Abstract
High-resolution, three-dimensional structures of the archetypal glycoside hydrolase family 16 (GH16) endo-xyloglucanases Tm-NXG1 and Tm-NXG2 from nasturtium (Tropaeolum majus) have been solved by x-ray crystallography. Key structural features that modulate the relative rates of substrate hydrolysis to transglycosylation in the GH16 xyloglucan-active enzymes were identified by structure–function studies of the recombinantly expressed enzymes in comparison with data for the strict xyloglucan endo-transglycosylase Ptt-XET16-34 from hybrid aspen (Populus tremula × Populus tremuloides). Production of the loop deletion variant Tm-NXG1-ΔYNIIG yielded an enzyme that was structurally similar to Ptt-XET16-34 and had a greatly increased transglycosylation:hydrolysis ratio. Comprehensive bioinformatic analyses of XTH gene products, together with detailed kinetic data, strongly suggest that xyloglucanase activity has evolved as a gain of function in an ancestral GH16 XET to meet specific biological requirements during seed germination, fruit ripening, and rapid wall expansion.
INTRODUCTION
The xyloglucan (XG) family of polysaccharides continues to receive considerable attention due to the key structural role these matrix polymers play in the cell walls of higher plants. Xyloglucans are major components of the primary wall of all dicots and some monocots (Carpita and McCann, 2000), where they bridge paracrystalline cellulose microfibrils by surface adsorption and chain intercalation (Pauly et al., 1999a), thereby modulating wall mechanical properties and affecting morphology (Rose and Bennett, 1999; Chanliaud et al., 2004; Cosgrove, 2005; Thompson, 2005; Brummell, 2006, and references therein). Indeed, based on the broad distribution of xyloglucans in vascular plants, it has been suggested that the evolution of xyloglucan as a cellulose binding polysaccharide may have conferred a particular advantage in the colonization of drier habitats (Popper and Fry, 2003, 2004). In addition to performing structural roles, xyloglucans are also found as the principal seed-storage polysaccharides in a number of land plants (Kooiman, 1960; Rao and Srivastava, 1973; Edwards et al., 1985; Wang et al., 1996; Buckeridge et al., 2000; Ren et al., 2004).
The primary structures of xyloglucans possess a common β(1→4)-d-glucan backbone that is regularly substituted with α(1→6)-d-xylopyranosyl residues. Two predominant xylosylation patterns have been observed in primary wall and storage xyloglucans (Vincken et al., 1997), both based on Glc4 backbone repeating structures: XXXG and XXGG (where X is [α-d-Xylp(1→6)]-Glcpβ(1→4) and G is Glcpβ(1→4), according to the nomenclature of Fry et al. [1993]). More rarely, Glc5- and Glc6-based backbone repeats have also been observed (Sims et al., 1996; Buckeridge et al., 1997; Tine et al., 2006). These basic motifs are further decorated with combinations of galactopyranose, fucopyranose, arabinofuranose, and O-acetyl residues, dependent on the tissue and species of origin, to produce a diversity of xyloglucan structures (see Hoffman et al., 2005, for a recent overview of xyloglucan phylogeny). The archetypal seed xyloglucan from Tamarindus indica is composed of XXXG, XXLG, XLXG, and XLLG oligosaccharides (where L is [β-d-Galp-(1→2)-α-d-Xylp(1→6)]-Glcpβ(1→4)) in the molar ratio 1.4:3:1:5.4 (York et al., 1990), while the majority of primary wall xyloglucans also contain XXFG (where F is L bearing an α-l-Fucp(1→2) substituent) (Carpita and McCann, 2000; Hoffman et al., 2005).
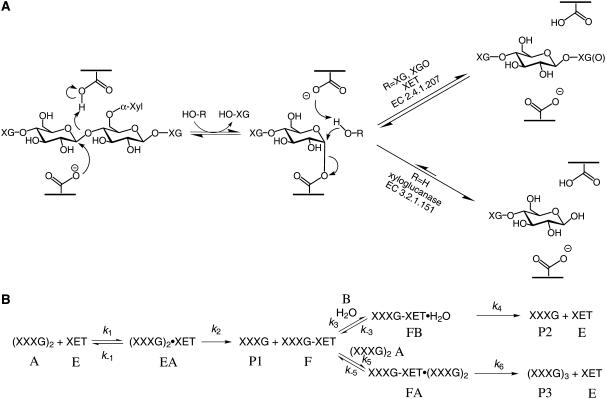
Considerable interest in the enzymes responsible for the metabolism of xyloglucans is sustained as a result of the physiological importance these polysaccharides have in cell wall morphogenesis. The nucleotide diphospho-sugar glycosyl transferases responsible for the anabolism of xyloglucan are beginning to be revealed (Reiter, 2002; Lerouxel et al., 2006), while recent three-dimensional structural analysis is highlighting the molecular details of xyloglucan depolymerization by microbial enzymes in glycoside hydrolase families (Henrissat and Davies, 1997) GH5 (Gloster et al., 2007), GH12 (Pauly et al., 1999b; Gloster et al., 2007), and GH74 (Yaoi et al., 2004; Martinez-Fleites et al., 2006). In plants, specific endolytic activity toward xyloglucan has only been demonstrated for members of glycoside hydrolase family GH16, encoded by the XTH (for xyloglucan endo-transglycosylase/hydrolase) gene subfamily (Rose et al., 2002). Like other GH16 enzymes, which include a range of endo-glucan and galactanases (Michel et al., 2001), the gene products of the XTH subfamily utilize a double displacement/retaining mechanism of glycosyl transfer (Figure 1).
Figure 1.
The Canonical Retaining Mechanism of Glycosyl Transfer Showing Product Partitioning between Hydrolysis and Transglycosylation.
(A) The chemical mechanism.
(B) Full kinetic scheme for the minimal xylogluco-oligosaccharide donor substrate XXXGXXXG under steady state conditions in the absence of products. Substrates (A = XXXGXXXG, B = H2O), enzyme intermediates (E, EA, F, FA, and FB), and products (P1 to P3) are denoted with uppercase letters, and individual rate constants (kn) are shown.
Salient features of the canonical mechanism (Sinnott, 1990) include two catalytic carboxylic amino acid side chains, one of which functions as a nucleophile attacking the C-1 (anomeric) carbon of an unsubstituted backbone glucose (G in the nomenclature of Fry et al. [1993]) to yield a covalent glycosyl enzyme intermediate (Vocadlo et al., 2001). Chain scission is promoted by proton donation from the protonated form of the second catalytic carboxylate to the departing oxygen atom. Once formed, the glycosyl enzyme can partition between two mechanistic pathways: it can be hydrolyzed (glycosyl transfer to water) to yield cleavage products, or it can be intercepted by an oligosaccharide or polysaccharide acceptor substrate to yield a transglycosylation product (glycosyl transfer to carbohydrate) (Cote and Tao, 1990; Sinnott, 1990; York and Hawkins, 2000). In either case, breakdown of the glycosyl enzyme is facilitated by the second catalytic carboxylate, which activates the glycosyl acceptor substrate through general base catalysis.
The majority of the XTH gene products for which full kinetic data exist have been shown to have very little or no detectable hydrolytic activity (see Supplemental Table 1 online) and are thus predominant or strict xyloglucan endo-transglycosylases (XET; EC 2.4.1.207) (Fry et al., 1992; Nishitani and Tominaga, 1992). Ptt-XET16-34 (previously Ptt-XET16A) from the hybrid aspen Populus tremula × Populus tremuloides is one such example that has undetectable xyloglucanase activity (EC 3.2.1.151; alt. xyloglucan endo-β-1,4-glucanase [XEG] or xyloglucan endo-hydrolase [XEH]) (Kallas et al., 2005). The three-dimensional structure of this enzyme has been solved (Johansson et al., 2004), and the catalytic mechanism has been probed using a range of synthetic xylogluco-oligosaccharides to elucidate details of enzyme–substrate interactions (Fauré et al., 2006). Intriguingly, xyloglucanase activity and gene sequence have been conclusively linked for only one member of GH16, Tm-NXG1 from Tropaeolum majus (nasturtium), which is a predominant endo-xyloglucanase that can also perform xyloglucan endo-transglycosylation at elevated concentrations of acceptor substrates (Edwards et al., 1986; de Silva et al., 1993; Fanutti et al., 1993, 1996; Chanliaud et al., 2004).
As a next step toward deciphering the determinants of xyloglucan transglycosylation versus hydrolysis in the plant XTH gene subfamily of GH16, we undertook a detailed structure–function study of Tm-NXG1 and the homologous Tm-NXG2. We report here the three-dimensional structures of Tm-NXG1 and Tm-NXG2 as examples of plant GH16 xyloglucanases and highlight key active site features that differ from the previously determined XET structure (Johansson et al., 2004). Detailed bioinformatic analysis was used to guide the construction of a hybrid enzyme that demonstrated intermediate kinetic activity, which effectively indicated that XET activity could be increased at the expense of xyloglucanase activity. These results are discussed in the context of the molecular evolution of GH16 xyloglucan-active enzymes to meet specific biological requirements for cell wall remodeling and storage polysaccharide mobilization.
RESULTS
Cloning of Tm-NXG2 cDNA and Mutagenesis to Produce Tm-NXG1 and the Deletion Mutant Tm-NXG1-ΔYNIIG
Tm-NXG2 (GenBank accession number X68255) represents a C-terminal sequence fragment that displays eight nucleotide polymorphisms relative to the full-length Tm-NXG1 sequence (GenBank accession number X68254), only one of which gives rise to variation in the protein primary structure: Met-129 in Tm-NXG1 is replaced by Leu in Tm-NXG2. In this study, a unique full-length cDNA was cloned from a pool obtained from the epicotyl of nasturtium seedlings that was 99.3% identical to Tm-NXG1 but that encoded the Tm-NXG2–like Leu-129 variant. To study both proteins, site-directed mutagenesis was used to introduce a L129M point mutation to produce a cDNA encoding Tm-NXG1 from the novel Tm-NXG2 sequence. The historical abbreviations NXG1 and NXG2 are retained throughout for direct comparison with the extensive literature record of these enzymes (Edwards et al., 1986; de Silva et al., 1993; Fanutti et al., 1993, 1996; Chanliaud et al., 2004). NXG1 has also been referred to as nXGase (for nasturtium xyloglucanase) (Chanliaud et al., 2004).
