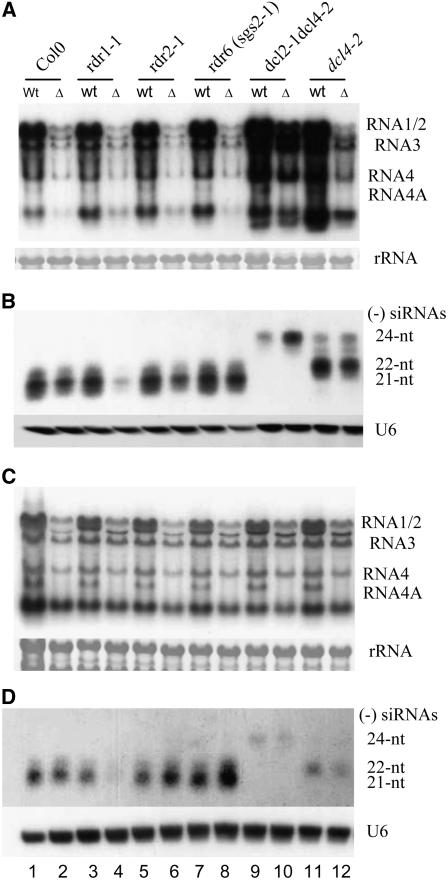
Production of CMV siRNAs Is Largely RDR1 Dependent
We next investigated a potential role of cellular RDRs in the biogenesis of viRNAs by challenging wild-type and rdr mutant plants with equal amounts of CMV and CMV-Δ2b virions. We found that the accumulation of viRNAs was significantly reduced in rdr1-1 plants compared with wild-type, rdr2-1, or rdr6 (sgs2-1) plants following infection with CMV-Δ2b (Figure 5B, compare lane 4 with lanes 2, 6, and 8). Similarly reduced accumulation of siRNAs was detected in the inoculated leaves at 5 DAI (Figure 5D) and in the upper systemically infected leaves at 14 DAI (Figure 5B), and this result was reproducible. Compared with infection with CMV, however, the accumulation of viral genomic and subgenomic RNAs remained as low in rdr1-1 plants as in wild-type, rdr2-1, and rdr6 plants following CMV-Δ2b infection (Figure 5A), indicating that the rdr1-1 mutation was insufficient to rescue the defect of CMV-Δ2b in systemic infection. Nevertheless, our results strongly indicate that CMV was specifically recognized by RDR1 and that the production of CMV siRNAs in wild-type, rdr2-1, or rdr6 (sgs2-1) plants infected with CMV-Δ2b was largely RDR1 dependent. It is unclear whether the low level of viRNAs detected in rdr1-1 plants infected with CMV-Δ2b was RDR-independent (e.g., primary siRNAs that are viral RdRP dependent) or dependent on one or more of the five remaining RDRs.
Figure 5.
RDR1-Dependent Production of CMV siRNAs in Arabidopsis.
Detection of CMV genomic/subgenomic RNAs ([A] and [C]) and (−)-strand CMV siRNAs ([B] and [D]) from the inoculated ([C] and [D]) and upper systemically infected leaves ([A] and [B]) of Arabidopsis seedlings of various genotypes as indicated at 5 d ([A] and [B]) and 14 d ([C] and [D]), respectively, after inoculation with CMV (wt) or CMV-Δ2b (Δ). 25S RNA served as a loading control. The CMV RNA species are indicated at right. The membrane was also probed for U6 RNA. Positions of the 21-, 22-, and 24-nucleotide (nt) siRNAs are indicated.
In contrast with infection with CMV-Δ2b, we detected similar accumulation levels of viRNAs in wild-type, rdr1-1, rdr2-1, and rdr6 (sgs2-1) plants infected wild-type CMV (Figure 5B, lanes 1, 3, 5, and 7). Thus, the presence of wild-type RDR1 in rdr2-1, rdr6 (sgs2-1), and wild-type plants did not result in higher production of viRNAs compared with rdr1-1 plants following CMV infection, suggesting a loss of the RDR1-dependent production of viRNAs when CMV 2b protein was expressed.
