Abstract
Lys-63–linked multiubiquitin chains play important roles in signal transduction in yeast and in mammals, but the functions for this type of chain in plants remain to be defined. The RING domain protein RGLG2 (for RING domain Ligase2) from Arabidopsis thaliana can be N-terminally myristoylated and localizes to the plasma membrane. It can form Lys-63–linked multiubiquitin chains in an in vitro reaction. RGLG2 has overlapping functions with its closest sequelog, RGLG1, and single mutants in either gene are inconspicuous. rglg1 rglg2 double mutant plants exhibit loss of apical dominance and altered phyllotaxy, two traits critically influenced by the plant hormone auxin. Auxin and cytokinin levels are changed, and the plants show a decreased response to exogenously added auxin. Changes in the abundance of PIN family auxin transport proteins and synthetic lethality with a mutation in the auxin transport regulator BIG suggest that the directional flow of auxin is modulated by RGLG activity. Modification of proteins by Lys-63–linked multiubiquitin chains is thus important for hormone-regulated, basic plant architecture.
INTRODUCTION
Ubiquitylation, the posttranslational attachment of ubiquitin to substrate proteins, proceeds through an enzymatic cascade of activation and transfer, which is conserved in all eukaryotes. Ubiquitin-activating enzyme (UBA or E1) activates ubiquitin by thioester formation between a catalytic Cys residue of E1 and the C-terminal Gly of ubiquitin. Activated ubiquitin is transferred to a ubiquitin-conjugating enzyme (UBC or E2), which associates with a ubiquitin ligase (E3). The E2-E3 complex confers substrate specificity and catalyzes isopeptide bond formation between the ɛ-amino group of internal Lys residues in the substrate and the ubiquitin C terminus. Ubiquitin ligases often contain so-called RING domains, which serve as docking sites for E2 association (Pickart, 2001). Comparison of yeast, animal, and plant ubiquitylation enzymes suggests orthology relationships among UBCs but identifies few clear orthologs among the numerous ligase proteins (Bachmair et al., 2001; Kosarev et al., 2002; Smalle and Vierstra, 2004; Kraft et al., 2005; Stone et al., 2005).
Ubiquitin can itself be a substrate for ubiquitylation; it has seven Lys residues that can serve as sites of modification (Peng et al., 2003; Pickart and Fushman, 2004; Hochstrasser, 2006). The most prominent chain type is formed by linkage of ubiquitin to Lys-48 of another ubiquitin moiety. Lys-48–linked chains target substrates for recognition and degradation by the proteasome. By contrast, Lys-63–linked chains confer a different, nonproteolytic fate onto a substrate. For instance, ubiquitin Lys-63 chains are involved in DNA repair (Hoege et al., 2002) and in receptor signaling (Chen, 2005) in animals. The formation of Lys-63–linked chains requires a heterodimeric E2 enzyme, formed by canonical E2 Ubc13 and another UBC fold protein, called Uev1A (for Ubiquitin enzyme variant 1A) in mammals and Mms2 (for Methyl methane sulfonate–sensitive2) in budding yeast (Hofmann and Pickart, 1999).
In plants, ubiquitylation is involved in many aspects of development and stress response (Schwechheimer and Schwager, 2004; Smalle and Vierstra, 2004; Lechner et al., 2006; Dreher and Callis, 2007). Of particular interest is the contribution to hormonal regulation. Response to the growth regulator auxin (Woodward and Bartel, 2005; Leyser, 2006; Quint and Gray, 2006; Teale et al., 2006; Dreher and Callis, 2007) requires the activity of an SCF (for Skp1, Cullin, F-box receptor)–type ubiquitin ligase (Dharmasiri and Estelle, 2004; Lechner et al., 2006; Parry and Estelle, 2006). Elements of auxin transport (Blakeslee et al., 2005; Paponov et al., 2005; Fleming, 2006; Kramer and Bennett, 2006) may depend on ubiquitin's role in endocytosis (Dupré et al., 2004; d'Azzo et al., 2005; Clague and Urbé, 2006).
Apical dominance, the growth-inhibitory effect of the main shoot on the initials of side shoots, is a classical trait governed by auxin (for recent reviews, see Leyser, 2005, 2006; Dun et al., 2006). Most mutants with decreased auxin content or sensitivity also display reduced apical dominance and therefore have a more bushy stature than wild-type plants. Basipetal transport of auxin from the main shoot is considered to be essential for apical dominance. Directional transport of auxin is also involved in many other differentiation processes, such as vascular tissue formation, and is a focus of intense research (Teale et al., 2006). It is mediated by both influx and efflux systems to channel auxin from cell to cell in a directional manner. Efflux depends on the group of PIN-FORMED (PIN) proteins, which determine auxin flow direction by their asymmetric distribution in the plasma membrane (Paponov et al., 2005; Kramer and Bennett, 2006).
In this work, we describe an Arabidopsis thaliana mutant that essentially lacks apical dominance and displays changes in auxin and cytokinin signaling. The mutant has defects in two sequence-related, membrane-associated RING domain proteins, RGLG1 and RGLG2 (for RING domain ligases 1 and 2). RGLG2 is shown to catalyze ubiquitin Lys-63 chain formation.
RESULTS
Identification of Arabidopsis Ubiquitin Ligases That Mediate Ubiquitin Lys-63 Chain Formation
A heterodimer consisting of Ubc13 and Mms2 is the only known UBC-class enzyme capable of forming the regulatory-type ubiquitin Lys-63 chains in animals and in yeast (Hofmann and Pickart, 1999). The two proteins have homologs in plants (Bachmair et al., 2001; Kraft et al., 2005). In particular, Arabidopsis has two homologs of Ubc13 (At UBC35 and At UBC36; called At UBC13A and At UBC13B, respectively, in Wen et al., 2006) and four homologs of Mms2, called MMZ1 to MMZ4 (for MMS zwei Homologe 1 to 4) in this article (for Arabidopsis Genome Initiative locus identifiers, see Methods).
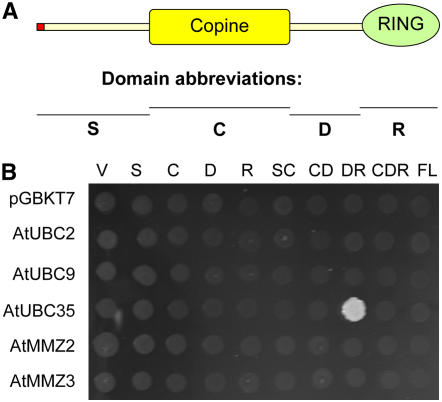
To search for interacting ubiquitin ligases, we performed a yeast two-hybrid experiment, using the heterodimeric MMZ2/At UBC35 complex as a bait (X.-J. Yin and A. Bachmair, unpublished data). In order to allow entry into the yeast nucleus, the C-terminal extension of MMZ2, which mediates membrane association (X.-J. Yin and A. Bachmair, unpublished data), was removed. MMZ2 was fused to the DNA binding domain, whereas At UBC35 was expressed as an unaltered protein. Two sequence-related RING domain proteins were identified in the screen. Their designations, RGLG1 and RGLG2 (At3g01650 and At5g14420, respectively), reflect this feature. RGLG1 and RGLG2 are each other's closest sequelogs, but they belong to a five-member family (Stone et al., 2005) (for gene designations, see Methods). Figure 1A shows the RGLG1 and RGLG2 domain structure, indicating that the proteins also contain a so-called copine (or von Willebrand factor type A) domain. No transmembrane domain was indicated by protein analysis programs such as pfam (www.sanger.ac.uk/Software/Pfam). However, fatty acid linkage to the N terminus, and protein–protein interactions, might direct these proteins to membranes (see below).
Figure 1.
Yeast Two-Hybrid Interactions of RGLG2.
(A) Domain structure of RGLG2 and abbreviations of RGLG2 parts used in (B). The red dot at the N terminus indicates that amino acid Gly at position 2 is a potential site of myristoylation. Copine indicates the copine (von Willebrand factor type A) domain, and RING indicates the RING domain.
(B) Yeast cells expressed various parts of RGLG2 in the activation domain vector pGAD424 and various UBC fold proteins fused to a GAL4 DNA binding domain. pGBKT7 is the empty vector, which was used for the expression of DNA binding domain fusions to At UBC2 (a homolog of yeast Rad6/Ubc2), At UBC9 (a homolog of At UBC8 and of yeast Ubc4), or At UBC35 (a homolog of yeast Ubc13) and of MMZ2 and MMZ3 (both are homologs of yeast Mms2). The medium contains no His but 5 mM 3-aminotriazole. Only At UBC35 and the DR domains of RGLG2 show interaction in the assay. V, no insert control of vector pGAD424; S, C, D, and R, parts of RGLG2 as indicated in (A); SC, CD, DR, and CDR, regions contained in vector pGAD424. FL indicates that the complete RGLG2 protein was contained in the vector.
RGLG2, which has a higher transcript level than RGLG1 (see below), was chosen to further investigate interactions with UBC-domain proteins. Figure 1B shows that a fragment of RGLG2 interacted with UBC35. The finding that full-length RGLG2 does not interact with UBC35 in this assay may indicate that additional proteins can influence E2 access, thereby modulating the activity of the complex (see Discussion). By contrast, the same RGLG2 fragment did not interact with two members of different UBC families, At UBC2 and At UBC9. Likewise, none of the tested fragments of RGLG2 interacted with MMZ2 or MMZ3.