Alignment of ∼130 full-length putative XTH gene sequences clearly indicated that the archetypal GH16 xyloglucanase, Tm-NXG1, contained unique loop extensions adjacent to the active site that are not present in the strict XETs from cauliflower (Brassica oleracea var botrytis) and hybrid aspen (Populus tremula × Populus tremuloides) (see Supplemental Figures 4 and 5 online). Neither Bob-XET16A nor Ptt-XET16-34 have detectable rates of xyloglucan hydrolysis using a sensitive reducing sugar assay (Henriksson et al., 2003; Kallas et al., 2005), whereas Tm-NXG1 has been demonstrated to be a predominant endo-hydrolase (EC 3.2.1.151) (Chanliaud et al., 2004) that nonetheless has some capacity to catalyze xyloglucan endo-transglycosylation (EC 2.4.1.207) (Fanutti et al., 1996). Therefore, deletion mutants of Tm-NXG1 were produced to test the hypothesis that the loop between β-strands β6 and β7, as well as the loop between β8 and β9, might be primary determinants affecting the hydrolysis-to-transglycosylation ratio in this enzyme family. Although recombinant expression of constructs containing deletion variants of the loop sequence joining β6 and β7 has been unsuccessful to date, the β8-β9 loop variant Tm-NXG1-ΔYNIIG was produced at levels similar to those of Tm-NXG1 and Tm-NXG2 (50 mg/L crude protein) in the methylotrophic yeast Pichia pastoris.
Catalytic Properties of the Recombinant Tm-NXG1, Tm-NXG2, and Tm-NXG1-ΔYNIIG
To resolve any potential ambiguity resulting from previous characterization of native purified enzymes (Edwards et al., 1986; Fanutti et al., 1993) in the context of the single amino acid polymorphism observed between Tm-NXG1 and Tm-NXG2 (de Silva et al., 1993), a detailed analysis of the activity of these enzymes was performed. These data, in turn, provided a basis for an analysis of the effects of loop truncation on the catalytic function of Tm-NXG1-ΔYNIIG.
Substrate Specificity
The activities of recombinantly produced and purified Tm-NXG1 and Tm-NXG2 were tested with a range of substituted and nonsubstituted β(1-3)- and β(1-4)-linked polysaccharides. Consistent with data reported >20 years ago (Edwards et al., 1986), these enzymes were specific xyloglucanases that exhibited no detectable activity on konjac glucomannan, Icelandic moss lichenan, barley (Hordeum vulgare) β-glucan, Avicel, and birchwood (Betula spp) xylan. A trace amount of activity was observed with hydroxyethyl cellulose, as was shown previously for an extracted and purified enzyme preparation (Edwards et al., 1986). The pH rate profile for the hydrolysis of xyloglucan by Tm-NXG1 was classically bell-shaped, with apparent kinetic pKa values of 4.3 ± 0.1 and 5.3 ± 0.1 (pH optimum, 4.8; see Supplemental Figure 1 online), similar to previous observations (Edwards et al., 1986). Tm-NXG2 exhibited an essentially identical pH rate profile (data not shown). Analysis of the recombinantly produced and purified enzymes yielded specific hydrolytic activities toward xyloglucan of 5.5 ± 0.5 mol XGO·min−1·mol−1 protein for Tm-NXG1 and 4.0 ± 0.3 mol XGO·min−1·mol−1 protein for Tm-NXG2 at the pH optimum (1 g/L xyloglucan). The hydrolytic activity of the loop deletion mutant Tm-NXG1-ΔYNIIG was significantly lower: 0.23 ± 0.058 mol XGO·min−1·mol−1 protein. Neither Tm-NXG1 nor Tm-NXG2 showed detectable transfer of xyloglucan to radiolabeled xylogluco-oligosaccharides under standard assay conditions (Fry et al., 1992; Henriksson et al., 2003; Kallas et al., 2005). Further comparative kinetic analyses of the three enzymes were performed using size-exclusion chromatography (SEC) and high-performance anion-exchange chromatography–pulsed-amperometric detection (HPAEC-PAD) assays.
Mode of Action
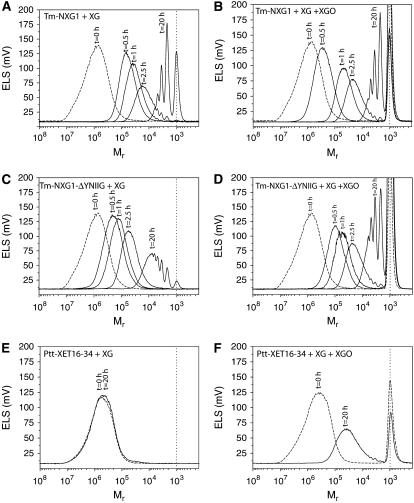
The depolymerization of xyloglucan by Tm-NXG1 was followed by SEC to confirm that this enzyme cleaved xyloglucan endolytically and to examine the effect of the addition of Glc4-based xylogluco-oligosaccharides (XGOGlc4) on the reaction. The Mr of XG rapidly decreased, with no initial production of xylogluco-oligosaccharides, upon incubation with Tm-NXG1 (Figure 2A), clearly indicating endo-xyloglucanase activity (EC 3.2.1.151). The addition of XGOGlc4 resulted in a reduction of depolymerization velocity (Figure 2B), indicating product inhibition of Tm-NXG1 and limited XET activity (EC 2.4.1.207). For comparison, XG was incubated with a true XET, Ptt-XET16-34 from the hybrid aspen, which has undetectable hydrolytic activity (Kallas et al., 2005; this work). In the absence of XGOGlc4, no change in the Mr distribution of XG was observed after overnight incubation (Figure 2E). Upon addition of XGOGlc4, an effective depolymerization of XG can be observed due to Ptt-XET16-34 activity (Figure 2F). SEC analysis indicated that Tm-NXG1-ΔYNIIG exhibited a qualitatively slower rate of XG hydrolysis than Tm-NXG1; after overnight incubation, a large amount of XG was not broken down to xylogluco-oligosaccharides (Figure 2C). However, addition of XGOGlc4 increased the effective rate of XG depolymerization by Tm-NXG1-ΔYNIIG, thus demonstrating improved XET activity relative to Tm-NXG1 (Figure 2D).
Figure 2.
Size-Exclusion Chromatography of the Products of Tm-NXG1, Tm-NXG1-ΔYNIIG, and Ptt-XET16-34 Operating on Xyloglucan Polysaccharide (Initial Mr 105 to 107) in the Absence and Presence of Xyloglucan Oligosaccharides (Mr 103).
Incubation times are indicated above each chromatogram. The vertical dotted line in all panels indicates the elution of Glc4-based XGOs. Protein concentrations were as follows: Tm-NXG1 (9 mg/L), Tm-NXG1-ΔYNIIG (10 mg/L), and Ptt-XET16-34 (10 mg/L). Other experimental conditions are described in Methods.
Action on a Minimal Donor Substrate, XGOGlc8
Current assays for xyloglucan-active enzymes visualize either XET activity (e.g., radiometric [Fry et al., 1992], dot-blot [Fry, 1997], and capillary electrophoresis [Saura-Valls et al., 2006] assays) or hydrolytic activity (e.g., reducing sugar quantitation and viscometry [Edwards et al., 1986]), but not both simultaneously. Consequently, an HPAEC-PAD–based assay was used to characterize Ptt-XET16-34 and the Tm-NXG variants, which allowed accurate quantitation of both XET and xyloglucanase activity under initial rate conditions. This assay utilizes a well-defined mixture of xylogluco-oligosaccharide donor substrates based on Glc8 backbones (XGOGlc8) that is readily obtained from endolysis of tamarind xyloglucan. For routine analysis, the XGOGlc8 mixture provides a convenient alternative to monodisperse donor substrates, including XXXGXXXG (Fanutti et al., 1996; Fauré et al., 2006), which are more difficult to obtain. Analysis of the rate of release of XGOGlc12 yields the transglycosylation (XET; EC 2.4.1.207) rate directly, while the hydrolytic (endo-xyloglucanase; EC 3.2.1.151) rate is obtained from calculations of the stoichiometry of XGOGlc4 production due to both transglycosylation and hydrolysis. Thus, under initial rate conditions in the absence of products, a strict XET will produce equimolar amounts of XGOGlc12 and XGOGlc4 from XGOGlc8, whereas a strict endo-xyloglucanase will generate two molar equivalents of XGOGlc4. The assay is generally applicable to all XETs and endo-xyloglucanases: XETs, in particular, cleave the xyloglucan backbone randomly (Steele et al., 2001), do not recognize the reducing ends of substrates (Lorences and Fry, 1993; Fanutti et al., 1996; Johansson et al., 2004), and have active sites that contain three positive and four negative backbone binding subsites (Johansson et al., 2004; this work).
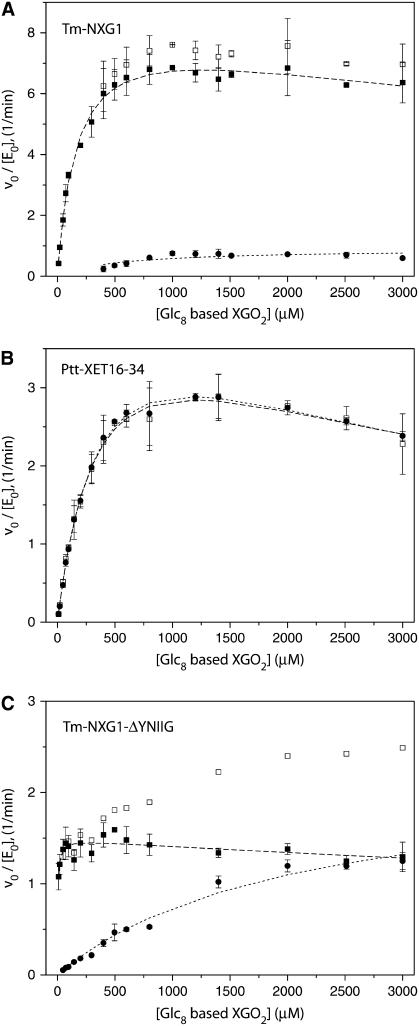
Initial rate kinetic analysis indicates that Tm-NXG1 is a predominant xyloglucan hydrolase with some capacity to catalyze xyloglucan endo-transglycosylation at increased substrate concentration. A small apparent transglycosylation rate leading to the production of XGOGlc12 and XGOGlc4 was observed at XGOGlc8 concentrations > 0.4 mM, which reached a maximum at 1 mM, as did the hydrolytic rate (Figure 3A). Notably, at these higher substrate concentrations, the observed ratio of transglycosylation to hydrolysis is nearly constant at 1:12. In stark contrast, Ptt-XET16-34 was shown to be an overwhelmingly predominant transglycosylase over the full concentration range from 10 μM to 3 mM XGOGlc8, in keeping with previous analyses (Kallas et al., 2005); under initial rate conditions, the stoichiometry of XGOGlc12 to XGOGlc4 production was 1:1 (Figure 3B). Remarkably, the deletion mutant Tm-NXG1-ΔYNIIG, which was predicted to produce a more XET-like enzyme relative to the wild type, exhibited increased transglycosylation ability and decreased hydrolytic ability, which become equivalent at high substrate concentrations (Figure 3C).
Figure 3.
Initial Rate Kinetics as a Function of XGOGlc8 Concentration.