DISCUSSION
The use of well-defined transgenic plant models based on silencing transgenes has greatly facilitated the identification and mechanistic analysis of VSRs (Roth et al., 2004). These systems assay for the suppression of RNA silencing of a transgene transcribed in the nucleus by a VSR often expressed from a nontargeted mRNA. However, VSRs target antiviral silencing induced by a replicating and movement-competent virus in infected hosts by expression from the infecting virus genome, which is also under the control of the induced antiviral silencing. We previously identified the dsRNA-siRNA pathway of Drosophila and Caenorhabditis elegans as the genetic target of the flock house virus VSR, because genetic defects in the RNAi pathway rescued the accumulation defects of a B2-deficient FHV (Li et al., 2002; Lu et al., 2005; Wang et al., 2006). However, similar genetic rescue of VSR-deficient mutant viruses has not been possible in plants until recently, because of the complex small RNA pathways controlled by four DCLs together with six RDRs. The recent availability of multiple DCL mutants of Arabidopsis allowed Voinnet and colleagues to demonstrate that p38 of TCV specifically inhibits the accumulation of the DCL4-dependent, 21-nucleotide class of viRNAs (Deleris et al., 2006).
In this study, we investigated the genetic mechanism involved in the induction and suppression of antiviral silencing during CMV infection in Arabidopsis plants. Our results demonstrate that 2b expressed from the CMV genome during infection potently inhibited the accumulation of the 21-, 22-, and 24-nucleotide CMV-specific siRNAs in Arabidopsis. We showed that the defect of CMV-Δ2b in the systemic infection was efficiently rescued in dcl2 dcl4 double mutant plants that are completely defective in the synthesis of both the 21- and 22-nucleotide viRNAs. These results indicate that the inhibitory effect of 2b on the accumulation of viRNAs, the 21- and 22-nucleotide classes in particular, plays a key role in both the suppression of antiviral silencing and the facilitation of systemic virus movement by 2b in infected plants. Recent studies have shown that DCL4-dependent, 21-nucleotide siRNAs and DCL3-dependent, 24-nucleotide siRNAs mediate non-cell-autonomous silencing and RdDM, respectively (Matzke and Birchler, 2005; Voinnet, 2005). Thus, the new activity of 2b in inhibiting the accumulation of three classes of siRNAs is consistent with previous observations that CMV 2b blocks both RdDM and the intercellular spread of RNA silencing in transgene-silencing models (Guo and Ding, 2002).
Analysis of both viral siRNA biogenesis and antiviral function showed that dcl4-1 is a significantly weaker allele than dcl4-2 in the production of the 21-nucleotide viRNAs, as found previously for the production of the 21-nucleotide trans-acting siRNAs (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). This is best illustrated by detection of abundant accumulation of 21-nucleotide siRNAs and potent antiviral silencing in dcl2-4 dcl4-1 plants but not in dcl2-1 dcl4-2 plants (Figure 4). Both dcl4-1 and dcl4-2 alleles contain a T-DNA inserted in the DCL4 gene. T-DNA insertion in dcl4-2 results in production of a chimeric mRNA with the sequence encoding the second dsRNA binding motif of DCL4 replaced with a 192-nucleotide segment of T-DNA-derived sequence (Xie et al., 2005). By contrast, dcl4-1 line contains a T-DNA inserted in an intron (Gasciolli et al., 2005), which may not completely eliminate expression of wild-type DCL4 protein.
We found that the incomplete loss-of-function allele of DCL4 exhibits two interesting properties. First, like wild-type DCL4 (Deleris et al., 2006), the weak dcl4-1 allele remained dominant over DCL2 because only very low levels of the 22-nucleotide viRNAs from wild-type DCL2 were detected in the dcl4-1 genetic background (Figures 4B and 4D, lanes 13 and 14). Second, the loss-of-function mutation in DCL3 associated with the dcl3-1 allele was severely inhibitory to both the production of the 21-nucleotide viRNAs and effective antiviral silencing mediated by the dcl4-1 allele (Figure 4). Thus, our results confirm an antiviral role of DCL3 against CMV suggested by previous studies (Bouche et al., 2006; Fusaro et al., 2006) and genetically place DCL3 upstream of DCL4 to enhance both the production of the 21-nucleotide viRNAs and antiviral silencing. This unique antiviral activity of DCL3, readily detectable only in the dcl4-1 background, may become critical in wild-type plants when DCL4 function is compromised due to interference by physiological, environmental, or viral factors such as p38 of TCV (Deleris et al., 2006). It is possible that DCL3 or the 24-nucleotide siRNAs produced by DCL3 is involved in the perception of the non-cell-autonomous silencing signal, thereby potentiating subsequent antiviral silencing by DCL4, as proposed previously for RDR6 (Schwach et al., 2005).