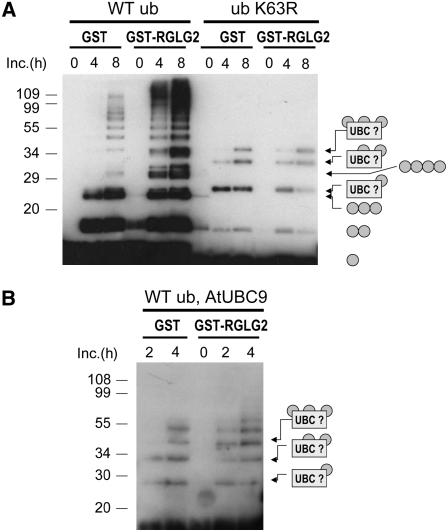
Yeast two-hybrid interaction with the ubiquitin-conjugating enzyme UBC35, and the presence of RING domains in RGLG1 and RGLG2, suggested that these proteins may act as ubiquitin ligases. We investigated this possibility for RGLG2 using in vitro chain-assembly assays. In the in vitro reaction, At UBC35 and At MMZ2 (with deleted C-terminal extension) are rather poor in the formation of ubiquitin chains. Chain-assembly activity in the presence of glutathione S-transferase (GST) (Figure 2A, lanes WT ub, GST) is similar to that without GST (data not shown). The chain linkage specified by At MMZ2/At UBC35 is through Lys-63, as the K63R mutation in ubiquitin abolishes chain formation (lanes ub K63R, GST). Weak bands seen in the ub K63R lanes are probably due to autoubiquitylation of one of the reaction components, presumably UBC35. The relatively low level of Lys-63 chain assembly by MMZ2/UBC35 is significantly stimulated by RGLG2 (lanes WT ub, GST-RGLG2). RGLG2 does not change the type of chain produced, as shown by the relative absence of chains assembled with ubiquitin K63R. In confirmation of a K63-specific reaction, replacement of wild-type ubiquitin by ubiquitin K48R, a variant with Lys-48 changed to Arg, does not prevent chain formation (see Supplemental Figure 1 online). In contrast with the reaction in the presence of MMZ2/UBC35, RGLG2 does not enhance to a similar extent the ubiquitylation by At UBC9 (a core UBC, belonging to the same class as yeast Ubc4; Figure 2B). The results shown in Figure 2 and Supplemental Figure 1 online suggested that Lys-63 chain formation is a major in vivo activity of RGLG2. However, participation in other ubiquitylation reactions remains a possibility that cannot be ruled out at this time.
Figure 2.
RGLG2 Is a Ubiquitin Ligase That Catalyzes the Formation of Ubiquitin Lys-63–Linked Chains.
(A) His6-MMZ2 core, At UBC35, human His6-UBA, human ubiquitin of either wild-type sequence, or variant ubiquitin with a Lys-to-Arg change at position 63 were incubated with either GST or GST fused to RGLG2. MMZ2/At UBC35 without GST-RGLG2 have weak activity to form ubiquitin chains (left three panels; WT ub, GST). The linkage type is revealed by the absence of these bands if ubiquitin K63R is used instead of ubiquitin (ub K63R, GST lanes). GST-RGLG2 enhances chain formation while maintaining the linkage specificity (lanes WT ub and GST-RGLG2 versus ub K63R and GST-RGLG2).
(B) RGLG2 does not enhance (auto)ubiquitylation by another UBC, At UBC9, to an extent comparable to the reaction in the presence of UBC35/MMZ2, indicating specificity for UBC35/MMZ2-dependent Lys-63 chain formation. Ubiquitin conjugates were visualized by protein gel blotting with anti-ubiquitin antibodies. The suggested identity of ubiquitin conjugates is shown at right. UBC? indicates proteins that are formed by attachment of single ubiquitin moieties, an apparent side reaction. Molecular mass marker positions and sizes (in kilodaltons) are indicated at left.
RGLG1 and RGLG2 Are Ubiquitously Expressed Membrane-Associated Proteins
According to microarray data, RGLG1 and RGLG2 mRNAs are rather abundant and ubiquitously expressed (with tissue-specific variations) (Kraft et al., 2005). To obtain a higher resolution picture, promoter regions of the two genes were fused to the β-glucuronidase (GUS) reporter and expressed in planta. Supplemental Figure 2 online shows a generally uniform expression of both reporter constructs, with stronger staining of the vasculature. RGLG2 expression is higher than RGLG1 expression in most tissues. One notable exception is the hypocotyl, where RGLG2 expression is very low.
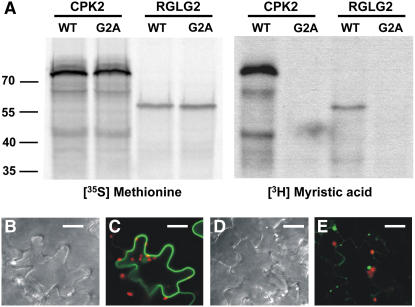
Because RGLGs have a predicted N-terminal myristoylation site, this issue was directly addressed by an in vitro reaction. RGLG2 and a variant with Gly-2 replaced by Ala (G2A) were expressed in a wheat germ–based in vitro translation system (Figure 3A). Protein kinase CPK2 (At3g10660), a known myristolyated protein, served as a positive control (Lu and Hrabak, 2002). Whereas the presence of radioactive Met led to the accumulation of a labeled protein for both wild-type RGLG2 and for the G2A mutant, the identical reaction in the presence of radioactive myristic acid allowed labeling of only wild-type RGLG2 but not of the mutant protein. We concluded that RGLG2 can be myristoylated in vitro and that the recognition motif for this posttranslational modification encompasses Gly at position 2, which is the predicted lipid attachment site. The presence of Gly-2 had a significant influence on the cellular localization of RGLG2 (Figures 3B to 3E). Whereas wild-type RGLG2, fused to a C-terminal green fluorescent protein (GFP) reporter, localized to the cell periphery upon tobacco (Nicotiana tabacum) agroinfection (Figure 3C), suggestive of membrane localization, the G2A mutant was much less abundant under the same conditions and accumulated in one or two aggregates per cell (Figure 3E). We concluded that the presence of an intact N terminus significantly influences RGLG2-GFP localization, consistent with an in vivo role for lipid modification.
Figure 3.
Modification of RGLG2 by Myristic Acid.
(A) RGLG2 (WT) and the RGLG2 variant with Gly-2 changed to Ala (G2A) were incubated with either radioactive Met (left panels) or radioactive myristic acid (right panels) in an in vitro reaction. Wild-type RGLG2, but not the G2A mutant, can be modified by myristoylation. Gly-2 is the predicted linkage point to the fatty acid, and its absence does not allow modification. Protein kinase CPK2 served as a positive control.
(B) to (E) Mutant and wild-type RGLG2 proteins fused to GFP were observed in tobacco leaf epidermis cells at 2 d after agroinfection. Whereas the wild-type protein localizes to the cell periphery (C), the G2A mutant RGLG2-GFP protein does not accumulate well and forms aggregates (E), supporting a role for the N-terminal myristic acid as a membrane anchor. (B) and (D) show phase contrast images of (C) and (E), respectively. GFP signals are in green, and chlorophyll fluorescence of chloroplasts is visible in red. Bars = 10 μm.
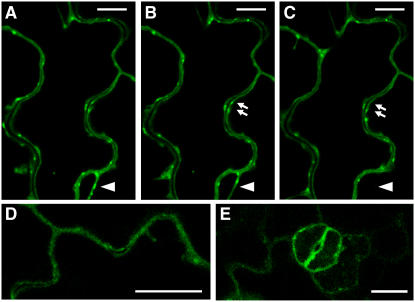
To study the intracellular localization in Arabidopsis, RGLG1 and RGLG2 proteins, fused to the C-terminal GFP reporter, were expressed in transgenic plants under the control of a cauliflower mosaic virus (CaMV) 35S-derived promoter. Confocal images in Figures 4A to 4C show the same frame of RGLG2-GFP in epidermis cells, with time differences of ∼8 s between Figures 4A and 4B and ∼30 s between Figures 4B and 4C. RGLG2 is predominantly localized at the plasma membrane. Figure 4 and images not shown here also suggested that RGLG2 can decorate membranes in transiently formed cytoplasmic strands (arrowheads in Figures 4A to 4C). Domains of increased RGLG2-GFP accumulation at certain areas of the cell membrane (e.g., arrows in Figures 4B and 4C) are also dynamic structures. These signals may result from vesicles close to the membrane or from protein accumulation in subdomains of the plasma membrane itself. Figures 4D and 4E document the presence of RGLG1-GFP on membranes (Figure 4D, leaf epidermis cell; Figure 4E, stomata). While the higher expression level of the available RGLG2-GFP expression lines allowed for better resolved signals, we saw no difference in localization between RGLG1 and RGLG2. This interpretation is consistent with the genetic data, which suggested redundant function for the two proteins (see below). The demonstration that the GFP fusion constructs used for imaging can complement phenotypes observed in mutants (see below) suggested that the observed intracellular distribution significantly overlaps the distribution of the authentic proteins.
Figure 4.
Localization of Ubiquitin Ligases RGLG1 and RGLG2.
(A) to (C) View of RGLG2-GFP as seen by confocal microscopy in a time series. Frame (B) was taken ∼8 s after frame (A), and the time difference between frames (B) and (C) is ∼30 s. The protein generally decorates the plasma membrane but is also present at the membrane of dynamically formed cytoplasm strands (arrowheads). Arrows show zones of transient RGLG2 accumulation.