Kinetics are shown for Tm-NXG1 (A), Tm-NXG1-ΔYNIIG (B), and Ptt-XET16-34 (C). Closed circles, rate of XGOGlc12 production due to transglycosylation (2 XGOGlc8 → XGOGlc12 + XGOGlc4); open squares, total rate of XGOGlc4 production; closed squares, corrected rate of XGOGlc4 production obtained by subtracting the contribution of XGOGlc4 release due to substrate transglycosylation. This observed rate is twice the actual catalytic rate, according to the stoichiometry of the hydrolysis reaction (XGOGlc8 → 2 XGOGlc4). Error bars indicate sd for triplicate (A), duplicate (B), and duplicate (C) measurements.
In most cases, the initial rates of XGOGlc8 hydrolysis and transglycosylation appear to essentially follow Michaelis-Menten–like kinetics, possibly involving varying degrees of substrate inhibition at high [XGOGlc8]. Ptt-XET16-34–catalyzed transglycosylation was previously shown to follow a ping-pong bi-bi mechanism with substrate inhibition by XXXG-based acceptors (Saura-Valls et al., 2006). By analogy with this and other transglycosidases and hydrolases that operate through the canonical retaining mechanism of glycosyl transfer (Sinnott, 1990), all GH16 xyloglucan-active enzymes can be expected to follow a similar reaction scheme, with modifications to include partitioning of the substrate between hydrolysis and transglycosylation (Figure 1).
The steady state kinetic equations describing the initial rate of formation of products from the two possible reaction pathways have been solved (see Supplemental Text online, Equations A and B); both equations are complex, containing four macroscopic rate constants and only a single independent variable, [XGOGlc8]. As a consequence, the equations are underdetermined, and meaningful fits to the data could not be obtained. Instead, the hydrolytic and transglycosylation rate data shown in Figure 3 were fit by the classical Michaelis-Menten expression, with or without a substrate inhibition parameter as appropriate (Brumer et al., 1999), to produce the apparent macroscopic kinetic constants shown in Table 1. Using this simplified treatment, the derived constants provide only a qualitative measure of the relative rates of enzymic transglycosylation and hydrolysis.
Table 1.
Apparent Kinetic Constants of Tm-NXG1, Ptt-XET16-34, and Tm-NXG1-ΔYNIIG on XGOGlc8 Obtained by Nonlinear Least-Squares Curve Fitting
| Hydrolysis
|
Transglycosylation
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Endo-Xyloglucanase | Km app(mM) | kcat app (min−1) | Kis app (mM) | (kcat/Km)app (mM−1 min−1) | Km app (mM) | kcat app (min−1) | Kis app (mM) | (kcat/Km)app (mM−1 min−1) |
| Tm-NXG1 | 0.08 ± 0.007 | 4.28 ± 0.14 | 9.4 ± 2.0 | 51 | 0.5 ± 0.25 | 0.89 ± 0.1 | — | 1.7 |
| Ptt-XET16-34 | — | — | — | — | 0.4 ± 0.01 | 4.80 ± 0.2 | 3.36 ± 0.35 | 12 |
| Tm-NXG1-ΔYNIIG | <0.01 | 0.74 ± 0.02 | 18.7 ± 7.5 | >74 | 2.0 ± 0.36 | 2.21 ± 0.2 | — | 1.1 |
Errors are sd values returned from the curve-fitting algorithm.
Although the initial rate equations for both transglycosylation to hydrolysis under steady state conditions have complex definitions, their ratio can be readily simplified to yield Equation 1 in terms of the microscopic rate constants shown in Figure 1B (see Supplemental Text online):
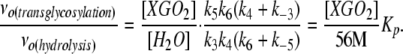 |
(1) |
Therefore, a partitioning constant, KP, can be defined as a concentration-independent parameter that reflects the ratio of the initial transglycosylation and hydrolysis rates [νo(transglycosylation)/νo(hydrolysis)]. KP is infinite for true XETs such as Ptt-XET16-34, implying that either k3 or k4 is effectively zero; water either does not bind productively or the complex is catalytically incompetent. By contrast, Tm-NXG1 exhibits a KP value of 11,500 ± 900, while the loop variant Tm-NXG1-ΔYNIIG has a KP value of 434,000 ± 2000 (see Supplemental Figures 2 and online). These values clearly highlight the significantly increased rate of transglycosylation to hydrolysis of Tm-NXG1-ΔYNIIG, which results from truncation of the loop connecting β-strands 8 and 9. Nevertheless, this effect is not simply due to a reduction of the hydrolytic activity; Tm-NXG1-ΔYNIIG shows an apparent 5.7-fold reduction of the hydrolytic rate at saturation compared with Tm-NXG1, as well as a doubling of the transglycosylation rate to one-half that observed for Ptt-XET16-34 (cf. data at [XGOGlc8] > 1 mM or apparent kcat(transglycosylation) values; Table 1).
Transfer to Cello-Oligosaccharides
Crude nasturtium XET enzyme extracts were recently demonstrated to use cellulose derivatives as alternative donor and acceptor substrates, together with xyloglucan oligosaccharides and polysaccharides (Mohand and Farkas, 2006). This study has confirmed that Tm-NXG1 possesses limited activity toward hydroxyethyl cellulose (Edwards et al., 1986), while Ptt-XET16-34 has been shown to use GGGGXXXG as a glycosyl donor with a fluorescent XXXG-based acceptor (Fauré et al., 2006). The ability to catalyze hetero-transglycosylation reactions, therefore, may be a feature of some GH16 XETs and xyloglucanases. To test the catalytic promiscuity of Tm-NXG1 and Tm-NXG1-ΔYNIIG, these enzymes were incubated in the presence of xyloglucan polysaccharide and a series of cello-oligosaccharide acceptors. The masses of the transfer products for cellobiose (XXXGGG, XLXGGG or XXLGGG, XLLGGG), cellotriose (XXXGGGG, XLXGGGG or XXLGGGG, XLLGGGG), and cellotetrose (XXXGGGGG, XLXGGGGG or XXLGGGGG, XLLGGGGG) were readily observed by electrospray ionization–time of flight–mass spectrometry (ESI-TOF MS; see Supplemental Table 3 online). Higher order xylogluco-oligosaccharide transglycosylation products (based on XGOGlc8) containing cello-oligosaccharides were also detected (data not shown). No transglycosylation products were detected when lactose (8 mM) was used as an acceptor substrate, thus indicating that glycosyl transfer most likely involved the equitorial C-4 hydroxy group on the nonreducing end of the cello-oligosaccharides. In comparison, Ptt-XET16-34 did not produce any hetero-transglycosylation products with cello-oligosaccharides or lactose, which indicated a much more strict substrate tolerance. The increased ability of Tm-NXG1-ΔYNIIG to catalyze transglycosylation relative to the wild-type enzyme (data not shown) was also reflected in an increased amount of obtained hetero-transglycosylation products produced by this mutant. Under preparative-scale conditions, hetero-transglycosylation was indeed significant; a reaction containing 25 mg of XG and 15 mg of cellobiose yielded 11 mg of pure XGOGlc4-cellobiose conjugates (XXXGGG, XLXGGG or XXLGGG, XLLGGG) and 6 mg of pure XGOGlc8-cellobiose conjugates after gel filtration through Bio-Gel P6.
Crystal Structures of Tm-NXG1 and Tm-NXG2
Overall Structure
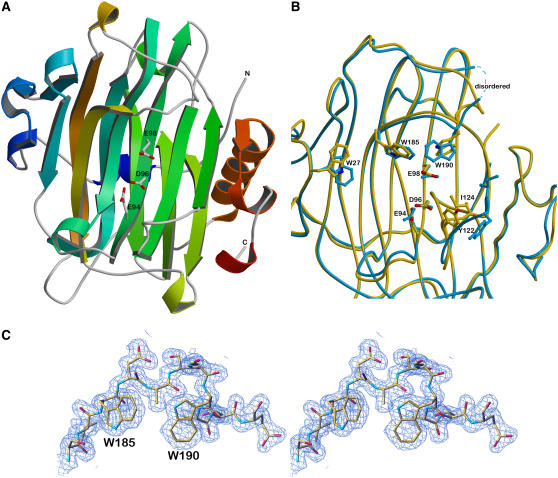
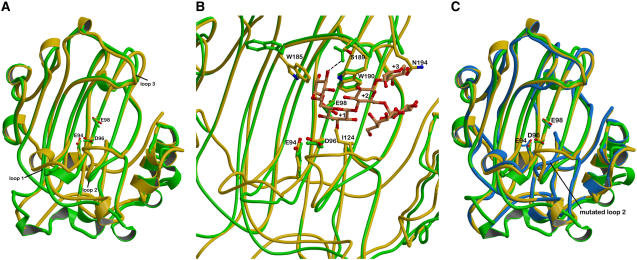
The crystal structures of Tm-NXG1 and Tm-NXG2 are illustrated in Figure 4A, and full crystallographic data are given in Table 2. The asymmetric unit of Tm-NXG1 crystals contained 443 water molecules, 4 glycerol molecules, and 3 copies of the protein molecule comprising 274 residues. This includes a three-residue extension (Ala-Tyr-Val), clearly defined by electron density at the N terminus, from the expression vector. Two hundred ninety-one water molecules and two copies of the enzyme were present in the asymmetric unit of Tm-NXG2, both including the 271 residues reported for the primary sequence of the mature protein (UniProt accession number Q07524). One loop was disordered in both molecules of the Tm-NXG2 crystal structure (residues Ala-191 to Tyr-197; Figure 4B). In accordance with the sequence similarity (39% identity; see Supplemental Figure 4 online), the overall structures are strikingly similar to that of their xyloglucan endo-transglycosylase relative, Ptt-XET16-34 (Protein Data Bank [PDB] identifier 1UN1). Both display the β-jellyroll fold typical for family GH16 enzymes and superimpose onto the structure of Ptt-XET16-34 with a root mean square deviation of 1.35 Å for Tm-NXG1 (241 Cα atoms) and 1.18 Å for Tm-NXG2 (221 Cα atoms). Like Ptt-XET16-34, the Tm-NXG structures contain the helical extension at the C terminus stabilized by two disulfide bonds, although the extremity of the C terminus clearly displays a different conformation in the two isoforms of Tm-NXG. Both Tm-NXG enzyme structures lack the glycosylation site reported for most of the known GH16 XTH gene family members (Campbell and Braam, 1999a). Glu-94 was identified as the catalytic nucleophile and Glu-98 as the catalytic acid/base in Tm-NXG1 by three-dimensional structural alignment with Ptt-XET16-34 (Johansson et al., 2004) and by mechanistic analogy with other GH16 enzymes (Planas, 2000).
Figure 4.
Three-Dimensional Structures of Tm-NXG1 and Tm-NXG2.
(A) Ribbon representation of the crystallographic structure of Tm-NXG1. The polypeptide chain is colored from blue (N terminus) to red (C terminus). The three strictly conserved amino acids forming the catalytic machinery are labeled.
(B) Superimposition of the structures of Tm-NXG1 (yellow) and Tm-NXG2 (blue) highlighting the different conformations of the loops that surround the active site groove. The catalytic residues, the three Trp residues that line the substrate binding cleft, and Tyr-122 and Ile-124 are labeled. Due to high disorder, the loop residues between Thr-192 and Lys-196 could not be modeled in the crystal structure of Tm-NXG2.