Most of the RNAi pathway genes isolated from Arabidopsis, such as RDR6, SDE3, SGS3, and AGO1, are required for silencing induced by sense RNA transgenes but not by inverted repeat RNA transgenes, indicating a role for these genes in an RDR-dependent de novo synthesis of dsRNA (Dalmay et al., 2000; Mourrain et al., 2000; Beclin et al., 2002). Such de novo dsRNA synthesis mediated by the host RDR pathway may play a unique role in antiviral silencing against CMV, because rdr6, sde3, sgs3, and ago1 mutants exhibit enhanced disease susceptibility to CMV but not to other (+)-strand RNA viruses examined, such as TCV, Tobacco rattle virus (TRV), Turnip mosaic virus, and a tobamovirus (Voinnet, 2005; Vaucheret, 2006). Recent studies showed that loss-of-function mutations in RDR1, RDR2, or RDR6 have no detectable impact on the production of viRNAs in Arabidopsis plants infected with TRV, TCV, and a tobamovirus (Blevins et al., 2006; Deleris et al., 2006). In this study, we also found no difference in the accumulation of viRNAs between the wild type and the three rdr mutants infected with wild-type CMV. However, the accumulation of viRNAs was markedly reduced in rdr1-1 plants compared with wild-type, rdr2-1, and rdr6 plants following infection with CMV-Δ2b. Thus, our results indicate that without interference of the 2b protein, most of the CMV siRNAs are RDR1 dependent, providing molecular evidence demonstrating a role for a host RDR in the biogenesis of virus-derived siRNAs. Recognition of CMV by the host RDR pathway is consistent with previous observations that Arabidopsis mutants defective for the host RDR pathway are hypersusceptible to CMV (Dalmay et al., 2000; Mourrain et al., 2000; Voinnet, 2005). It remained to be determined whether other RDRs, RDR6 in particular, contribute to the biogenesis of the viRNAs detected in rdr1-1 plants infected with CMV-Δ2b and whether genetic rescue of CMV-Δ2b infection occurs after the inactivation of additional RDRs.
Based on these findings, we propose that CMV infection results in the synthesis of CMV-specific dsRNA not only by the viral RdRP but also by the host RDR, leading to the production of primary and secondary viRNAs. RDR1 plays a key role in the production of viRNAs, probably because it is inducible by SA (Yu et al., 2003) and pathogen infection triggers SA production. It is possible that one or more of those genes, such as SGS3 (Mourrain et al., 2000), AGO1 (Morel et al., 2002), or SDE3 (Dalmay et al., 2001), essential for RDR-dependent silencing amplification, also contributes to the biogenesis of CMV secondary siRNAs. These RDR-dependent viral secondary siRNAs may act cell-autonomously to destroy viral genomic and subgenomic RNAs in the infected cells where these siRNAs are made. More importantly, they may have potential to act non-cell-autonomously, as shown for the transgene-derived secondary siRNAs (Dunoyer and Voinnet, 2005), to spread outside of the vasculature where CMV is unloaded to initiate replication in the upper systemically infected leaves (Havelda et al., 2003; Deleris et al., 2006). Application of exogenous SA enhances virus resistance possibly by amplifying the antiviral role of RDR1 and any other components in the antiviral silencing pathway that respond to SA (Ji and Ding, 2001; Yu et al., 2003).