(D) and (E) RGLG1-GFP seen at the plasma membrane. The localization of RGLG1-GFP and RGLG2-GFP seems identical.
Images show leaf epidermis ([A] to [D]) and stomata (E). Bars = 10 μm.
Inactivation of Both RGLG1 and RGLG2 Results in the Abolishment of Apical Dominance and the Alteration of Normal Leaf Phyllotaxy
T-DNA insertion alleles in the genes RGLG1 (SALK_011892) and RGLG2 (SALK_062384) were characterized from populations of transformed lines (Alonso et al., 2003). Supplemental Figure 3 online presents a characterization of the mutant alleles and indicates that neither mutant produced full-length RGLG transcript. Together with the findings (presented below) that both insertion alleles were recessive, these data strongly suggested that these alleles display a loss of function. Plants homozygous for either the rglg1 or rglg2 knockout mutation were phenotypically similar to wild-type plants (data not shown), supporting the notion that both genes perform identical functions. However, double mutant combinations encompassing rglg1 and rglg2 insertion alleles showed remarkable developmental alterations.
Plants carrying the homozygous rglg1 mutation in the hemizygous rglg2/RGLG2 background (Figure 5B, left plant) were only slightly smaller than wild-type plants (Figure 5A, left plant), whereas the reverse combination (rglg1/RGLG1 rglg2/rglg2) displayed reduced apical dominance (Figure 5B, right plant). The stronger effect of the loss of both RGLG2 copies and one RGLG1 copy, compared with the reverse combination, is consistent with a redundant function but a higher expression level of RGLG2 compared with RGLG1. The homozygous rglg1 rglg2 double mutant showed a complete loss of apical dominance, lacking normal dominance of the central apical meristem, and the development of a central inflorescence (Figure 5C). All double mutants grown to maturity had >30 shoots of comparable length, suggesting that the loss of apical dominance trait has 100% penetrance. By contrast, the rglg1 rglg2 double mutant expressing RGLG2-GFP under the control of the CaMV 35S promoter looked similar to the wild-type plant (Figure 5A, right plant versus left plant). rglg1 rglg2 plants containing an RGLG1-GFP expression construct also looked like wild-type plants (data not shown), demonstrating the functional equivalence of both proteins.
Figure 5.
Phenotypes of rglg1 rglg2 Mutant Plants.
(A) A wild-type Arabidopsis plant (left) next to an rglg1rglg2 double homozygous plant transformed with a RGLG2-GFP fusion gene under the control of the CaMV 35S promoter (as used in Figure 4).
(B) An rglg1/rglg1 rglg2/RGLG2 plant (left) looks very similar to the wild type, whereas an rglg1/RGLG1 rglg2/rglg2 mutant plant (right) shows reduced apical dominance.
(C) A mature rglg1 rglg2 double mutant plant without a complementing RGLG construct.
(D) and (E) Ten-day-old seedlings of rglg1 rglg2 double mutant (E) or wild-type (D) genotype.
(F) and (G) At age 3 weeks, differences in leaf morphology become more pronounced ([G], mutant; [F], wild type).
(H) to (J) rglg1 rglg2 plants at different stages of development.
(H) After the emergence of elongated leaves, small leaves emerge to give the plant a bushy appearance.
(I) Early shoots of rglg1 rglg2 double mutants are often very short, leaving the siliques almost directly connected to the rosette.
(J) Shoots of rglg1 rglg2 double mutants have few cauline leaves and axillary meristems.
(K) and (L) Cleared and counterstained leaves show thicker and less branched vasculature of rglg1 rglg2 (L) versus wild-type (K) plants.
(M) and (N) Altered phyllotaxy of rglg1 rglg2 mutants. rglg1 rglg2 mutant plants form leaves at roughly opposite sides, then at a right angle to the previously formed leaf pair (N), whereas wild-type plants produce a spiral arrangement of leaf primordia with approximately three leaves per round (M).
Bars in (A) to (C) = 2 cm, those in (D) to (J) = 0.3 cm, those in (K) and (L) = 0.1 cm, and those in (M) and (N) = 0.4 mm.
Figures 5E, 5G, and 5H to 5J show different stages in the rglg1 rglg2 double mutant life cycle. Although the overall growth of young rglg1 rglg2 seedlings closely resembled that of wild-type plants (Figure 5D, wild-type, versus Figure 5E, mutant), the early rosette leaves were more elongated (Figure 5F, wild type, versus Figures 4G and 4H, mutant) and darker (Figures 5H and 5I). These early rosette leaves senesce after ∼4 to 6 weeks of growth (visible in Figure 5J). The first emerging shoots were short and terminated by one or few flowers yielding fertile siliques (Figure 5I). Later developing shoots were longer (Figures 5C and 5J). Another difference between the wild type and the rglg1 rglg2 mutant was leaf shape and architecture. Mutant leaves were more oblong and often asymmetric and had reduced numbers of veins (Figure 5K, wild-type leaf; Figure 5L, mutant leaves). Figures 5M and 5N show sections through the shoot meristem region, demonstrating characteristic alterations in phyllotaxy. Whereas leaf initials in the wild type formed a spiral, with an ∼137.5° angle between leaf initials (Figure 5M), the rglg1 rglg2 mutant formed two initials opposite to each other and then switched by 90° to form the next pair of opposing leaves (Figure 5N). Thus, rglg1 rglg2 plants often had a decussate pattern of leaf emergence, instead of the spiral pattern of wild-type Arabidopsis. Additional phenotypes of aerial plant parts, in particular an increased cell size, are shown in Supplemental Figure 4 online (differences in cell size are also visible in Figures 5M versus 5N).
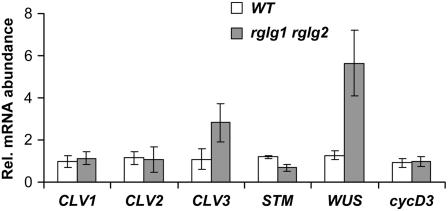
Abolishment of the development of the primary shoot meristem and changes in leaf phyllotaxy in the rglg1 rglg2 double mutant coincided with changes in the expression of genes acting in the control of apical stem cell maintenance. We observed a significant change in the level of WUSCHEL (WUS; At2g17950) mRNA, which was approximately fourfold higher in the mutants (Figure 6). However, mRNA levels of other genes of the WUS regulon, including CLAVATA1 (CLV1; encoded by At1g75820) and CLV2 (At1g65380), were unchanged. CLV3 (At2g27250) (Williams and Fletcher, 2005; Kondo et al., 2006), the gene encoding a peptide ligand of the CLV1/2 membrane receptor, was elevated by more than twofold. Its increased expression would actually downregulate WUS in the wild type, which was obviously not the case in rglg1 rglg2. On the other hand, WUS negatively regulates the abundance of the SHOOT MERISTEMLESS (STM; At1g62360) transcript in the wild type, which was also the case in the double mutant (Figure 6). These measurements suggested changes in signal transduction between CLV3 perception at the membrane and WUS expression. A mechanistic resolution of the role of RGLGs in the regulation of stem cell maintenance and differentiation in the apical meristem remains a task for further studies. As documented in Supplemental Figure 5 online, rglg1 rglg2 mutants also showed impaired circadian clock function.
Figure 6.
rglg1 rglg2 Mutant Plants Misexpress Genes of the Apical Meristem.
Apical meristem regions of 24-d-old plantlets were used for RNA preparation, reverse transcription, and real-time PCR quantification. rglg1 rglg2 double mutant plants contain approximately four times more WUS transcript, approximately three times as much CLV3 transcript, and approximately half the amount of STM mRNA, whereas other gene expression profiles, such as those of CLV1, CLV2, and cycD3, remain unchanged.
Roots of wild-type and mutant plants were similar. We found a moderate (14%) reduction in the main root length of plantlets grown on plates (in one experiment, root length at 4 d after germination was 1.65 ± 0.05 cm for the wild type and 1.42 ± 0.04 cm for the double mutant). Reduced root growth in the presence of the auxin naphthylacetic acid (NAA) at 85 nM concentration maintained the percentile difference, and root curvature in a standard gravitropism assay was apparently unchanged (data not shown).
The rglg1 rglg2 Mutation Causes Alterations in the Expression of Genes Controlled by Auxin and Cytokinin Hormone Levels
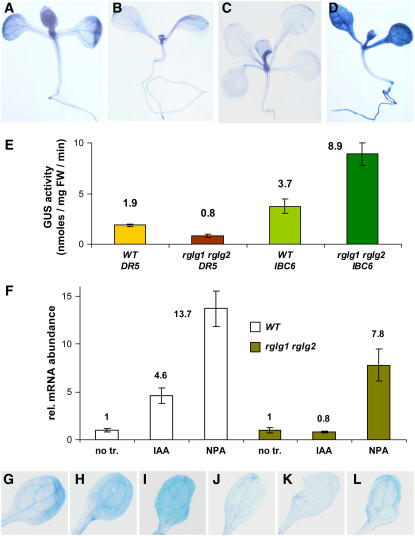
The bushy stature, altered phyllotaxy, and other phenotypes led us to test auxin- and cytokinin-responsive reporter constructs, which were crossed into the rglg1 rglg2 background. Interestingly, the auxin-responsive gene construct DR5:GUS was expressed twofold lower than in the wild type (Figures 7B versus 7A, and Figure 7E), whereas the cytokinin-responsive promoter:GUS fusion IBC6:GUS directed an approximately twofold higher expression in the rglg1 rglg2 mutant than in the wild type (Figures 7D versus 7C, and Figure 7E). A similar situation was also found for endogenous genes by measuring mRNA levels of the auxin-inducible IAA1 (At4g14560) and the cytokinin-responsive IBC7 (At1g10470; data not shown). These data suggested that the rglg1 rglg2 mutation could alter either spatial distribution or the synthesis or metabolism of one of these hormones, which would ultimately cause a change in the normal auxin–cytokinin balance.