(C) Wall-eyed stereo view of the final 2Fo-Fc electron density map displayed at a 1σ level around the loop containing Trp-185 and Trp-190 in the crystal structure of Tm-NXG1.
Table 2.
Data Collection and Refinement Statistics for the Tm-NXG Variants
| Data Statistics | Tm-NXG1 | Tm-NXG2 | Tm-NXG1-ΔYNIIG |
|---|---|---|---|
| European Synchrotron Research Facilities beamline | ID23-1 | ID23-1 | ID23-1 |
| Space group | P31 | P6 | P6 |
| Unit cell parameters | 116.11 Å | 152.12 Å | 153.89 Å |
| 116.11 Å | 152.12 Å | 153.89 Å | |
| 63.1 Å | 83.2 Å | 83.82 Å | |
| Resolution | 1.8 | 2.3 | 2.0 |
| No. of observations | 396,325 | 224,293 | 834,857 |
| No. unique | 87,640 | 47,776 | 76,393 |
| Rsym (%) | 12.3 (65)a | 10.6 (60) | 11.3 (48) |
| I/σ (I) | 9.8 (2.2) | 10.9 (2.5) | 18.7 (4.7) |
| Redundancy | 4.5 (4.4) | 4.7 (4.7) | 10.9 (5.4) |
| Completeness (%) | 99.4 (98.9) | 98.3 (99.1) | 100.0 (100.0) |
| Molecular replacement model used | Tm-NXG2 | 1UN1 | Tm-NXG1 |
| R factor (%) | 44.3 | 48.6 | 65.5 |
| Correlation | 37.3 | 26.9 | 33.8 |
| Molecules/asymmetric unit | 3 | 2 | 2 |
| Refinement | |||
| R factor (%) | 17.6 | 16.7 | 19.2 |
| R free (%) | 19.9 | 19.6 | 21.2 |
| Bond length deviation from ideal | 0.007 Å | 0.024 Å | 0.022 Å |
| Torsion angle deviation from ideal | 1.25° | 2.1° | 1.9° |
Values in parentheses are for the resolution bins 1.90 to 1.80 Å, 2.42 to 2.30 Å, and 2.11 to 2.00 Å for Tm-NXG1, Tm-NXG2, and Tm-NXG1-ΔYNIIG, respectively.
Structural Comparison of Tm-NXG1 and Tm-NXG2
Although the primary sequences of Tm-NXG1 and Tm-NXG2 differ only by one amino acid (Met-129 in Tm-NXG1 is Leu-129 in Tm-NXG2; see Supplemental Figure 4 online), the two isoforms display considerable differences in the substrate binding, active site cleft formed by the concave surface of the β-jellyroll. In particular, three loops were appreciably different between Tm-NXG1, Tm-NXG2, and Ptt-XET16-34: loop 1, Asn-84 to Asp-93; loop 2, Glu-117 to Gly-126; and loop 3, Trp-190 to Tyr-197 (Figures 4B and 5A). The largest differences are localized in loops 2 and 3 on either side of the positive subsites (nomenclature of Davies et al. [1997]). Except for these two loops, the structures of the two isoforms of Tm-NXG are almost identical, with a root mean square deviation value of 0.55 Å for 259 Cα atoms. In both Tm-NXG1 and Tm-NXG2, loop 2 is involved in crystal contacts to a neighboring molecule. It is noteworthy that this loop is followed closely by the sole amino acid difference between the two isoforms (Met-129/Leu-129). Loop 3 is disordered in Tm-NXG2, while it is stabilized by crystal contacts in the Tm-NXG1 structure (Figure 4). Interestingly, this loop is involved in binding of the oligosaccharide ligand in the XLLG-XET complex structure (PDB identifier 1UMZ). The conformation of the corresponding amino acids in the Ptt-XET16-34 crystal structures (PDB identifiers 1UN1 and 1UMZ) is again different from those observed in the individual Tm-NXG isoforms (Figure 5). Due to the high degree of structural similarity between the Tm-NXG1 and Tm-NXG2 structures, subsequent structural comparisons with Ptt-XET16-34 will primarily refer to the Tm-NXG1 structure, which was determined to a higher resolution.
Figure 5.
Structural Comparison of Ptt-XET16-34 and Tm-NXG1.
(A) Ribbon representation of the superimposition of Ptt-XET16-34 (green) onto Tm-NXG1 (yellow) illustrating the structural differences that appear mainly in loops 1 through 3 and in the C-terminal region. The catalytic machinery is labeled.
(B) Close-up view into the positive subsites of Ptt-XET16-34. The superimposition was performed using the complex structure of Ptt-XET16-34 with bound XLLG (PDB identifier 1UMZ). Asp-178 in Ptt-XET16-34 forms a hydrogen bond to a xylosyl branch of the bound oligosaccharide. Ser-189 in Tm-NXG1 (or Tm-NXG2) is too distant to be able to form an equivalent bond. Gly-183 is replaced by Asn-194 in Tm-NXG1, which would collide with bound sugar in this loop conformation. Ile-124, which is part of the critical loop insertion in Tm-NXG1, would collide with the glucose unit bound in subsite +1 of the acceptor binding cleft.
(C) Ribbon representation of the superimposition of Tm-NXG1-ΔYNIIG (blue) onto Ptt-XET16-34 (green) and Tm-NXG1 (yellow) illustrating that the Cα trace of the truncated loop 2 in Tm-NXG1-ΔYNIIG is now closer to that of Ptt-XET16-34. The catalytic machinery is labeled.
Structural Comparison of Tm-NXG1 and Ptt-XET16-34
In addition to the aforementioned loop variants, an N-terminal extension, a C-terminal truncation, three small loop truncations, and a shortening of the C-terminal α-helix were observed in the Tm-NXG enzymes relative to Ptt-XET16-34 (see Supplemental Figure 4 online). Superimposition of either of the Tm-NXG isoforms onto the XLLG-XET complex structure indicates that loop 2 interferes with substrate binding. For both Tm-NXG structures, a collision between Ile-124 and the glucose unit in the +1 binding site would occur if the loops were indeed fixed in that position. Another remarkable difference between Tm-NXG1 and Ptt-XET16-34 occurs in loop 3. In this case, there are no insertions or deletions, but the sequence 178DWATRGG184 in Ptt-XET16-34 is replaced by 189SWATENG195 in Tm-NXG1 (see Supplemental Figure 4 online). This loop, disordered in Tm-NXG2, has a different conformation in Tm-NXG1 compared with that observed in Ptt-XET16-34. Within this loop, Trp-179 in Ptt-XET16-34 and the equivalent Trp-190 in the Tm-NXG enzymes form the platform for hydrophobic stacking interactions with the glucose unit bound in subsite +1. In Ptt-XET16-34, Asp-178 is involved in a hydrogen bond to the xylosyl branch of the sugar bound in the +1 binding site of the XLLG-XET complex. This residue is replaced by a Ser in Tm-NXG1, whose side chain would be too distant to form the same hydrogen bond. Furthermore, Gly-183 in Ptt-XET16-34 is replaced by Asn-194 in Tm-NXG1, and its side chain would collide with bound oligosaccharide in the observed loop conformation.
Crystal Structure of the Loop 2 Truncation Mutant, Tm-NXG1-ΔYNIIG
The asymmetric unit of Tm-NXG1-ΔYNIIG crystals contained 235 water molecules and two copies of the protein molecule comprising 272 residues. This includes a one-residue extension (Val), clearly defined by electron density at the N terminus, arising from the expression construct. The root mean square deviation value for the superimposition of Tm-NXG1 with Tm-NXG1-ΔYNIIG is 0.44 Å for 264 Cα atoms and 1.0 Å for 225 Cα atoms with Ptt-XET16-34. As expected, Tm-NXG1-ΔYNIIG closely resembles Tm-NXG1, except for the region around the loop 2 insertion, where the Cα trace of Tm-NXG1-ΔYNIIG intimately follows that of Ptt-XET16-34 (Figure 5C).
Phylogenetic Analysis of XTH Gene Products
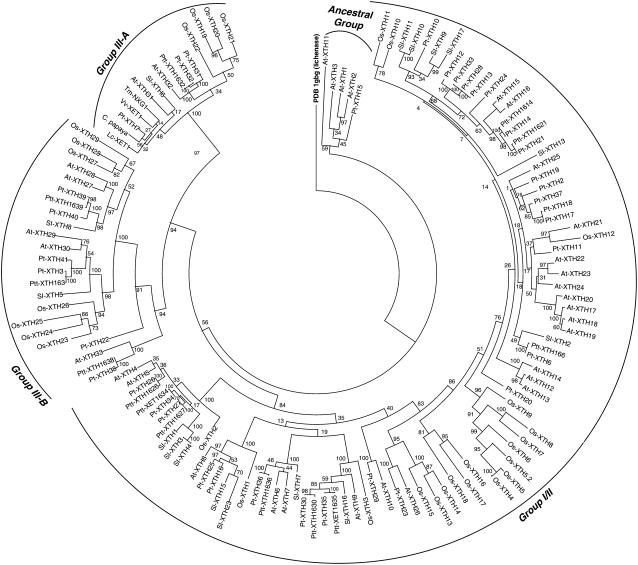
XTH gene products from GH16 have historically been classified into three major phylogenetic subgroups (groups I to III) on the basis of sequence similarity (reviewed in Campbell and Braam, 1999a). The obvious question thus arises whether the current phylogenetic classification can be used to predict biochemical function in the XTH subfamily. A burgeoning amount of genomic data, while providing unprecedented insights into the sequence diversity of XTH gene products, is at the same time blurring the distinction between the traditional subgroups. In particular, comparison of XTH genes in the Arabidopsis thaliana and rice (Oryza sativa) genomes has indicated that there is no longer a clear division between the previously distinct groups I and II (Yokoyama et al., 2004). XTH gene products are small (∼33 kD) and display a high degree of sequence similarity, so it is likely that as the number of available XTH gene sequences continues to rise, the current subgroup definitions will require revision. Therefore, a new bioinformatic analysis of this GH16 subfamily was performed to provide an updated phylogenetic framework in which to discuss the present three-dimensional structure-function analysis of Tm-NXG1, the archetypal mixed-function xyloglucanase/XET from the historical group III (Edwards et al., 1986; de Silva et al., 1993; Fanutti et al., 1993, 1996; Chanliaud et al., 2004). Approximately 130 full-length protein sequences were used to derive a phylogenetic tree from a structure-based sequence alignment and reconstruction using a maximum likelihood method (Guindon and Gascuel, 2003), including all Arabidopsis, rice, and black cottonwood genomic sequences, all tomato (Solanum lycopersicum) and hybrid aspen EST sequences, and selected individual sequences from ripening fruits and germinating seeds (specified in Methods).