We further propose that 2b is able to inhibit the RDR-dependent production of secondary CMV siRNAs, which are inducible by SA and have potential to act non-cell-autonomously. This hypothesis is based on our findings that in infected Arabidopsis, expression of 2b was associated with drastically reduced accumulation of CMV siRNAs (Figure 1A), most of the CMV siRNAs produced without 2b interference were RDR1 dependent (Figure 5B), and the RDR1-dependent production of CMV siRNAs was undetectable in the presence of 2b (Figure 5B). Our hypothesis also is consistent with a previous finding that 2b does not inhibit the accumulation of primary siRNAs derived from an inverted repeat RNA (Qi et al., 2004) and explains why 2b is a potent inhibitor of both non-cell-autonomous silencing and the SA-mediated virus resistance (Ji and Ding, 2001; Guo and Ding, 2002). Two possible biochemical mechanisms could explain how 2b inhibits the RDR-dependent production of secondary CMV siRNAs. First, the direct interaction between 2b and AGO1 observed recently was shown to inhibit the slicing activity of AGO1 (Zhang et al., 2006), which explains why 2b is able to suppress RNA silencing initiated by siRNAs (Qi et al., 2004) and why high levels of viRNAs inhibit the systemic infection of CMV-Δ2b but not of CMV (Figure 2). Such a direct interaction may also inhibit the biogenesis of secondary siRNAs, because AGO1 is essential for RDR-dependent silencing amplification. Second, 2b may directly inhibit the production of secondary CMV siRNAs by binding to siRNAs and its dsRNA precursor, as shown for B2 of Flock house virus (Lu et al., 2005; Chao et al., 2005). The 2b protein encoded by Tomato aspermy virus, which belongs to the same Cucumovirus genus as CMV, indeed binds to dsRNA and siRNA, and the crystal structure of 2b in complex with siRNA was solved recently (J.B. Ma, F. Li, D.J. Patel, and S.W. Ding, unpublished data).
We found that CMV infection resulted in higher accumulation levels and induced more severe disease symptoms in both dcl2-1 dcl4-2 and dcl2-1 dcl3-1 dcl4-2 plants than in wild-type plants (Figures 2A and 3B), as reported recently for TCV, TRV, and other more virulent CMV isolates (Bouche et al., 2006; Deleris et al., 2006; Fusaro et al., 2006). However, CMV-Δ2b induced similar EDS phenotypes in these Arabidopsis mutants that in fact occurred at least 1 week earlier than after CMV infection (Figures 2A and 3B). Therefore, our results show that, first, 2b was dispensable for eliciting disease symptoms. This finding argues against a direct role for VSRs in virus virulence (Mallory et al., 2002; Kasschau et al., 2003; Chen et al., 2004; Dunoyer et al., 2004; Mlotshwa et al., 2005; Zhang et al., 2006; Lewsey et al., 2007) but supports the hypothesis that symptom expression is complex and may result from interference of host physiological and developmental processes by virus replication and infection (Matthews, 1991; Poethig et al., 2006). Second, 2b exhibited a novel activity to repress disease symptom development at the early stages of infection. It is not clear at present whether 2b represses disease development in these mutant plants by a mechanism similar to or distinct from silencing suppression. Nevertheless, if suppression of early disease development also occurs in CMV infection of wild-type plants, this new activity of 2b should facilitate virus propagation and dissemination, consistent with the notion that requisition of VSR genes represents an evolutionary adaptation of viruses to the host RNAi-mediated immunity (Li and Ding, 2006).
METHODS
Plant Materials
Mutant lines for Arabidopsis thaliana rdr1-1, rdr2-1, rdr6 (sgs2-1), dcl2-1, dcl3-1, dcl4-2, dcl2-1 dcl3-1, dcl2-4 dc3-1, dcl2-1 dcl4-2, dc2-4 dcl4-1, dcl3-1 dcl4-2, dcl3-1 dcl4-1, dcl2-1 dcl3-1 dcl4-2, and dcl2-4 dcl3-1 dcl4-1 were described previously (Mourrain et al., 2000; Xie et al., 2004, 2005; Gasciolli et al., 2005; Henderson et al., 2006). To ensure synchronous germination and development of Arabidopsis, seeds were first vernalized at 4°C in the dark for 5 d prior to transferring to a growth room for germination. Plants were maintained at 24°C and 10-h-light/14-h-dark cycles.