Figure 7.
The Expression of Hormone-Responsive Genes Is Changed in rglg1 rglg2 Mutant Plants.
(A) to (E) The auxin-inducible reporter construct DR5:GUS shows lowered GUS enzyme activity, as shown by staining ([A], wild-type plant; [B], mutant plant) and by enzyme activity measurement (E). Similarly, the cytokinin-inducible reporter construct IBC6:GUS is increased, as shown by staining ([C], wild-type plant; [D], mutant plant) and by enzyme activity measurement (E). FW, fresh weight.
(F) Wild-type and mutant seedlings were harvested for quantification of IAA1 mRNA by real-time PCR. The effect of auxin was determined after growth for 15 h on plates containing 1 μM indole acetic acid (IAA). For treatment with auxin transport inhibitor, seeds were germinated on plates containing 10 μM NPA. Controls contained neither compound. Auxin treatment does not increase the level of IAA1 mRNA in mutant seedlings, and the response to NPA is smaller than in the wild type. no tr., control without added substance; IAA, IAA added; NPA, NPA added.
(G) to (L) Staining of DR5:GUS plants confirms the results shown in (F). Seedlings were transferred at 5 d after germination from normal agar growth medium to plates containing no drug ([G] and [J]), the auxin NAA (85 nM; [H] and [K]), or NPA (10 μM; [I] and [L]). After 15 h, seedlings were harvested and stained for GUS activity.
(G) to (I) The GUS transgene was in the wild-type background.
(J) to (L) The GUS transgene was in the rglg1 rglg2 mutant background.
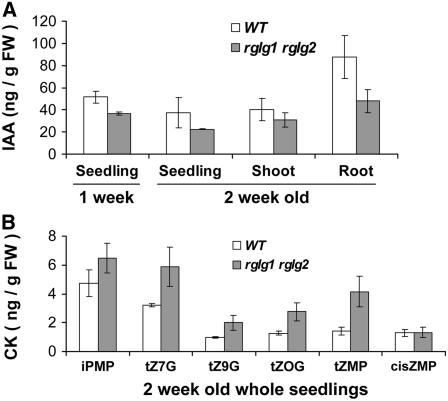
To support this hypothesis, auxin and cytokinin contents were directly measured in rglg1 rglg2 plants and compared with the wild-type levels. Auxin (IAA) concentration was almost twofold lower in roots of 2-week-old mutant seedlings (Figure 8A). A still unresolved issue is that shoot morphology was apparently more perturbed than root morphology (see above), although the reduction in auxin levels was more pronounced in roots than in shoots. However, rglg1 rglg2 mutants also differed from wild-type plants in cytokinin content. Figure 8B, and Supplemental Figures 6 and 7 online, show values for different cytokinin nucleotides and conjugates. Cytokinin differences were more pronounced in shoots. Generally, the observed changes in hormone concentration correlated well with the changes in hormone-responsive transcription activities and therefore were consistent with the hypothesis that RGLG proteins influence primarily hormone concentrations (rather than transcriptional responses to hormones).
Figure 8.
Auxin and Cytokinin Levels Are Altered in rglg1 rglg2 Plants.
(A) IAA levels in wild-type and rglg1 rglg2 double mutant seedlings aged 1 or 2 weeks were determined. Mutants have lower auxin content. The difference seems to be more pronounced in roots. FW, fresh weight.
(B) Several different cytokinins are elevated in rglg1 rglg2 double mutant plants. iPMP, isopentenyladenine riboside-5′-monophosphate; tZ7G, trans-zeatin-7-glucoside; tZ9G, trans-zeatin-9-glucoside, tZOG, trans-zeatin-O-glucoside; tZMP, trans-zeatin-riboside 5′-monophosphate; cisZMP, cis-zeatin-riboside-5′-monophosphate; CK, cytokinin.
We also investigated whether the difference in auxin response can be rescued by the addition of exogenous auxin. We used expression of the auxin-responsive gene IAA1 as a biological readout. Figure 7F indicates that a 15-h incubation in the presence of auxin (IAA at 1 μM) did not lead to the expected increased expression of IAA1. Moreover, the auxin transport inhibitor naphthylphthalamic acid (NPA; at 10 μM) also did not lead to the same increase in auxin response as in wild-type plants. This finding was confirmed by exposure of DR5:GUS plantlets to auxin (in this experiment, the more long-lived NAA was applied at 85 nM for 15 h) and auxin transport inhibitor (NPA at 10 μM) prior to GUS staining (Figures 7G to 7L). Characteristically, this phenotype implicates auxin transport capacity as a target of RGLG activity.
Relationship to Known Auxin Response Mutants
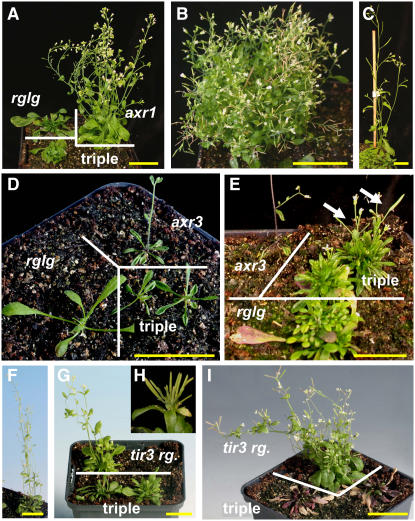
The changes in auxin levels and response led us to test genetic interactions with known components of auxin response. According to current knowledge, the auxin signaling that converges on the transcription regulation of primary response genes uses the F-box protein TRANSPORT INHIBITOR RESPONSE1 (TIR1) as an auxin receptor (Dharmasiri et al., 2005; Kempinski and Leyser, 2005; Tan et al., 2007). Auxin binding confers increased TIR1 affinity to AUX/IAA-type transcriptional effectors, channeling them into degradation by ubiquitylation via an SCF-type ubiquitin ligase (Dharmasiri and Estelle, 2004). Mutation axr1-12 (Leyser et al., 1993) decreases SCFTIR1 ligase activity, whereas axr3-1 (Rouse et al., 1998) renders an SCFTIR1 substrate more resistant to degradation. To examine how RGLG1 and RGLG2 interact with this SCF-TIR1 pathway, triple mutants axr1-12 rglg1 rglg2 and axr3-1 rglg1 rglg2 were made (Figure 9). Mutant plants axr1-12 rglg1 rglg2 had an approximately additive phenotype. They were bushy and displayed the reduced fertility of axr1-12 plants (Figure 9B). axr3-1 plants usually form one shoot, supported by a small rosette (Figures 9D and 9E, axr3). rglg1 rglg2 seems to be epistatic regarding this shoot phenotype, because the triple mutant axr3-1 rglg1 rglg2 was bushy, formed several shoots, and was generally more similar to the rglg1 rglg2 parent (Figure 9E, triple). Leaf shape in the rosette, however, resembled the axr3-1 mutant (Figure 9D, axr3 versus triple). Generally, the growth habit of both triple mutants was consistent with a model in which RGLG1 and RGLG2 influence primarily auxin hormone levels, whereas the SCF-TIR1 pathway translates hormone levels into transcriptional changes.
Figure 9.
Triple Mutant Combinations with Mutations in Auxin Response.
(A) to (E) Mutations in the SCF-TIR1 pathway indicate that rglg1 rglg2 acts additive with, or partially suppresses, mutations in this pathway.
(A) axr1-12 mutation in combination with rglg1 rglg2. Plants are 6 weeks old. rglg indicates genotype rglg1 rglg2, and axr1 indicates plant homozygous for allele axr1-12. Triple indicates the triple mutant axr1-12 rglg1 rglg2.
(B) Mature axr1-12 rglg1 rglg2 plants have a bushy stature and reduced fertility, like the axr1-12 mutant.
(C) Columbia wild-type plant, age-matched with (A) for comparison.
(D) and (E) An axr3-1 mutant (axr3), which has increased apical dominance and produces only one shoot, was grown in the same pot as an rglg1 rglg2 mutant (rglg) and triple mutants axr3-1 rglg1 rglg2 (triple). The leaf form of triple mutants at age 3.5 weeks (D) resembles that of the axr3-1 single mutant, but shoot emergence is delayed. At age 8 weeks (E), several shoots (white arrows) emerge from the triple mutant axr3-1 rglg1 rglg2.
(F) to (I) Interaction between BIG (allele tir3-1) and RGLG1 RGLG2.
(F) tir3-1 mutant at age 6 weeks.
(G) Triple mutant tir3-1 rglg1 rglg2 at age 6 weeks (triple) compared with a plant that is homozygous for mutations tir3-1 rglg2 and heterozygous at the RGLG1 locus (tir3 rg.).
(H) Siliques of these latter plants often occur in bundles.
(I) Similar to (G), but plants at age 9 weeks. Note that the triple mutant tir3-1 rglg1 rglg2 died at the time of expected shoot emergence.
Bars = ∼2 cm.