Using a structurally characterized bacterial lichenase as an outlier, the tree forms two predominant branches with significant bootstrap values (Figure 6). The largest cluster affirmed previous observations about the merging of groups I and II (Yokoyama et al., 2004). However, this large cluster can, in fact, be subdivided into a number of smaller, statistically significant clades of protein sequences. To avoid further obfuscation, we will nonetheless refer to this ensemble broadly as group I/II. The second main branch encompasses fewer sequences and coincides with the historical group III. This analysis indicates that group III is not monolithic, as commonly represented (Campbell and Braam, 1999a; Yokoyama et al., 2004; Saladié et al., 2006), but can instead be subdivided into two predominant clades. For the purposes of this work, the first clade, which includes Tm-NXG1, will be designated group III-A. Group III-B includes, notably, the Sl-XTH5 and At-XTH27 gene products, which are XETs (Campbell and Braam, 1999b; Saladié et al., 2006). A small outlying group, close to the root and comprising five sequences (At-XTH1 to At-XTH3, At-XTH11, and Pt-XTH15), was also observed. Bootstrap values indicate that these sequences form an intermediate, possibly ancestral, clade between group I/II and group III. At-XTH11 is a notably curious sequence that is likely to encode a catalytically inactive enzyme; the catalytic acid/base and nucleophile-helper residues are mutated to noncarboxylic acid residues (Cys and Gln, respectively), and loop 3 is truncated by nine amino acid residues. Removal of this sequence significantly increases the bootstrap value for this clade.
Figure 6.
Unrooted Phylogenetic Tree of ∼130 Full-Length XTH Gene Products and Bacillus licheniformis Lichenase (PDB identifier 1GBG; GenPept Accession Number CAA40547).
Bootstrap values from 100 maximum likelihood resamplings are indicated.
Groups III-A and III-B, as well as the putative ancestral group, have been analyzed in detail by the combination of primary sequence alignment with structure–activity data for Tm-NXG1 and Ptt-XET16-34. All sequences of Group III-A contain similar extensions of both loop 1 and loop 2 (see Supplemental Figure 5 online), identified in this work to be critical for the endo-xyloglucanase activity of Tm-NXG1. In addition, most of the sequences of group III-A also contain the Ser-189 and Asn(Glu)-194 substitutions in loop 3 found in Tm-NXG1. Sequences of members belonging to group III-B feature a loop 1 extension of the same length as that observed in group III-A, but with a notably different amino acid composition (see Supplemental Figure 5 online). More importantly, although this group has a short insertion in loop 2 relative to Ptt-XET16-34, it is four residues shorter than Tm-NXG1 and lacks the sterically critical Ile-124 (Figures 4 and 5). Furthermore, two residues in loop 3 (Ser-189 and Asn-194 in Tm-NXG1) of group III-B members bear stronger resemblance to those observed in Ptt-XET16-34 than those in Tm-NXG1. None of the enzymes from group III-B, to our knowledge, has been shown to have endo-xyloglucanase activity (see Supplemental Table 1 online). Sequences of members of the putative ancestral group display the shorter loops 1 and 2 and similar loop 3 residues typical of Ptt-XET16-34. Altogether, this analysis suggests that all members of group III-B and the putative ancestral group may be strict, or at least predominant, XETs, while mixed-function xyloglucanase/XETs (e.g., Tm-NXG1) are found only in group III-A. Therefore, we favor the abandonment of the historical group divisions, which in the context of existing biochemical data have no predictive value and are potentially misleading. However, we also note that any group nomenclature is likely to be tenuous until a significant number of sequences allows a stable phylogeny to be constructed (especially in group I/II), which necessarily requires the generation of more plant whole-genome sequences than those available at present.
DISCUSSION
What Determines the Fate of the Glycosyl Enzyme Intermediate in the XTH Subfamily of GH16?
The potential to catalyze xyloglucan endo-transglycosylation (EC 2.4.1.207) or endo-hydrolysis (EC 3.2.1.151) is a key feature of the retaining catalytic mechanism used by plant xyloglucan-active enzymes from GH16 (Sinnott, 1990). Our present structural and biochemical analysis indicates that the extension of loop 2 in Tm-NXG1 and Tm-NXG2 is a determinant of endo-xyloglucanase activity in XTH gene products. The strict transglycosylase Ptt-XET16-34 from hybrid aspen lacks this extension, which the three-dimensional structure of Tm-NXG1 shows is capable of interacting with substrate bound in the positive subsites of the enzyme, potentially modulating the binding of xylogluco-oligosaccharide acceptor substrates. Truncation of this loop to produce a hybrid, Ptt-XET16-34–like enzyme does in fact boost the rate of transglycosylation slightly, while hydrolytic activity is diminished (Figure 3). The variation in the length of loop 2 is clearly an important difference between the XETs and the xyloglucanases in GH16, but our inability to generate a strict transglycosylase from a predominant hydrolase indicates that this is not the only determinant.
Another obvious difference that distinguishes Tm-NXG1 and Tm-NXG2 from Ptt-XET16-34 is the presence of a three-residue extension of loop 1. We have been unsuccessful in expressing Tm-NXG1 variants that possess a truncation of this loop, either alone or in combination with loop 2 truncation, but can nonetheless infer from recent literature data that loop 1 alone is not likely to affect the transglycosylation/hydrolysis ratio. The tomato Sl-XTH5 gene is abundantly expressed during fruit ripening (Saladié et al., 2006) and, like Tm-NXG1, possesses the longer loop 1 variant (see Supplemental Figure 5 online). Viscometric and HPLC-based assays recently indicated that Sl-XTH5 is a predominant XET; under identical assay conditions, Tm-NXG1 exhibited predominant or exclusive endo-xyloglucanase activity (Saladié et al., 2006). Similarly, early work by Campbell and Braam (1999b) demonstrated that recombinant Arabidopsis At-XTH27 (EXGT-A3) was likewise a XET, devoid of hydrolytic activity. Unfortunately, the assays employed in these studies do not allow quantitative comparison of the transglycosylation and hydrolysis rates of these enzymes in terms of turnover numbers; nonetheless, they show qualitatively that the longer loop 1 observed in Sl-XTH5 and At-XTH27 (see Supplemental Figure 5 online) does not confer significant hydrolytic activities to these enzymes. Instead, our evidence suggests that the extension of loop 2 in Tm-NXG1–like enzymes is a major structural change responsible for the XET-to-xyloglucanase switch in GH16, as indicated by our mutagenesis and quantitative kinetic analyses. It is apparent, however, that a number of smaller, currently unexplored sequence variations (see Supplemental Figures 4 and 5 online) also conspire to dictate the fate of the glycosyl enzyme intermediate (Figure 1). Thus, further structure–function studies are clearly needed to unlock all of the secrets necessary to fully convert a xyloglucanase into a XET or vice versa.
The Evolution of Divergent Xyloglucan Activities in GH16
In view of the entire GH16 family, these phylogenetic analyses raise a fundamental evolutionary question. Because the ancestral enzyme of GH16 is likely to be a hydrolytic laminarinase (Barbeyron et al., 1998; Michel et al., 2001), is the ancestor of the XTH gene family a hydrolytic or transglycosylating enzyme? Our phylogenetic tree, based on maximum likelihood reconstructions, indicates that both main branches contain predominant XETs, while predominant xyloglucan hydrolases only appear in group III-A. Closer examination indicates that the putative ancestral group, which contains only sequences with XET-like loops, is likely to have emerged earlier than the Tm-NXG1–containing group III-A. Furthermore, the observation that group III-A is not very divergent (between 60 and 70% pairwise sequence identity) provides additional support for the hypothesis that the emergence of Tm-NXG–like enzymes is a relatively recent event. Moreover, Tm-NXG–like enzymes are rare, and their presence appears to be limited to germinating seeds, ripening fruits, and some fast-growing tissues (vide infra). This rareness is also exemplified, for instance, by a census of the Arabidopsis XTH family: from >30 homologous enzymes, only 2 (At-XTH31 and At-XTH32) are predicted to have xyloglucanase activity by our analysis (Figure 6).
Perhaps the strongest argument for the evolution of GH16 xyloglucanases from XETs involves a common structural characteristic that distinguishes the XTH gene family from all other GH16 enzymes. As illuminated by the first XET three-dimensional structure (Johansson et al., 2004), these enzymes have diverged from their hydrolytic cousins, the lichenases and laminarinases, by extension of the C terminus, thereby increasing the protein–substrate interactions in the positive subsites. Like the true XETs, Tm-NXG1 also has the C-terminal loop extension, which provides an extra β-strand extending the length of the substrate binding cleft. Therefore, it is highly unlikely that Tm-NXG1–like enzymes have emerged directly from a hydrolytic ancestor that lacks this C-terminal extension. Indeed, here, we show that in Tm-NXG1, the additional hydrolytic activity is principally due to subtle changes in a surface loop that interacts with the positive subsites, probably affecting acceptor substrate binding. Altogether, these arguments favor the conclusion that xyloglucan endo-transglycosylation is the ancestral activity, which predates xyloglucanase activity arising from a secondary divergence in (historical) group III.
Biological Implications
Analysis of the tissue-specific expression of a range of GH16 XTH genes has been performed for a variety of species, including Arabidopsis, Populus, and rice, whose complete genomes have been sequenced (Yokoyama and Nishitani, 2001; Yokoyama et al., 2004; Aspeborg et al., 2005; Geisler-Lee et al., 2006). Recent comprehensive work by Braam and coworkers using XTH:GUS gene fusions has further illuminated the temporal and spatial expression of these genes in Arabidopsis (Becnel et al., 2006). With regard to the biological role of group III-A enzymes, which may represent a specific subgroup of predominant xyloglucanases in GH16, some interesting patterns emerge. The tissue-specific expression of two nasturtium XTH genes, Tm-NXG1 (group III-A) and Tm-XET1 (group I/II; GenBank accession number L43094), has been studied previously (Rose et al., 1996). XET1, which is 88% identical and 94% similar to the nonhydrolytic Ptt-XET16-34 (Kallas et al., 2005) at the protein level, exhibits a systemic expression pattern, with the exception of germinating seed cotyledons. Tm-NXG1, on the other hand, is expressed exclusively in this tissue, from which the active enzyme has been isolated (Edwards et al., 1986). Furthermore, Tm-NXG1 homologs are expressed specifically in a range of ripening fruits, including Litchi (Lu et al., 2006), papaya (Carica papaya) (GenBank accession number AY032600; R. Haireen, Z.M. Ali, R. Othman, and H. Lazan, unpublished data), grape (Vitis vinifera) (Ishimaru and Kobayashi, 2002), and apricot (Prunus armeniaca) (GenBank accession number AY578086; F. Geuna, R. Banfi, and D. Bassi, unpublished data). These observations are consistent with a general role of Tm-NXG–like enzymes in the degradation of xyloglucan during seed germination, on the one hand, and fruit ripening, on the other.