Viruses and Infection Assays
The wild-type Q strain of CMV and the two 2b-deletion mutants used in this study were described previously (Ding et al., 1995b; Li et al., 2002). Virions were propagated in Nicotiana glutinosa plants and purified essentially by the method of (Peden and Symons, 1973). Wild-type and mutant Arabidopsis plants (∼4 weeks old) were mock-inoculated or inoculated with CMV or CMV-Δ2b at a concentration of 20 μg/mL. Inoculated leaves were harvested at 5 DAI, and upper systemically infected leaves were harvested at both 5 and 14 DAI from pools of 16 to 24 plants.
RNA Gel Blot Analyses
RNA gel blot analyses of low and high molecular weight RNAs were performed with 10 and 5 μg of total RNA, respectively, as described previously (Guo and Ding, 2002). Small RNAs were separated by electrophoresis on 16% polyacrylamide gels and blotted on membranes. The blot hybridization was performed at 38°C for 12 to 16 h in PerfectHyb Plus buffer (Sigma-Aldrich), and the probes used were DNA oligonucleotides end-labeled with [γ-32P]ATP by T4 polynucleotide kinase (New England Biolabs) and purified through MicroSpin G-25 columns (Amersham) according to the manufacturers' recommendations. The blot was washed once with 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.2% SDS for 30 min and twice with 0.2× SSC and 0.1% SDS for 20 min at 48°C. For repeated hybridization, the membrane was stripped twice with 0.5% SDS and 20 mM EDTA for 30 min at 80°C. For detection of CMV siRNAs, a mixture of seven DNA oligonucleotides corresponding to the (+)-strand of CMV RNA3 (nucleotides 1 to 40, 241 to 280, 741 to 780, 1041 to 1080, 1341 to 1380, 1641 to 1680, and 2131 to 2170) was used. The same procedure was employed to detect U6 and miR173. High molecular weight RNA gel blots were probed with radiolabeled DNAs corresponding to the 3′ terminal 340 nucleotides of CMV RNA 2 by random priming reactions (Promega) in the presence of [α-32P]dCTP.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Genetic Requirements for DCLs in the Production of CMV siRNAs.
Supplementary Material
Acknowledgments
This article is dedicated to the memory of Robert H. Symons, who passed away on October 4, 2006. We thank Jim Carrington, Steve Jacobsen, and Herve Vaucheret for their gifts of mutant Arabidopsis seeds, Olivier Voinnet for sharing data prior to publication, and Hailing Jin, Xuemei Chen, Jian-Kang Zhu, and members of the Ding laboratory for stimulating discussions. This work was supported by U.S. Department of Agriculture National Research Initiative Grants 2005-35319-15331 and 2005-34399-16077.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Shou-Wei Ding (dingsw@ucr.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Anandalakshmi, R., Pruss, G.J., Ge, X., Marathe, R., Mallory, A.C., Smith, T.H., and Vance, V.B. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2005). RNA silencing. Trends Biochem. Sci. 30 290–293. [DOI] [PubMed] [Google Scholar]
- Beclin, C., Berthome, R., Palauqui, J.C., Tepfer, M., and Vaucheret, H. (1998). Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of nonviral (trans)genes. Virology 252 313–317. [DOI] [PubMed] [Google Scholar]
- Beclin, C., Boutet, S., Waterhouse, P., and Vaucheret, H. (2002). A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12 684–688. [DOI] [PubMed] [Google Scholar]
- Blevins, T., Rajeswaran, R., Shivaprasad, P.V., Beknazariants, D., Si-Ammour, A., Park, H.S., Vazquez, F., Robertson, D., Meins, F., Jr., Hohn, T., and Pooggin, M.M. (2006). Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34 6233–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche, N., Lauressergues, D., Gasciolli, V., and Vaucheret, H. (2006). An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti, G., Voinnet, O., Li, W.X., Ji, L.H., Ding, S.W., and Baulcombe, D.C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chao, J.A., Lee, J.H., Chapados, B.R., Debler, E.W., Schneemann, A., and Williamson, J.R. (2005). Dual modes of RNA-silencing suppression by flock house virus protein B2. Nat. Struct. Mol. Biol. 12 952–957. [DOI] [PubMed] [Google Scholar]