By contrast, the triple mutant combination tir3-1 rglg1 rglg2 showed considerable interaction between RGLG1/RGLG2 and TIR3. TIR3 (=BIG; At3g02260) is a large membrane-associated protein and a regulator of auxin transport (Gil et al., 2001). Mutants with genotype tir3-1 rglg2 (both alleles homozygous) rglg1/RGLG1 showed some phenotypic similarity with tir3-1 axr1-12 plants (Gil et al., 2001), being more bushy than tir3-1 plants and often producing siliques in bundles (Figure 9G, tir3 rg., and Figures 9H and 9I). Triple mutants tir3-1 rglg1 rglg2 had low viability, accumulated anthocyanin, and all plants died before shoot emergence (Figure 9I, triple), suggesting that RGLG1/2 and BIG impinge on the same essential process, auxin transport.
RGLG1 and RGLG2 Influence Auxin Transport Protein Abundance
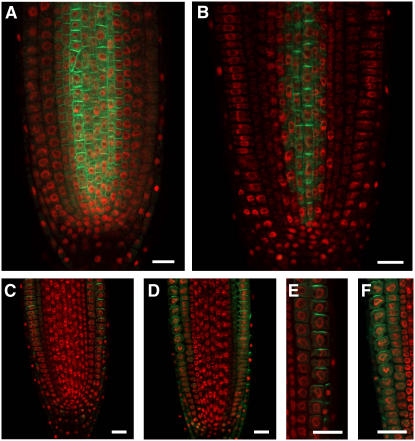
To further define the influence of RGLG proteins on auxin transport, we investigated members of the PIN group of membrane proteins, which are known to mediate auxin transport processes (Figure 10) (Teale et al., 2006). Visualization of these proteins as fusions to GFP has previously allowed detailed studies of their abundance and distribution. Functional PIN1 promoter:PIN1-GFP and PIN2 promoter:PIN2-GFP constructs were crossed into the rglg1 rglg2 background to observe their distribution in mutant plants. Interestingly, the PIN1 construct showed decreased abundance, whereas PIN2 abundance seemed only marginally affected. The overall intracellular distribution of both PIN isoforms was apparently maintained, although polar localization of PIN2 appeared less pronounced in the mutant (Figures 10E versus 10F). Decreased abundance of auxin transport proteins certainly contributes to the suggested decrease in auxin transport capacity. Interestingly, the influence of RGLGs on PIN1 may be based on physical interaction, because RGLG2 and PIN1 proteins interact in the yeast two-hybrid assay (see Supplemental Figure 8 online).
Figure 10.
RGLG1/2 Influence the Abundance of the PIN1-GFP Fusion Protein.
Mutant ([B], [D], and [F]) and wild-type ([A], [C], and [E]) plants expressing PIN1-GFP ([A] and [B]) or PIN2-GFP ([C] to [F]) proteins under the control of their natural promoters were analyzed by confocal microscopy. Compared with the wild type, the abundance of PIN1-GFP is reduced in rglg1 rglg2 roots (B), whereas the abundance of PIN2-GFP is much less affected ([C] to [F]) Nuclei were stained with 4′,6-diamidino-2-phenylindole (shown in red). Bars = 20 μm.
DISCUSSION
Unlike chains formed by linkage of the ubiquitin C terminus to Lys-48 of another ubiquitin moiety, which mediate proteasomal turnover, those formed by linkage via Lys-63 do not generally lead to substrate degradation. Instead, ubiquitin Lys-63 chains may have roles in cellular signaling. In this work, we present the characterization of RGLG1 and RGLG2, two membrane-associated ubiquitin Lys-63 chain–forming enzymes from Arabidopsis.
Double mutant Arabidopsis plants rglg1 rglg2 differ slightly from wild-type plants at the early seedling stage (Figures 5D versus 5E), indicating that the genes play only a minor role in early development. This may result from the fact that Arabidopsis has three additional RGLG2-related genes (Stone et al., 2005). Postembryonal development of rglg1 rglg2 plants, however, differs significantly from that of wild-type plants in a variety of traits. Many of these traits are known to be critically influenced by the plant growth regulator auxin (reviewed in Woodward and Bartel, 2005; Leyser, 2006; Quint and Gray, 2006; Teale et al., 2006). In particular, auxin influences apical dominance (Leyser, 2005, 2006), phyllotaxy (Reinhardt et al., 2003; Jönsson et al., 2006; Smith et al., 2006), leaf venation (Fukuda, 2004), and circadian rhythms (Hanano et al., 2006), all of which differ in the rglg1 rglg2 mutants. To establish how auxin responses and concentrations are changed in mutant plants, transcription of hormone-responsive genes and hormone concentrations were determined. rglg1 rglg2 plants have lower auxin (IAA) concentrations (Figure 8). Likewise, auxin-responsive genes display lower expression levels (Figure 7). In contrast with the wild type, the mutant also does not adapt transcription of auxin-responsive genes after exogenous application of auxin (Figures 7F to 7L; data not shown).
The best understood module of auxin response regulates transcription. The SCF-TIR1 pathway consists of a soluble auxin receptor, TIR1 (Dharmasiri et al., 2005; Kempinski and Leyser, 2005; Tan et al., 2007), which binds to transcriptional regulators (AUX/IAA proteins) in an auxin-dependent way to mediate their turnover through ubiquitylation by ubiquitin ligase SCFTIR1 (reviewed in Dharmasiri and Estelle 2004; Parry and Estelle 2006). We made combinations of two SCF-TIR1 pathway mutants, axr1-12 and axr3-1, with rglg1 rglg2 mutants (Figure 9). Phenotypic analysis shows that mutations in RGLG1/2 are additive with, or partially suppress the effects of, mutations in the SCF-TIR1 pathway. The most plausible interpretation of this result is that RGLG1 and RGLG2 proteins are primarily elements involved in adjusting intracellular auxin levels, whereas the SCF-TIR1 pathway is responsible for transcriptional responses to the intracellular auxin concentration. Therefore, RGLG1 and RGLG2 define hormone regulatory elements distinct from the SCF-TIR1 pathway. This interpretation is supported by the fact that all known elements of the SCF-TIR1 pathway are soluble, whereas RGLG1/2 are membrane-associated (Figures 3 and 4). By contrast, anthocyanin accumulation and the death of the triple mutant tir3-1 rglg1 rglg2 at later stages of development indicate that the respective proteins impinge on the same process(es) (Figure 9). The large membrane-associated protein, BIG/TIR3, was described as a regulator of polar auxin transport (Gil et al., 2001).
Although mutations in BIG are pleiotropic (Kanyuka et al., 2003; Desgagné-Penix et al., 2005; López-Bucio et al., 2005; Kasajima et al., 2007), many phenotypes can be linked to auxin transport defects. In particular, the tir3-1 mutation was shown to reduce GA sensitivity (Desgagné-Penix et al., 2005). However, a link was previously found between auxin transport and GA signaling (Fu and Harberd, 2003). Furthermore, GA defects are not normally linked to cell death, so that we provisionally attribute the lethality of the triple mutant rglg1 rglg2 tir3-1 to auxin transport defects, and not to interference with GA hormonal pathways by either tir3-1 or rglg1 rglg2 mutations.
Interestingly, a number of results can be explained most easily by the assumption that rglg1 rglg2 mutants are compromised in auxin transport. First, auxin concentration, which was measured separately in shoots and roots (Figure 8), is more significantly decreased in roots, which are a sink tissue for auxin, and at least partially depend on auxin import from shoots. Second, pharmacological inhibition of auxin transport has a lower effect on transcription in rglg1 rglg2 mutants than in wild-type plants (Figures 7F to 7L), which may be expected if the auxin transport capacity is already lowered by the mutations in RGLG1 and RGLG2. Third, triple mutants with inactive RGLG1, RGLG2, and BIG die during somatic development (Figures 9F to 9I), which may indicate that auxin transport needs at least one of the accessory factors, RGLG1, RGLG2, or BIG, in order to allow plant survival. Fourth, an influence of RGLG proteins on auxin transport capacity is also suggested by the finding that a protein fusion between the auxin transport component PIN1 and GFP, expressed from the PIN1 promoter on a transgene, has reduced abundance in mutant plants (Figure 10). The related PIN2 protein, however, seems only marginally affected (Figure 10). PIN family auxin transport proteins are regulated by a network of currently incompletely defined feedback loops that include the assessment of auxin concentration (Vieten et al., 2005; Sauer et al., 2006). The contribution of RGLGs to PIN1 regulation may, at least in part, be based on direct physical contact, because RGLG2 and PIN1 can interact in the yeast two-hybrid system (see Supplemental Figure 8 online).
The rglg1 rglg2 mutants also have higher cytokinin concentrations (Figure 8; see Supplemental Figures 6 and 7 online), and cytokinin-responsive genes display a higher expression than in wild-type plants (Figure 7). Furthermore, the upregulation of the gene WUS (Figure 6), which encodes a critical component of a stem cell maintenance regulatory loop in the apical meristem region, may be a consequence of the cytokinin responsiveness of the WUS-STM regulon (Mizuno, 2004; Leibfried et al., 2005). Similarly, suppression of the shoot phenotype of the axr3-1 mutation by rglg1 rglg2 in the triple mutant (Figure 9) may be connected to increased cytokinin in the rglg double mutant background, because many phenotypes of the axr3-1 mutation can be partially suppressed by exogenous supply of cytokinin to axr3-1 single mutants (Leyser et al., 1996). The changes in cytokinin could be either a direct result of the absence of RGLG1 and RGLG2 in the mutant or an indirect consequence of differences in auxin concentration. This is because there is a crosstalk between auxin and cytokinin, so that mutants in auxin response can also have changes in cytokinin (Nordström et al., 2004b; Rashotte et al., 2005; Tanaka et al., 2006).