Two Arabidopsis XTH gene products are also found in group III-A: At-XTH31 (65% identical and 85% similar protein sequence to Tm-NXG1) is predicted to have a role in seed germination (Aubert and Herzog, 1996) and root morphogenesis (Aubert and Herzog, 1996; Becnel et al., 2006), whereas At-XTH32 (68% identical and 81% similar to Tm-NXG1) is expressed primarily in the shoot apex (Becnel et al., 2006). The observation of the expression of these genes in expanding tissues is not inconsistent with a potential role in xyloglucan degradation allowing rapid wall extension. Xyloglucanase activity is not likely to be a ubiquitous requirement for wall degradation in processes such as fruit ripening, however (Brummell, 2006); the Tm-NXG1 homolog Sl-XTH6 (group III-A) was not observed in EST libraries from ripening tomato, whereas the group III-B XET encoded by Sl-XTH5 was implicated in fruit development (Saladié et al., 2006).
As a notable side activity, it is currently unclear whether the ability of Tm-NXG1 to perform hetero-transglycosylation to cello-oligosaccharide acceptors has biological relevance. Indeed, it is likely that the in vitro cross-conjugation of xyloglucan and linear β-glucans observed in crude nasturtium extracts (Mohand and Farkas, 2006) is due, at least in part, to Tm-NXG1, based on our results. However, in the germinating nasturtium seed, the effective physiological concentrations of internal XG bonds and water are likely to be much higher than the concentration of nonreducing ends of other β-glucans, thus implying that the effective rate of hetero-transglycosylation would be low. Similarly, a barley XET isozyme was recently shown to have very low hetero-transglycosylation activity using alternating XG:β-glucan and β-glucan:XG donor/acceptor pairs in vitro (Hrmova et al., 2007). Although there is currently no evidence for the existence of analogous block copolysaccharide structures in muro (Hrmova et al., 2007), the predominance of (1,3:1,4)-β-glucan over XG in barley primary cell wall raises the possibility that hetero-transglycosylation is kinetically feasible in that system. However, it should be borne in mind that promiscuous glycosyl transfer by retaining glycosidases to nonnatural acceptors is well documented, including for endo-(xylo)glucanases (York and Hawkins, 2000).
In this context, it is interesting that Tm-NXG1 is a comparatively feeble xyloglucanase, with a vo/[E]t value of 5.5 ± 0.5 min−1 at the pH optimum and 1 g/L tamarind xyloglucan. Under similar conditions and an identical substrate concentration, a strict xyloglucanase from Paenibacillus pabuli (GH5, retaining) has a vo/[E]t value of 8700 ± 220 min−1, while a nonspecific endo-glucanase from Bacillus licheniformis (GH12, retaining) has a vo/[E]t value of 54 ± 3 min−1 (Gloster et al., 2007). Similarly, the inverting microbial xyloglucanases from GH74 have catalytic rates of at least 5 to 10 times greater than that of Tm-NXG1 (Irwin et al., 2003; Martinez-Fleites et al., 2006). Therefore, the poor hydrolytic activity of Tm-NXG1 may reflect the different physiological requirements for xyloglucan degradation by plants and microorganisms. During germination, it may be anticipated that the developing embryo requires metered, continuous release of sugar in the confines of the seed, whereas in rapidly expanding tissues, xyloglucan degradation must not be so rapid as to lead to irreparable wall loosening. On the other hand, the ability to rapidly degrade available carbohydrate sources would confer a selection advantage to microbes in more competitive environments. Tm-NXG1 nonetheless exhibits a rate of transglycosylation that is only fivefold lower than that of the strict transglycosylase Ptt-XET16-34 over a wide concentration range (Figure 3, Table 2). This supports bioinformatic analyses that indicate that mixed-function xyloglucanases/XETs may have evolved from true XETs; xyloglucanase activity has resulted from a gain of function in GH16, where glycosyl transfer to water has been favored by extension of specific active site loops. It is apparent that additional comprehensive biochemical studies are required to further illuminate the catalytic diversity hidden within this enzyme family.
METHODS
Material Sources
Ultrapure water was used in all experiments and refers to water purified on a Milli-Q system (Millipore) with a resistivity of ρ > 18.2 MΩ·cm. Polysaccharides were obtained from the following sources: konjac glucomannan, Icelandic moss lichenan, tamarind xyloglucan, and barley (Hordeum vulgare) β-glucan (all from Megazyme); Avicel and hydroxyethyl cellulose (both from Fluka); and birchwood xylan (from Sigma-Aldrich). Xylogluco-oligosaccharides based on Glc4, Glc8, and Glc12 backbones (XGOGlc4, XGOGlc8, and XGOGlc12, respectively) were produced as reported by Martinez-Fleites et al. (2006). These samples represent mixtures of variably galactosylated oligosaccharides arising from the natural distribution of XXXG, XLXG, XXLG, and XLLG in tamarind kernel xyloglucan. The XGOGlc8 preparation was contaminated with 0.075% XGO and 0.5% XGOGlc12 (HPAEC-PAD analysis; vide infra); in all calculations of enzyme activities, these background values were subtracted. The average molecular weight of XGOGlc8 (Mr 2530) was calculated from the composition of oligosaccharides XXXG, XLXG, XXLG, and XLLG (15:7:32:46) in tamarind xyloglucan.
Protein Analytical Methods
Proteins were analyzed by SDS-PAGE using an Xcell II Mini Cell (Novex). Precast NuPAGE gradient gels (4 to 12%) were purchased from Invitrogen. Molecular mass standards (LMW calibration kit) were obtained from Amersham Biosciences. Protein gels were stained by Coomassie Brilliant Blue R 250. Protein concentrations were determined spectrophotometrically by the Bradford method using BSA as a standard. DNA sequencing was performed by the Sanger method using an ABI Prism 377 DNA sequencer (Perkin-Elmer). The pH rate profiles of Tm-NXG1 and Tm-NXG2 were determined in duplicate using the method described by Nelson (1944) with XG (1 g/L) as substrate. The following 100 mM buffer systems were used: sodium acetate, pH 4.0 to 5.75; sodium phosphate, pH 6.0 and pH 6.5. The unfolding temperature of proteins was determined by the fluorescence-based thermal shift assay of Pantoliano et al. (2001), as described by Kallas et al. (2005). ESI-MS analysis of proteins was performed as described by Sundqvist et al. (2007).
Carbohydrate Analytical Methods
XET and xyloglucanase activities were measured with the colorimetric assay according to Sulova et al. (1995) and the radioactive assay devised by Fry et al. (1992). Activities on soluble polysaccharides other than xyloglucan were determined at concentrations of 0.5 g/L and an enzyme concentration of 0.02 g/L at 30°C. Activity on Avicel was measured on a 0.5 g/L suspension. Aliquots were taken at 10, 30, 60, and 130 min, and reducing sugars were quantified using the method of Nelson (1944). A six-point standard curve was made with glucose in the range 20 to 150 mg/L.
HPAEC-PAD analysis of xylogluco-oligosaccharides was performed using a Dionex Carbopac PA100 column on a Dionex ICS-3000 HPLC system controlled by Chromelion software, version 6.80 (Dionex). An optimized gradient program was used for simultaneous analysis of all xylogluco-oligosaccharides (XGOGlc4, Glc4-based xylogluco-oligosaccharides; XGOGlc8, Glc8-based xylogluco-oligosaccharides; and XGOGlc12, Glc12-based xylogluco-oligosaccharides). Solvent A was water; solvent B was 1 M sodium hydroxide; solvent C was 1 M sodium acetate. The gradient program was as follows: 0 to 3 min, 100 mM sodium hydroxide and 40 mM sodium acetate; 3 to 18 min, 100 mM sodium hydroxide and a linear gradient of 40 to 300 mM sodium acetate; 18 to 19 min, 500 mM sodium hydroxide and 500 mM sodium acetate. The system was then reequilibrated for 4 min with the initial conditions prior to the next injection. For kinetic experiments, standard curves were made for XGO (0 to 50 μM, 13 points) and the transglycosylation product XGOGlc12 (Glc12-based xylogluco-oligosaccharides; 0 to 50 μM, 13 points). Chromelion (version 6.80; Dionex) HPLC software was used to fit a cubic function to data from samples analyzed in triplicate.
Time Dependence of the Tm-NXG–Catalyzed Depolymerization of Xyloglucan
Samples (250 μL total volume) containing XG (1 g/L) and Tm-NXG1 (9 mg/L), Ptt-XET16-34 (10 mg/L), or Tm-NXG1-ΔYNIIG (10 mg/L) in sodium acetate buffer (25 mM, pH 5.0) were incubated without or with XGO (0.35 g/L, 0.0035 g/L in the case of Tm-NXG1-ΔYNIIG) at 30°C for 30, 60, or 150 min and overnight. The reactions were immediately frozen, lyophilized, and redissolved in 250 μL of DMSO prior to SEC analysis as described by Brumer et al. (2004). Control samples were treated under identical conditions but without enzyme.
Initial Rate Kinetics with XGOGlc8 Xylogluco-Oligosaccharides
For each substrate concentration (10 to 3000 μM, 19 points), 20 μL of XGOGlc8 dilutions was prewarmed for 5 min at 30°C. Enzyme solution in 25 mM sodium acetate buffer, pH 5.0 (5 μL), was added and the reaction was incubated at 30°C; after 7 min, 10 μL of sodium hydroxide (2.5 M) was added to stop the reaction. Enzyme activity was adjusted so that the substrate (XGOGlc8) turnover was in all cases <10% of the initial concentration and typically <1%. All samples were made in duplicate. Samples were kept at 4°C for not longer than 6 h until analysis by HPAEC-PAD.
Transfer of Xyloglucan to Cello-Oligosaccharide Acceptor Substrates
For analytical reactions, Tm-NXG1 (0.02 g/L) was incubated for 18 h with XG (1 g/L) and lactose (8 mM), cellobiose (8 mM), cellotriose (8 mM), or cellotetraose (8 mM) in 16 mM sodium acetate buffer, pH 5.0. Samples (25 μL total volume) were analyzed by HPAEC-PAD and ESI-TOF-MS (Brumer et al., 2004). For preparative-scale synthesis of transfer products, Tm-NXG1-ΔYNIIG (0.01 g/L) was incubated with cellobiose (3.6 M) and XG (2 g/L) in 16 mM sodium acetate buffer, pH 5.0 (12 mL), for 24 h. Products were purified by the gel-filtration method described by Martinez-Fleites et al. (2006) and identified by ESI-TOF-MS.