- Chapman, E.J., Prokhnevsky, A.I., Gopinath, K., Dolja, V.V., and Carrington, J.C. (2004). Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Li, W.X., Xie, D., Peng, J.R., and Ding, S.W. (2004). Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell 16 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. (2005). MicroRNA biogenesis and function in plants. FEBS Lett. 579 5923–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101 543–553. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Horsefield, R., Braunstein, T.H., and Baulcombe, D.C. (2001). SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris, A., Gallego-Bartolome, J., Bao, J., Kasschau, K.D., Carrington, J.C., and Voinnet, O. (2006). Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313 68–71. [DOI] [PubMed] [Google Scholar]
- Ding, S.W., Anderson, B.J., Haase, H.R., and Symons, R.H. (1994). New overlapping gene encoded by the cucumber mosaic-virus genome. Virology 198 593–601. [DOI] [PubMed] [Google Scholar]
- Ding, S.W., Li, W.X., and Symons, R.H. (1995. a). A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J. 14 5762–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.W., Rathjen, J.P., Li, W.X., Swanson, R., Healy, H., and Symons, R.H. (1995. b). Efficient infection from cDNA clones of cucumber mosaic cucumovirus RNAs in a new plasmid vector. J. Gen. Virol. 76 459–464. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., Lecellier, C.H., Parizotto, E.A., Himber, C., and Voinnet, O. (2004). Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dunoyer, P., and Voinnet, O. (2005). The complex interplay between plant viruses and host RNA-silencing pathways. Curr. Opin. Plant Biol. 8 415–423. [DOI] [PubMed] [Google Scholar]
- Fusaro, A.F., Matthew, L., Smith, N.A., Curtin, S.J., Dedic-Hagan, J., Ellacott, G.A., Watson, J.M., Wang, M.B., Brosnan, C., Carroll, B.J., and Waterhouse, P.M. (2006). RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7 1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli, V., Mallory, A.C., Bartel, D.P., and Vaucheret, H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15 1494–1500. [DOI] [PubMed] [Google Scholar]
- Guo, H.S., and Ding, S.W. (2002). A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952. [DOI] [PubMed] [Google Scholar]
- Havelda, Z., Hornyik, C., Crescenzi, A., and Burgyan, J. (2003). In situ characterization of cymbidium ringspot tombusvirus infection-induced posttranscriptional gene silencing in Nicotiana benthamiana. J. Virol. 77 6082–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I.R., Zhang, X., Lu, C., Johnson, L., Meyers, B.C., Green, P.J., and Jacobsen, S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38 721–725. [DOI] [PubMed] [Google Scholar]
- Herr, A.J. (2005). Pathways through the small RNA world of plants. FEBS Lett. 579 5879–5888. [DOI] [PubMed] [Google Scholar]
- Ji, L.H., and Ding, S.W. (2001). The suppressor of transgene RNA silencing encoded by Cucumber mosaic virus interferes with salicylic acid-mediated virus resistance. Mol. Plant Microbe Interact. 14 715–724. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., and Carrington, J.C. (1998). A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 95 461–470. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4 205–217. [DOI] [PubMed] [Google Scholar]
- Lakatos, L., Csorba, T., Pantaleo, V., Chapman, E.J., Carrington, J.C., Liu, Y.P., Dolja, V.V., Calvino, L.F., Lopez-Moya, J.J., and Burgyan, J. (2006). Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos, L., Szittya, G., Silhavy, D., and Burgyan, J. (2004). Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewsey, M., Robertson, F.C., Canto, T., Palukaitis, P., and Carr, J.P. (2007). Selective targeting of miRNA-regulated plant development by a viral counter-silencing protein. Plant J. 50 240–252. [DOI] [PubMed] [Google Scholar]
- Li, F., and Ding, S.W. (2006). Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60 503–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.W. (2001). Roles of Tomato Aspermy Virus 2b in Plant Hypersensitive Resistance and in Post-Transcriptional Gene Silencing Defense. PhD dissertation (Singapore: National University of Singapore).