How might RGLG1/2 exert their functions? Interaction partners, protein structure, and in vitro activity of these proteins hint at a possible mode of action. Both proteins interact in a yeast two-hybrid assay with UBC35, a plant homolog of the yeast ubiquitin conjugation enzyme Ubc13. The latter protein forms a heterodimer with Mms2 to catalyze the formation of ubiquitin Lys-63–linked chains (Hofmann and Pickart, 1999). In the presence of ubiquitin, ubiquitin-activating enzyme, UBC35, and the Arabidopsis Mms2 homolog MMZ2, RGLG2 enhances the formation of Lys-63–linked ubiquitin chains (Figure 2).
An interesting facet of RGLG activity is that, in the yeast two-hybrid system, UBC35 does not interact with the full-length RGLG2 protein but only with a fragment that contains the RING domain (Figure 1). We interpret this finding to indicate that, in vivo, interaction of RGLG2 with UBC35/MMZ2, and therefore enzyme activity, is regulated by additional proteins. The recent finding that another yeast two-hybrid binding partner of RGLG2 can stimulate its in vitro activity is consistent with this conjecture (X.-J. Yin and A. Bachmair, unpublished data).
While we cannot rule out that, together with other UBC enzymes, RGLGs may form ubiquitin linkage types other than via Lys-63, the data presented clearly show that RLGL2 ubiquitylation in the presence of UBC35 and MMZ2 is specific for ubiquitin Lys-63 linkages (Figure 2; see Supplemental Figure 1 online). The robustness of the in vitro reaction, and yeast two hybrid data (Figure 1), furthermore suggest that Lys-63 chain formation is the major activity of these ubiquitin ligases. Also consistent with this interpretation is the finding that Arabidopsis mutants in both MMZ1 and MMZ2, UBC cofactors essential for Lys-63 chain formation, display the similar phenotype of reduced apical dominance (X.-J. Yin and A. Bachmair, unpublished data), implying ubiquitin Lys-63 chain formation in the regulation of plant shape.
RGLG1 and RGLG2 have a copine domain (also called a von Willebrand factor type A domain) positioned approximately in the middle of the protein (Figure 1A). Copine domains can exist in two alternative conformations, and conformational switches have been investigated at the genetic and structural levels (Mould et al., 2003; Barton et al., 2004). Therefore, a conformational switch could be part of RGLG activity.
Ubiquitin Lys-63 chain–forming enzymes that operate with or in membrane-associated protein complexes have been identified in several regulatory contexts in animals and in yeast. For instance, some membrane proteins require ubiquitin Lys-63 chain attachment, instead of the usual monoubiquitylation, for endocytosis and/or recycling (reviewed in Dupré et al., 2004; d'Azzo et al., 2005; Clague and Urbé, 2006). Furthermore, signaling from membrane receptors may require ubiquitin Lys-63 chain formation. The cascade from the interleukin-1 receptor to the transcriptional activator nuclear factor-κB is best characterized and involves autoubiquitylation of a Lys-63 chain–specific ubiquitin ligase, TRAF6 (Chen, 2005; Krappmann and Scheidereit, 2005; Lamothe et al., 2007). It remains to be determined whether RGLG1/2 have a role in the internalization of membrane proteins, in signal transduction, or in other processes, such as the modification of membrane protein activity.
This work highlights the role of two ubiquitin ligases as membrane-associated elements of hormone adjustment in plants. One of these ligases was shown to form ubiquitin Lys-63 chains. The ligases are not part of the known SCF-TIR1 pathway and influence the levels of the plant hormones auxin and cytokinin. The membrane localization, triple mutant analysis, and additional results suggest that they may influence auxin transport.
METHODS
Growth Conditions, Plant Lines, and Transgenes
Arabidopsis thaliana plants were grown on soil in controlled-environment rooms under long-day conditions (10 h of light/6 h of day-extension illumination/8 h of dark) or on Murashige and Skoog agar under true long days (16 h of light/8 h of dark). Arabidopsis ecotype Columbia was the accession used in this study. Mutants rglg1 (SALK_011892; accessible via http://signal.salk.edu/cgi-bin/tdnaexpress), rglg2 (SALK_062384), axr3-1 (N57504), axr1-12 (N3076), and tir3-1 (N3928) were obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/). The Arabidopsis plants harboring promoter:luciferase (LUC) constructs, CIRCADIAN CLOCK–ASSOCIATED1 (CCA1):LUC and COLD- AND CIRCADIAN-REGULATED2 (CCR2; also termed At GRP7):LUC, were kindly provided by F. Nagy (Biological Research Center, Szeged, Hungary). Arabidopsis plants harboring DR5:GUS and PIN1:PIN1-GFP were described previously (Ulmasov et al., 1997; Benková et al., 2003). PIN2:PIN2-GFP will be described in detail elsewhere (P. Wolff and K. Palme, unpublished data). Plants expressing IBC6:GUS were a kind gift of J. Kieber (University of North Carolina). DR5:GUS, IBC6:GUS, CCA1:LUC, CCR2:LUC, PIN1-GFP, and PIN2-GFP constructs were introgressed into the rglg1 rglg2 double mutant for analysis. Other transgenes used were constructed as detailed below and in the Supplemental Methods online. Binary plasmids were introduced into Agrobacterium tumefaciens strain C58C1 (pGV2260) and transformed by floral dip. Plasmids p3RGLG1-GFP and p3RGLG2-GFP were introduced into rglg1/RGLG1 rglg2/rglg2 plants for subsequent complementation tests, and all other constructs were transformed into Columbia. p3RGLG2-GFP and a G2A version of the plasmid were used for agroinfiltration of tobacco (Nicotiana tabacum) leaves according to Bucher et al. (2003).
Yeast Two-Hybrid Analysis
Standard methods were applied for yeast two-hybrid analysis using strain PJ69-4A (James et al., 1996). Vectors pGAD424 and pGBKT7 were from Clontech.
In Vitro Ubiquitylation Assays
Proteins for assays were expressed in Escherichia coli and purified prior to use as detailed in the Supplemental Methods online. Standard ubiquitylation assays were performed in 20 μL volume containing human His6-E1 at a concentration of 0.1 μM and human ubiquitin (either wild type or K63R mutant) at 117 μM. The other protein reactants were at 0.45 μM. The reaction buffer consisted of 1.5 mM DTT, 65 mM Tris-HCl, pH 7.6, 5 mM MgCl2, 0.75 mM EDTA, 110 mM NaCl, and 15% (w/v) glycerol. Energy was supplied by 2 mM ATP plus regenerating system (10 mM phosphocreatine, 3.5 units/mL creatine kinase, and 0.3 unit/mL inorganic pyrophosphatase). Reactions were incubated at 37°C for the indicated times (up to 8 h). Aliquots (6 μL) were withdrawn at the indicated times, and the reaction was stopped with 24 μL of β-mercaptoethanol–containing sample buffer. Samples were size-fractionated on 13.5% SDS-PAGE gels, transferred to membranes, and detected using affinity-purified anti-ubiquitin antibody (conj8-2 from rabbit) or anti-GST antibody (SC-459 from rabbit; Santa Cruz). Detection used secondary antibody (donkey anti-rabbit) linked to horseradish peroxidase (NA944V; GE Healthcare) and the ECL system (GE Healthcare).
In Vitro Myristoylation
In vitro myristoylation was performed essentially as described by Lu and Hrabak (2002), using the TNT-coupled wheat germ extract system (Promega) with protein sequences cloned into vector pBAT.
Construction of Plasmids for Plant Transformation
PCR for cloning purposes was performed using platinum Pfx DNA polymerase (Invitrogen), and constructs were verified by DNA sequencing of critical regions. To obtain the 35S:RGLG1-GFP construct, cDNA clone BX824117 (Génoscope cDNA; http://www.genoscope.cns.fr, accessed via http://signal.salk.edu/cgi-bin/tdnaexpress) was cleaved with EcoRI, treated with Klenow enzyme, and digested with PstI. The purified 1.1-kb fragment was ligated with pBluescript II SK+ (digested with SmaI and PstI) to obtain pSK-RGLG1A. The second EcoRI-PstI fragment of RGLG1 cDNA (0.4 kb long) was ligated to pBluescript II SK-RGLG1A to form pBluescript II SK-RGLG1B. After the ligation with oligonucleotides 406A (5′-TCGACATGGGAGGAGGG-3′) and 407A (5′-AATTCCCTCCTCCCATG-3′) as adaptors, the full-length RGLG1 cDNA was inserted into pBluescript II SK+ vector and named pSK-RGLG1. A fragment of RGLG1, amplified with primers 413B (5′-GAAACCGGTGCAGAGTGGATCATCA-3′) and 414B (5′-CGGGATCCCTGCAGCATATGGATTATAAGGTAAAGCTTGATTCTGGTCT-3′) and pBluescript II SK-RGLG1, was digested with AgeI and BamHI. The purified fragments were ligated to give pSK-RGLG1PNPB. An EGFP fragment was inserted into pSK-RGLG1PNPB to obtain pSK-RGLG1-GFP. Thereafter, the RGLG1-GFP fragment from pSK-RGLG1-GFP was introduced into p3, a pBI-based binary vector containing a 35S promoter with triple enhancer and a 35S terminator sequence. p3 was formed by insertion of a p2RT HindIII fragment into the unique HindIII site of p35H (Schlögelhofer and Bachmair, 2002). To make the 35S:RGLG2-GFP construct, an EcoRI-BamHI fragment from cDNA clone pda08576 (RIKEN) was ligated into pBluescript II SK vector to form pSK-RGLG2A. After the insertion of oligonucleotides 392A (5′-TCGACATGGGGACAGGG-3′) and 393A (5′-AATTCCCTGTCCCCATG-3′) as adaptors, RGLG2 full-length cDNA was cloned into pBluescript II SK+ vector and named pSK-RGLG2. A partial RGLG2 fragment was amplified with oligonucleotides 353A (5′-CCGGAATTCACAAGCTCTGCTTCTGATAACC-3′) and 417B (5′-CGGGATCCCTGCAGCATATGGATTATAAGGTAGAGCTTTATTCTTGTCT-3′). This fragment and pSK-RGLG2 were digested with BglII and BamHI and ligated together to form pSK-RGLG2PNPB. An EGFP fragment was ligated with pSK-RGLG2PNPB to form pSK-RGLG2-GFP. The RGLG2-GFP fragment from pSK-RGLG2-GFP was introduced into the p3 binary vector. A mutant version of p3-RGLG2-GFP with a G2A change was made by exchange of an NcoI-BglII fragment in pSK-RGLG2-GFP, followed by introduction of the insert into p3 as described for pSK-RGLG2-GFP. The mutated NcoI-BglII fragment was created by digestion of a PCR fragment, generated with primers 352A (5′-AGAACTGCAGTTAGTAGAGCTTTATTCTTGTCT-3′) and 843A (5′-GGATGTCGACCATGGCGACAGGGAATTCTAAAGA-3′) and RGLG2 cDNA as a template.