Bioinformatic Analysis
Bioinformatic analysis was performed on ∼130 full-length protein sequences that reflected the diversity of XETs and xyloglucanases in GH16. These included all genome-derived sequences from Arabidopsis thaliana (33 sequences) and Oryza sativa (29 sequences) and individual sequences from Carica papaya (GenPept accession number AAK51119; R. Haireen, Z.M. Ali, R. Othman, and H. Lazan, unpublished data), Tropaeolum majus (Tm-NXG1) (de Silva et al., 1993), Litchi chinensis (Lc-XET1) (Lu et al., 2006), and Vitis labrusca × Vitis vinifera (Vv-XET1) (Ishimaru and Kobayashi, 2002), selected from curated Carbohydrate-Active Enzyme database entries (http://www.cazy.org). Full-length Solanum lycopersicum ESTs (16 sequences) were selected from those reported by Saladié et al. (2006). Thirteen full-length Populus tremula × Populus tremuloides ESTs (Nishikubo et al., 2007) and 36 Populus trichocarpa genomic sequences (Geisler-Lee et al., 2006) were from PopulusDB (http://poppel.fysbot.umu.se/) and the Joint Genome Initiative P. trichocarpa genome (Tuskan et al., 2006) release version 1.0 (http://genome.jgi-psf.org/poplar), respectively. After removal of predicted signal peptides (Bendtsen et al., 2004), protein sequence alignments were performed using MAFFT with the iterative refinement method and the scoring matrix Blosum62 (Katoh et al., 2002). The multiple alignment of these sequences was manually refined using Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) on the basis of the superposition of the crystal structures of Tm-NXG1 (this work) and of Ptt-XET16-34 (1UN1; Johansson et al., 2004). Sequence alignments were rendered with ESPript (Gouet et al., 1999). Phylogenetic trees were derived from this refined alignment using the maximum likelihood method with the program PhyML (Guindon and Gascuel, 2003). The lichenase from Bacillus licheniformis (PDB identifier 1GBG) was used as an outgroup. The reliability of the trees was tested by bootstrap analysis using 100 resamplings of the data set. Trees were displayed with MEGA 3.1 (Kumar et al., 2004).
Cloning of Tm-NXG2 cDNA from Nasturtium
Nasturtium (Tropaeolum majus cv Scarlett Gleam) seeds were obtained from Weibulls Trädgård. Total RNA was extracted from epicotyls (leaves removed) at 4 d after germination with the RNeasy Plant Mini kit (Qiagen) according to the manufacturer's protocol and frozen in liquid nitrogen. Total RNA was extracted, and cDNA was produced with the ThermoScript kit (Invitrogen) with gene-specific primer Tm-NXG-F1 (see Supplemental Table 4 online). Primer design was based on GenBank sequence X68254. The resulting cDNA was further amplified with Taq DNA polymerase (Fermentas), using Tm-NXG-F2 and Tm-NXG-R2 as primers (see Supplemental Table 4 online). The PCR product was cloned directly into the pGEM T Easy (Promega) vector system, and 10 positive clones were sequenced. Full-length Tm-NXG2 (corresponding to GenBank accession number X68255) was amplified by PCR using a proofreading DNA polymerase (Pfu; Stratagene), the forward primer Tm-NXG-F2, and the reverse primer Tm-NXG-R2. The amplified fragment was cloned into the Pichia pastoris expression vector pPIC9 (Invitrogen).
Mutagenesis of Tm-NXG2 to Tm-NXG1 and Preparation of Tm-NXG1-ΔYNIIG
To obtain the full-length Tm-NXG1 (GenBank accession number X68254) cDNA sequence, the Tm-NXG2 L129M mutant was produced using the method published by Nourizad et al. (2003). Two different DNA fragments were amplified from the pPIC9-Tm-NXG2 plasmid with Pfu (Stratagene) proofreading DNA polymerase and either primers Tm-NXG-F2 and 5′biotin-Tm-NXG2-L129M-R1 or 5′biotin-Tm-NXG2-L129M-F1 and Tm-NXG-R2 (see Supplemental Table 4 online). The biotinylated fragments were immobilized, denatured, and reannealed, and the full-length Tm-NXG1 was amplified with the outer primers Tm-NXG-F2 and Tm-NXG-R2. The amplified fragment was cloned into pPIC9 (Invitrogen), sequenced, and expressed in P. pastoris. To prepare the deletion variant Tm-NXG1-ΔYNIIG by removal of residues 122 through 126 in the loop bridging β-strands 8 and 9, primer pairs Tm-NXG-F2/5′biotin-Tm-NXG-ΔYNIIG-R1 and Tm-NXG-R2/5′biotin-Tm-NXG-ΔYNIIG-F1 were used (see Supplemental Table 4 online), according to the same procedure.
Expression and Affinity Protein Purification of Tm-NXG1, Tm-NXG2, and Tm-NXG1-ΔYNIIG
Tm-NXG1, Tm-NXG2, and Tm-NXG1-ΔYNIIG were cloned into the pPIC9 vector and expressed in the methylotrophic yeast P. pastoris (strain GS115) under the control of the alcohol oxidase promoter essentially as described previously (Henriksson et al., 2003; Kallas et al., 2005). After 4 to 5 d of induction at 22°C, cultures producing Tm-NXG1 contained typically 50 mg/L total protein, at which time cells were removed by centrifugation (2800g, 20 min, 4°C). Remaining cell debris was removed by filtration through a Mini Capsule filter with a 0.45-μm pore size Versapor (Pall Europe) membrane. An affinity purification method was developed by coupling XGO-NH2 (Greffe et al., 2005) to 6 mL of HiTrap NHS-activated HP medium (GE Healthcare) (column dimensions: diameter, 1.4 cm; height, 3 cm). The column material was pretreated according to the manufacturer's instructions. The coupling solution (12 mL), containing 235 μmol of XGO-NH2 (four times excess relative to the NHS-activated groups), was pumped through the column overnight. The column was subsequently washed according to the manufacturer's instructions. Cell-free culture filtrate (500 mL) was loaded directly onto a 6-mL XGO-affinity column after equilibration with 25 mM sodium acetate, pH 5.0. After washing (10 column volumes, 25 mM sodium acetate, pH 5.0), bound protein was eluted with 80% ethylene glycol in 25 mM sodium acetate, pH 5.0, in 10 column volumes in 10-mL fractions. Enzyme activity was visualized with the colorimetric assay. Active fractions were pooled, and the buffer was exchanged with 25 mM sodium acetate, pH 5.0 (3 × 15 mL), by concentration on the Millipore Centricon Plus 20 Biomax PES (MWCO 5000; Millipore). To avoid potential cross-contamination, each protein variant was purified on new column material.
ESI-MS analysis of the purified proteins confirmed the expected molecular masses: for Tm-NXG1, 31,548 D calculated, 31,548 D found; for Tm-NXG2, 31,530 D calculated, 31,531 D found; for Tm-NXG1-ΔYNIIG, 30,988 D calculated, 30,989 D found. Due to the cloning strategy used and incomplete cleavage of the α-factor secretion signal peptide, six additional amino acids (protein sequence EAEAYV) remained on the N terminus of all proteins (reflected in the calculated molecular masses). The protein-unfolding temperatures were tested with a range of different buffer conditions and additives (see Supplemental Table 2 online). The observed unfolding temperatures for Tm-NXG2 were consistently 1 to 2°C lower than those for Tm-NXG1 for all conditions tested.
Crystallization of Tm-NXG1, Tm-NXG2, and Tm-NXG1-ΔYNIIG
The protein solutions of Tm-NXG1, Tm-NXG2, and Tm-NXG1-ΔYNIIG (all in 25 mM sodium acetate buffer, pH 5.5) were concentrated to 15.0 g/L, 6.2 g/L, and 15.0 g/L, respectively, with Centricon devices (cutoff of 10 kD). Initial crystallization trials were performed with MDL (for Molecular Dimensions Limited) and Decode Genetics (Wizard 1 &2; Emerald Biostructures) commercial kits for Tm-NXG1 and with the MDL and Nextal Cation screen for Tm-NXG2 and Tm-NXG1-ΔYNIIG (i.e., a total of 192 trials each on two 96-multiwell crystallization plates from Corning). These trials were set up using a Cartesian crystallization robot, mixing 300 nL of protein with 150 nL of reservoir solution. All crystallization trials were performed at 19°C.
First crystals of Tm-NXG1 and Tm-NXG2 were obtained in the conditions MDL2 (22) and Cation screen (91), respectively. These conditions were subsequently optimized on Linbro plates, using the vapor diffusion method in hanging drops by mixing equal amounts (2 μL) of protein solution with optimized reservoir solution. The solutions for the optimized conditions of Tm-NXG1 contained 15 to 20% polyethylene glycol 20,000 and 100 mM HEPES buffer, pH 7.5; those for Tm-NXG2 contained 2 to 3 M potassium acetate, 2% 2,5-methylpentane-diol, and 100 mM Tris buffer, pH 8.5. First crystals of Tm-NXG1-ΔYNIIG were obtained in the condition MDL1 (12), which contained 1.4 M sodium acetate and 100 mM sodium cacodylate buffer, pH 6.5. Hexagonal plates grew directly to a suitable size for x-ray diffraction and were used directly for the subsequent data collection.
Data Collection, Molecular Replacement, and Structure Refinement of Tm-NXG1, Tm-NXG2, and Tm-NXG1-ΔYNIIG
The x-ray diffraction data of Tm-NXG1, Tm-NXG2, and Tm-NXG1-ΔYNIIG crystals were collected at 100 K at the European Synchrotron Research Facilities beamline ID23 EH1 using an ADSC Quantum 4R CCD detector. The crystals were flash-frozen in a liquid nitrogen stream with 15% (v/v) glycerol as cryoprotectant, using nylon cryoloops of 0.1 mm diameter. The wavelength of the synchrotron x-rays was 0.933 Å. The crystals were rotated through 120° with a 0.5° oscillation range per frame. The raw data were processed using the program MOSFLM (Powell, 1999) and then merged and scaled using the program SCALA (Collaborative Computational Project, Number 4, 1994). Further data collection statistics are provided in Table 2.
Molecular replacement with the program AMoRe (Navaza, 1994) in the resolution range 9 to 3.1 Å with the model of Ptt-XET16-34 (PDB identifier 1UN1) gave two solutions (two molecules in the asymmetric unit) for Tm-NXG2, with an overall correlation coefficient of 0.27 and an R factor of 48.6%. The Matthews coefficient (Matthews, 1968) was calculated to be VM = 4.50, and the crystals contained 72.7% solvent. The residue changes corresponding to the Tm-NXG2 primary sequence were determined using the program TURBO (Roussel and Cambillau, 1991). Water molecules were added with wARP (Perrakis et al., 1997). Graphic inspection of the polypeptide chain and the water molecules was subsequently performed with TURBO. The refinement was performed using REFMAC5, part of the CCP4 package (Collaborative Computational Project, Number 4, 1994). The stereochemistry of the final structures was evaluated using PROCHECK (Laskowski et al., 1993). The refined coordinates of Tm-NXG2 were then used as a search model for Tm-NXG1. In the resolution range 9 to 2.8 Å, three solutions, corresponding to the three molecules in the asymmetric unit, led to a correlation coefficient of 0.37 and an R factor of 44.3%. The Matthews coefficient was calculated to be VM = 2.64, and the crystals contained 53.0% solvent. The refined coordinates of Tm-NXG1 were in turn used as a search model for Tm-NXG1-ΔYNIIG. In the resolution range 9 to 3.5 Å, two solutions with space group P6 led to a correlation coefficient of 0.65 and an R factor of 33.8%. The Matthews coefficient was calculated to be VM = 4.62, and the crystals contained 73.4% solvent. The same model construction, water addition, and refinement procedure were then applied to both Tm-NXG1 and Tm-NXG1-ΔYNIIG, as described above for Tm-NXG2. All further refinement statistics are provided in Table 2. The coordinates of the crystallographic structures of Tm-NXG1, Tm-NXG1-ΔYNIIG, and Tm-NXG2 have been deposited with the Protein Data Bank (PDB identifiers 2UWA, 2UWB, and 2UWC, respectively). Protein cartoon representations were drawn with Molscript (Kraulis, 1991) and rendered with Raster3D (Merritt and Murphy, 1994). Electron density representations were prepared using TURBO (Roussel and Cambillau, 1991).