- Li, H.W., Li, W.X., and Ding, S.W. (2002). Induction and suppression of RNA silencing by an animal virus. Science 296 1319–1321. [DOI] [PubMed] [Google Scholar]
- Li, W.X., and Ding, S.W. (2001). Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12 150–154. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., and Carrington, J.C. (2000). Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Maduro, M., Li, F., Li, H.W., Broitman-Maduro, G., Li, W.X., and Ding, S.W. (2005). Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436 1040–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucy, A.P., Guo, H.S., Li, W.X., and Ding, S.W. (2000). Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19 1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Ely, L., Smith, T.H., Marathe, R., Anandalakshmi, R., Fagard, M., Vaucheret, H., Pruss, G., Bowman, L., and Vance, V.B. (2001). HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Reinhart, B.J., Bartel, D., Vance, V.B., and Bowman, L.H. (2002). A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA 99 15228–15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, R.E.F. (1991). Plant Virology. (San Diego, CA: Academic Press).
- Matzke, M.A., and Birchler, J.A. (2005). RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6 24–35. [DOI] [PubMed] [Google Scholar]
- Mayers, C.N., Palukaitis, P., and Carr, J.P. (2000). Subcellular distribution analysis of the cucumber mosaic virus 2b protein. J. Gen. Virol. 81 219–226. [DOI] [PubMed] [Google Scholar]
- Meister, G., and Tuschl, T. (2004). Mechanisms of gene silencing by double-stranded RNA. Nature 431 343–349. [DOI] [PubMed] [Google Scholar]
- Merai, Z., Kerenyi, Z., Kertesz, S., Magna, M., Lakatos, L., and Silhavy, D. (2006). Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 80 5747–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa, S., Schauer, S.E., Smith, T.H., Mallory, A.C., Herr, J.M., Jr., Roth, B., Merchant, D.S., Ray, A., Bowman, L.H., and Vance, V.B. (2005). Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell 17 2873–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, J.B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101 533–542. [DOI] [PubMed] [Google Scholar]
- Palukaitis, P., and Garcia-Arenal, F. (2003). Cucumoviruses. Adv. Virus Res. 62 241–323. [DOI] [PubMed] [Google Scholar]
- Peden, K.W., and Symons, R.H. (1973). Cucumber mosaic virus contains a functionally divided genome. Virology 53 487–492. [DOI] [PubMed] [Google Scholar]
- Poethig, R.S., Peragine, A., Yoshikawa, M., Hunter, C., Willmann, M., and Wu, G. (2006). The function of RNAi in plant development. Cold Spring Harb. Symp. Quant. Biol. 71 165–170. [DOI] [PubMed] [Google Scholar]
- Qi, Y., and Hannon, G.J. (2005). Uncovering RNAi mechanisms in plants: Biochemistry enters the foray. FEBS Lett. 579 5899–5903. [DOI] [PubMed] [Google Scholar]
- Qi, Y., Zhong, X., Itaya, A., and Ding, B. (2004). Dissecting RNA silencing in protoplasts uncovers novel effects of viral suppressors on the silencing pathway at the cellular level. Nucleic Acids Res. 32 e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, B.M., Pruss, G.J., and Vance, V.B. (2004). Plant viral suppressors of RNA silencing. Virus Res. 102 97–108. [DOI] [PubMed] [Google Scholar]
- Schwach, F., Vaistij, F.E., Jones, L., and Baulcombe, D.C. (2005). An RNA-dependent RNA polymerase prevents meristem invasion by Potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy, D., Molnar, A., Lucioli, A., Szittya, G., Hornyik, C., Tavazza, M., and Burgyan, J. (2002). A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H. (2006). Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 20 759–771. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2005). Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 6 206–220. [DOI] [PubMed] [Google Scholar]
- Wang, X.H., Aliyari, R., Li, W.X., Li, H.W., Kim, K., Carthew, R., Atkinson, P., and Ding, S.W. (2006). RNA interference directs innate immunity against viruses in adult Drosophila. Science 312 452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Tzfira, T., Gaba, V., Citovsky, V., Palukaitis, P., and Gal-On, A. (2004). Functional analysis of the cucumber mosaic virus 2b protein: Pathogenicity and nuclear localization. J. Gen. Virol. 85 3135–3147. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Wilken, A., and Carrington, J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, M., Peragine, A., Park, M.Y., and Poethig, R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D., Fan, B., MacFarlane, S.A., and Chen, Z. (2003). Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant Microbe Interact. 16 206–216. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Yuan, Y.R., Pei, Y., Lin, S.S., Tuschl, T., Patel, D.J., and Chua, N.H. (2006). Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.