Confocal Laser Scanning Microscopy
Localization of fluorescent protein–tagged proteins in fresh tissue samples was performed using either Zeiss LSM 510 Meta (Carl Zeiss) or Leica SP2 AOBS (Leica). GFP was excited with the argon laser at 488 nm, and the emitted fluorescence was detected between 505 and 520 nm. Bright-field images were recorded by a transmission detector. Samples for Figure 10 were fixed with 4% formaldehyde and mounted in Prolong Gold antifade reagent containing 4′,6-diamidino-2-phenylindole (Molecular Probes) at age 7 d. Representative frames were selected and processed with Adobe Photoshop.
GUS Assays and Microscopy Analysis
GUS enzyme activity was determined by homogenization of 20 mg of tissue in 200 μL of extraction buffer (50 mM NaPO4 pH 7, 10 mM 2-mercaptoethanol, 10 mM EDTA, 0.1% SDS, and 0.1% Triton X-100). After centrifugation at 4°C, aliquots of the supernatant were used for incubation in 450 μL of assay buffer (50 mM NaPO4, pH 7, 5 mM DTT, 1 mM EDTA, and 2 mM para-nitro-phenylglucuronide [Sigma-Aldrich]). Upon incubation at 37°C, aliquots were withdrawn regularly to determine hydrolysis rates. The reaction was terminated by supplementing the aliquots with 8 volumes of 0.4 M Na2CO3. Absorbance measurement at 405 nm was used for calculations (extinction coefficient of para-nitrophenol, 18,000) (Gallagher, 1992). For GUS staining, seedlings were placed in staining solution (100 mM phosphate buffer, pH 7.0, 10 mM EDTA, 2 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 0.1% Triton X-100, 5 mM K3Fe[CN]6, and 0.5 mM K4Fe[CN]6), vacuum-infiltrated for 10 min, and incubated at 37°C overnight. After staining, the tissues were fixed overnight in ethanol:acetic acid (9:1) solution at room temperature. Subsequent incubation in 90% ethanol for 30 min was followed by 70% ethanol incubation overnight. Stained seedlings were examined. To prepare tissue for histological analysis, tissues were passed through an ethanol series and embedded in paraffin. Sections (10 μm) were then cut from the embedded tissue, the wax was removed, and the tissues were stained with 0.1% toluidine blue (Fluka).
Real-Time Quantitative RT-PCR
At least two independent RNA preparations were used to carry out triplicate reactions for each primer pair. Shoot apical region tissue (all of the leaves were removed) of 24-d-old Columbia wild-type and rglg1 rglg2 homozygous plants was used for real-time PCR analysis. cDNA synthesis was performed in a 20-μL reaction using 1.5 μg of DNase I–treated total RNA by the reverse transcription system (Invitrogen). The cDNA reaction mixture was diluted 1:10 using RNase-free water, and 2 μL was taken for real-time amplification. Amplifications were performed on 96-well plates in a 20-μL reaction volume. All reactions were performed independently three times on iCycler (Bio-Rad). For all samples, cDNA levels were normalized using an eIF4A control. Primers used are detailed in the Supplemental Methods online. Each primer pair generated a fragment of ∼150 bp.
Determination of Phytohormone Concentrations
Phytohormone concentrations were measured essentially as described by Ljung et al. (2005), Nordström et al. (2004a), and Edlund et al. (1995), as detailed in the Supplemental Methods online.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes identified and other major genes mentioned in this article are as follows: RGLG1 (At3g01650), RGLG2 (At5g14420), RGLG3 (At5g63970), RGLG4 (At1g79380), RGLG5 (At1g67800), MMZ1 (At1g23260), MMZ2 (At1g70660) MMZ3 (At2g36060), MMZ4 (At3g52560), At UBC35 (At1g78870), and At UBC36 (At1g16890).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. In Vitro Ubiquitylation Assay with Ubiquitin Variants.
Supplemental Figure 2. Promoter:GUS Fusions to Assess Expression Domains of RGLG1 and RGLG2.
Supplemental Figure 3. Characterization of T-DNA Insertions in Genes RGLG1 and RGLG2.
Supplemental Figure 4. rglg1 rglg2 Mutants Display Differences in Leaf Cell Differentiation and Have Larger Cells.
Supplemental Figure 5. The Circadian Clocks of rglg1 rglg2 Mutant and Wild-Type Plants Have Different Speeds.
Supplemental Figure 6. Alterations in Cytokinin Levels of rglg1 rglg2 Plants.
Supplemental Figure 7. rglg1 rglg2 Mutant Plants Differ from Wild-Type Plants in Cytokinin Content.
Supplemental Figure 8. PIN1 Interacts with RGLG2 in the Yeast Two-Hybrid Assay.
Supplemental Methods.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Cecile Pickart (1954–2006). We thank Joe Kieber, Peter Schlögelhofer, Susanne Stary, Guido Jach, and Ferenc Nagy for clones and plant lines; Rainer Franzen, Rolf-Dieter Hirtz, Coral Freialdenhoven, Kerstin Luxa, and Michaela Lehnen for technical support; Maret Kalda for photography; Hans Sommer for the yeast two-hybrid library; the consortium of RIKEN, Génoscope, and SALK for cDNAs and plant lines; and Robert Cohen, Csaba Koncz, and George Coupland for comments on the manuscript. S.V. was supported by a training grant from the National Institute of Environmental Health Sciences (Grant T32 ES-07141), and M.T. was supported by the Austrian Science Foundation FWF (Grant P19825-B12). Work in A.B.'s laboratory was supported by the Max Planck Society and by the Deutsche Forschungsgemeinschaft (Grant Ba 1158/3-1).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Andreas Bachmair (bachmair@mpiz-koeln.mpg.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bachmair, A., Novatchkova, M., Potuschak, T., and Eisenhaber, F. (2001). Ubiquitylation in plants: A post-genomic look at a post-translational modification. Trends Plant Sci. 6 463–470. [DOI] [PubMed] [Google Scholar]
- Barton, S.J., Travis, M.A., Askari, J.A., Buckley, P.A., Craig, S.E., Humphries, M.J., and Mould, A.P. (2004). Novel activating and inactivating mutations in the integrin β1 subunit A domain. Biochem. J. 380 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Blakeslee, J.J., Peer, W.A., and Murphy, A.S. (2005). Auxin transport. Curr. Opin. Plant Biol. 8 494–500. [DOI] [PubMed] [Google Scholar]
- Bucher, E., Sijen, T., de Haan, P., Goldbach, R., and Prins, M. (2003). Negative-strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genome position. J. Virol. 77 1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.J. (2005). Ubiquitin signaling in the NF-κB pathway. Nat. Cell Biol. 7 758–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague, M.J., and Urbé, S. (2006). Endocytosis: The DUB version. Trends Cell Biol. 16 551–559. [DOI] [PubMed] [Google Scholar]
- d'Azzo, A., Bongiovanni, A., and Nastasi, T. (2005). E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6 429–441. [DOI] [PubMed] [Google Scholar]
- Desgagné-Penix, I., Eakanunkul, S., Coles, J.P., Phillips, A.L., Hedden, P., and Sponsel, V.M. (2005). The auxin transport inhibitor response 3 (tir3) allele of BIG and auxin transport inhibitors affect the gibberellin status of Arabidopsis. Plant J. 41 231–242. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., and Estelle, M. (2004). Auxin signaling and regulated protein degradation. Trends Plant Sci. 9 302–308. [DOI] [PubMed] [Google Scholar]
- Dreher, K., and Callis, J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun, E.A., Ferguson, B.J., and Beveridge, C.A. (2006). Apical dominance and shoot branching. Divergent opinions or divergent mechanisms? Plant Physiol. 142 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré, S., Urban-Grimal, D., and Haguenauer-Tsapis, R. (2004). Ubiquitin and endocytic internalization in yeast and animal cells. Biochim. Biophys. Acta 1895 89–111. [DOI] [PubMed] [Google Scholar]
- Edlund, A., Eklöf, S., Sundberg, B., Moritz, T., and Sandberg, G. (1995). A microscale technique for gas chromatography-mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol. 108 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, A.J. (2006). Plant signalling: The inexorable rise of auxin. Trends Cell Biol. 16 397–402. [DOI] [PubMed] [Google Scholar]
- Fu, X., and Harberd, N.P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 740–743. [DOI] [PubMed] [Google Scholar]
- Fukuda, H. (2004). Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 5 379–391. [DOI] [PubMed] [Google Scholar]
- Gallagher, S.R. (1992). GUS Protocols (San Diego, CA: Academic Press).
- Gil, P., Dewey, E., Friml, J., Zhao, Y., Snowden, K.C., Putterill, J., Palme, K., Estelle, M., and Chory, J. (2001). BIG: A calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 15 1985–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano, S., Domagalska, M., Nagy, F., and Davis, S.J. (2006). Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 11 1381–1392. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (2006). Lingering mysteries of ubiquitin-chain assembly. Cell 124 27–34. [DOI] [PubMed] [Google Scholar]
- Hoege, C., Pfander, B., Moldovan, G.L., Pyrowolakis, G., and Jentsch, S. (2002). RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419 135–141. [DOI] [PubMed] [Google Scholar]
- Hofmann, R.M., and Pickart, C.M. (1999). Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96 645–653. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson, H., Heisler, M., Shapiro, B.E., Meyerowitz, E.M., and Mjolsness, E. (2006). An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA 103 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanyuka, K., Praekelt, U., Franklin, K.A., Billingham, O.E., Hooley, R., Whitelam, G.C., and Halliday, K.J. (2003). Mutations in the huge Arabidopsis gene BIG affect a range of hormone and light responses. Plant J. 35 57–70. [DOI] [PubMed] [Google Scholar]
- Kasajima, I., Ohkama-Ohtsu, N., Ide, Y., Hayashi, H., Yoneyama, T., Suzuki, Y., Naito, S., and Fujiwara, T. (2007). The BIG gene is involved in regulation of sulfur deficiency-response genes in Arabidopsis thaliana. Physiol. Plant. 129 351–363. [Google Scholar]
- Kempinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. [DOI] [PubMed] [Google Scholar]
- Kondo, T., Sawa, S., Kinoshita, A., Mizuno, S., Kakimoto, T., Fukuda, H., and Sakagami, Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313 845–848. [DOI] [PubMed] [Google Scholar]
- Kosarev, P., Mayer, K.F.X., and Hardtke, C.S. (2002). Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 3: RESEARCH0016.1–RESEARCH0016.12. [DOI] [PMC free article] [PubMed]
- Kraft, E., Stone, S.L., Ma, L., Su, N., Gao, Y., Lau, O.-S., Deng, X.-W., and Callis, J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139 1597–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, E.M., and Bennett, M.J. (2006). Auxin transport: A field in flux. Trends Plant Sci. 11 382–386. [DOI] [PubMed] [Google Scholar]
- Krappmann, D., and Scheidereit, C. (2005). A pervasive role of ubiquitin conjugation in activation and termination of IκB kinase pathways. EMBO Rep. 6 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamothe, B., Besse, A., Campos, A.D., Webster, W.K., Wu, H., and Darnay, B.G. (2007). Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of IκB kinase activation. J. Biol. Chem. 282 4102–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, E., Achard, P., Vansiri, A., Potuschak, T., and Genschik, P. (2006). F-box proteins everywhere. Curr. Opin. Plant Biol. 9 631–638. [DOI] [PubMed] [Google Scholar]
- Leibfried, A., To, J.P.C., Busch, W., Stehling, S., Kehle, A., Demar, M., Kieber, J.J., and Lohmann, J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438 1172–1175. [DOI] [PubMed] [Google Scholar]
- Leyser, O. (2005). The fall and rise of apical dominance. Curr. Opin. Genet. Dev. 15 468–471. [DOI] [PubMed] [Google Scholar]
- Leyser, O. (2006). Dynamic integration of auxin transport and signalling. Curr. Biol. 16 R424–R433. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., Lincoln, C.A., Timpte, C., Lammer, D., Turner, J., and Estelle, M. (1993). Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364 161–164. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10 403–413. [DOI] [PubMed] [Google Scholar]
- Ljung, K., Hull, A.K., Celenza, J., Yamada, M., Estelle, M., Normanly, J., and Sandberg, G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio, J., Hernández-Abreu, E., Sánchez-Calderón, L., Pérez-Torres, A., Rampey, R.A., Bartel, B., and Herrera-Estrella, L. (2005). An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol. 137 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S.X., and Hrabak, E.M. (2002). An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol. 128 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, T. (2004). Plant response regulators implicated in signal transduction and circadian rhythm. Curr. Opin. Plant Biol. 7 499–505. [DOI] [PubMed] [Google Scholar]
- Mould, A.P., Barton, S.J., Askari, J.A., McEwan, P.A., Buckley, P.A., Craig, S.E., and Humphries, M.J. (2003). Conformational changes in the integrin βA domain provide a mechanism for signal transduction via hybrid domain movement. J. Biol. Chem. 278 17028–17035. [DOI] [PubMed] [Google Scholar]
- Nordström, A., Tarkowski, P., Tarkowska, D., Dolezal, K., Åstot, C., Sandberg, G., and Moritz, M. (2004. a). Derivatization for LC-electrospray ionisation-MS: A tool for improving reversed-phase separation and ESI responses of bases, ribosides, and intact nucleotides. Anal. Chem. 76 2869–2877. [DOI] [PubMed] [Google Scholar]
- Nordström, A., Tarkowski, P., Tarkowska, D., Norbaek, R., Åstot, C., Dolezal, K., and Sandberg, G. (2004. b). Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 101 8039–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov, I.A., Teale, W.D., Trebar, M., Blilou, I., and Palme, K. (2005). The PIN auxin efflux facilitators: Evolutionary and functional perspectives. Trends Plant Sci. 10 170–177. [DOI] [PubMed] [Google Scholar]
- Parry, D., and Estelle, M. (2006). Auxin receptors: A new role for F-box proteins. Curr. Opin. Cell Biol. 18 152–156. [DOI] [PubMed] [Google Scholar]
- Peng, J., Schwartz, D., Elias, J.E., Thoreen, C.C., Cheng, D., Marsischky, G., Roelofs, J., Finley, D., and Gygi, S.P. (2003). A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21 921–926. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70 503–533. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M., and Fushman, D. (2004). Polyubiquitin chains: Polymeric protein signals. Curr. Opin. Chem. Biol. 8 610–616. [DOI] [PubMed] [Google Scholar]
- Quint, M., and Gray, W.M. (2006). Auxin signaling. Curr. Opin. Plant Biol. 9 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte, A.M., Chae, H.S., Maxwell, B.B., and Kieber, J.J. (2005). The interaction of cytokinin with other signals. Physiol. Plant. 123 184–194. [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279 1371–1373. [DOI] [PubMed] [Google Scholar]
- Sauer, M., Balla, J., Luschnig, C., Wisniewska, J., Reinöhl, V., Friml, J., and Benková, E. (2006). Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20 2902–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlögelhofer, P., and Bachmair, A. (2002). A test of fusion protein stability in the plant Arabidopsis thaliana reveals degradation signals from ACC synthase and from the plant N-end rule pathway. Plant Cell Rep. 21 174–179. [Google Scholar]
- Schwechheimer, K., and Schwager, K. (2004). Regulated proteolysis and plant development. Plant Cell Rep. 23 353–364. [DOI] [PubMed] [Google Scholar]
- Smalle, J., and Vierstra, R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Smith, R.S., Guyonmac'h, S., Mandel, T., Reinhardt, D., Kuhlemeier, C., and Prusinkiewicz, P. (2006). A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA 103 1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., Hauksdóttir, H., Troy, A., Herschleb, J., Kraft, E., and Callis, J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, X., Calderon-Villalobos, L.I.A., Sharon, M., Zheng, C., Robinson, C.V., Extelle, M., and Zheng, N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645. [DOI] [PubMed] [Google Scholar]
- Tanaka, M., Takei, K., Kojima, M., Sakakibara, H., and Mori, H. (2006). Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 45 1028–1036. [DOI] [PubMed] [Google Scholar]
- Teale, W.D., Paponov, I.A., and Palme, K. (2006). Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7 847–859. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten, A., Vanneste, S., Wisniewska, J., Benková, E., Benjamins, R., Beeckman, T., Luschnig, C., and Friml, J. (2005). Functional redundancy of PIN proteins is accomplished by auxin-dependent cross-regulation of PIN expression. Development 132 4521–4530. [DOI] [PubMed] [Google Scholar]
- Wen, R., Newton, L., Li, G., Wang, H., and Xiao, W. (2006). Arabidopsis thaliana UBC13: Implication of error-free DNA damage tolerance and Lys63-linked polyubiquitylation in plants. Plant Mol. Biol. 61 241–253. [DOI] [PubMed] [Google Scholar]
- Williams, L., and Fletcher, J.C. (2005). Stem cell regulation in the Arabidopsis shoot apical meristem. Curr. Opin. Plant Biol. 8 582–586. [DOI] [PubMed] [Google Scholar]
- Woodward, A.W., and Bartel, B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.