Accession Numbers
Accession numbers for the sequences described in this article are as follows: Bacillus licheniformis lichenase (PDB 1GBG, GenPept CAA40547), Bob-XET16A (GenPept AAO00727), unpublished sequence from Carica papaya (GenBank AY032600), Lc-XET1 (GenPept ABK30788), Ptt-XET16-34 (PDB 1UN1 and 1UMZ, GenPept AAN87142), unpublished sequence from Prunus armeniaca (GenBank AY578086), Sl-XTH5 (GenPept AAS46240), Sl-XTH6 (GenPept AAS46242), Tm-NXG1 (PDB 2UWA, GenBank X68254, GenPept CAA48324), Tm-NXG1-ΔYNIIG (PDB 2UWB), Tm-NXG2 (PDB 2UWC, GenBank X68255, GenPept CAA48325), Tm-XET1 (GenBank L43094), and Vv-XET1 (GenPept BAB78506).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. pH Rate Profile of Tm-NXG1.
Supplemental Figure 2. Ratio of Initial Rates of Transglycosylation to Hydrolysis Catalyzed by Tm-NXG1 as a Function of [XGO2].
Supplemental Figure 3. Ratio of Initial Rates of Transglycosylation to Hydrolysis Catalyzed by Tm-NXG1-ΔYNIIG as a Function of [XGO2].
Supplemental Figure 4. Full-Length Protein Sequence Alignment of Tm-NXG1, Tm-NXG2, Bob-XET16A, and Ptt-XET16-34.
Supplemental Figure 5. Partial Protein Sequence Alignment of Historical Group III Gene Products and Ptt-XET16-34.
Supplemental Table 1. Heterologously Expressed Members of the XTH Gene Family.
Supplemental Table 2. Unfolding Temperature of Tm-NXG1 and Tm-NXG2.
Supplemental Table 3. Hetero-Transglycosylation Products Obtained from Xyloglucan and Cello-Oligosaccharides.
Supplemental Table 4. Primers Used for Amplifying Tm-NXG DNA Fragments.
Supplemental Text. Kinetic Derivations.
Supplementary Material
Acknowledgments
M.J.B. thanks the Swedish Biofiber Materials Center for predoctoral funding. H.B. (VR Rådsforskare) acknowledges funding from the Swedish Research Council (Vetenskapsrådet) and is grateful to Pedro Coutinho and Bernard Henrissat (Architecture et Fonction des Macromolécules Biologiques–Centre National de la Recherche Scientifique, Marseille, France) for a stimulating visit to Marseille, including preliminary bioinformatic analysis of GH16 XETs. M.C. was supported by a grant from the Conseil Générale in France, Région Bretagne. We are also indebted to the European Synchrotron Research Facilities (Grenoble, France) for beamtime allocation (BAG-MX485) for this project and to the staff on beamline ID23-EH1 for technical assistance during data collection. We also thank Björn Sundberg, Nobuyuki Nishikubo, and Ewa Mellerowicz (Umeå Plant Science Center) for access to the hybrid aspen sequences prior to publication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Harry Brumer III (harry@biotech.kth.se).
Online version contains Web-only data.
References
- Aspeborg, H., et al. (2005). Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol. 137 983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert, D., and Herzog, M. (1996). A new cDNA encoding a xyloglucan endo-transglycosylase-related polypeptide (AtXTR8) preferentially expressed in seedling, root and stem of Arabidopsis thaliana. Plant Sci. 121 187–196. [Google Scholar]
- Barbeyron, T., Gerard, A., Potin, P., Henrissat, B., and Kloareg, B. (1998). The kappa-carrageenase of the marine bacterium Cytophaga drobachiensis. Structural and phylogenetic relationships within family-16 glycoside hydrolases. Mol. Biol. Evol. 15 528–537. [DOI] [PubMed] [Google Scholar]
- Becnel, J., Natarajan, M., Kipp, A., and Braam, J. (2006). Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol. Biol. 61 451–467. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340 783–795. [DOI] [PubMed] [Google Scholar]
- Brumer, H., Sims, P.F.G., and Sinnott, M.L. (1999). Lignocellulose degradation by Phanerochaete chrysosporium: Purification and characterization of the main alpha-galactosidase. Biochem. J. 339 43–53. [PMC free article] [PubMed] [Google Scholar]
- Brumer, H., Zhou, Q., Baumann, M.J., Carlsson, K., and Teeri, T.T. (2004). Activation of crystalline cellulose surfaces through the chemoenzymatic modification of xyloglucan. J. Am. Chem. Soc. 126 5715–5721. [DOI] [PubMed] [Google Scholar]
- Brummell, D.A. (2006). Cell wall disassembly in ripening fruit. Funct. Plant Biol. 33 103–119. [DOI] [PubMed] [Google Scholar]
- Buckeridge, M.S., Crombie, H.J., Mendes, C.J.M., Reid, J.S.G., Gidley, M.J., and Vieira, C.C.J. (1997). A new family of oligosaccharides from the xyloglucan of Hymenaea coubaril L. (Leguminosae) cotyledons. Carbohydr. Res. 303 233–237. [DOI] [PubMed] [Google Scholar]
- Buckeridge, M.S., dos Santos, H.P., and Tine, M.A.S. (2000). Mobilisation of storage cell wall polysaccharides in seeds. Plant Physiol. Biochem. 38 141–156. [Google Scholar]
- Campbell, P., and Braam, J. (1999. a). Xyloglucan endotransglycosylases: Diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 4 361–366. [DOI] [PubMed] [Google Scholar]
- Campbell, P., and Braam, J. (1999. b). In vitro activities of four xyloglucan endotransglycosylases from Arabidopsis. Plant J. 18 371–382. [DOI] [PubMed] [Google Scholar]
- Carpita, N., and McCann, M. (2000). The cell wall. In Biochemistry and Molecular Biology of Plants, B. Buchanan, W. Gruissem, and R. Jones, eds (Somerset, NJ: John Wiley & Sons), pp. 52–108.
- Collaborative Computational Project, Number 4 (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Chanliaud, E., de Silva, J., Strongitharm, B., Jeronimidis, G., and Gidley, M.J. (2004). Mechanical effects of plant cell wall enzymes on cellulose/xyloglucan composites. Plant J. 38 27–37. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6 850–861. [DOI] [PubMed] [Google Scholar]
- Cote, G.L., and Tao, B.Y. (1990). Oligosaccharide synthesis by enzymatic transglycosylation. Glycoconj. J. 7 145–162. [Google Scholar]
- Davies, G.J., Wilson, K.S., and Henrissat, B. (1997). Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321 557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva, J., Jarman, C.D., Arrowsmith, D.A., Stronach, M.S., Chengappa, S., Sidebottom, C., and Reid, J.S.G. (1993). Molecular characterization of a xyloglucan-specific endo-(1→4)-beta-d-glucanase (xyloglucan endotransglycosylase) from nasturtium seeds. Plant J. 3 701–711. [PubMed] [Google Scholar]
- Edwards, M., Dea, I.C.M., Bulpin, P.V., and Reid, J.S.G. (1985). Xyloglucan (amyloid) mobilization in the cotyledons of Tropaeolum majus L seeds following germination. Planta 163 133–140. [DOI] [PubMed] [Google Scholar]
- Edwards, M., Dea, I.C.M., Bulpin, P.V., and Reid, J.S.G. (1986). Purification and properties of a novel xyloglucan-specific endo-(1→4)-β-d-glucanase from germinated nasturtium seeds (Tropaeolum majus L.). J. Biol. Chem. 261 9489–9494. [PubMed] [Google Scholar]
- Fanutti, C., Gidley, M.J., and Reid, J.S.G. (1993). Action of a pure xyloglucan endo-transglycosylase (formerly called xyloglucan-specific endo-(1–4)-beta-d-glucanase) from the cotyledons of germinated nasturtium seeds. Plant J. 3 691–700. [DOI] [PubMed] [Google Scholar]
- Fanutti, C., Gidley, M.J., and Reid, J.S.G. (1996). Substrate subsite recognition of the xyloglucan endo-transglycosylase or xyloglucan-specific endo-(1->4)-beta-d-glucanase from the cotyledons of germinated nasturtium (Tropaeolum majus L) seeds. Planta 200 221–228. [DOI] [PubMed] [Google Scholar]
- Fauré, R., Saura-Valls, M., Brumer, H., Planas, A., Cottaz, S., and Driguez, H. (2006). Synthesis of a library of xylogluco-oligosaccharides for active-site mapping of xyloglucan endo-transglycosylase. J. Org. Chem. 71 5151–5161. [DOI] [PubMed] [Google Scholar]
- Fry, S.C. (1997). Novel ‘dot-blot’ assays for glycosyltransferases and glycosylhydrolases: Optimization for xyloglucan endotransglycosylase (XET) activity. Plant J. 11 1141–1150. [Google Scholar]
- Fry, S.C., Smith, R.C., Renwick, K.F., Martin, D.J., Hodge, S.K., and Matthews, K.J. (1992). Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 282 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, S.C., et al. (1993). An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plant. 89 1–3. [Google Scholar]
- Geisler-Lee, J., et al. (2006). Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 140 946–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloster, T.M., et al. (March 21, 2007). Characterisation and 3-D structures of two distinct bacterial xyloglucanases from families GH5 and GH12. J. Biol. Chem. http://dx.doi.org/10.1074/jbc.M700224200. [DOI] [PubMed]
- Gouet, P., Courcelle, E., Stuart, D.I., and Metoz, F. (1999). ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 15 305–308. [DOI] [PubMed] [Google Scholar]
- Greffe, L., Bessueille, L., Bulone, V., and Brumer, H. (2005). Synthesis, preliminary characterization, and application of novel surfactants from highly branched xyloglucan oligosaccharides. Glycobiology 15 437–445. [DOI] [PubMed] [Google Scholar]
- Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52 696–704. [DOI] [PubMed] [Google Scholar]
- Henriksson, H., Denman, S.E., Campuzano, I.D.G., Ademark, P., Master, E.R., Teeri, T.T., and Brumer, H. (2003). N-linked glycosylation of native and recombinant cauliflower xyloglucan endotransglycosylase 16A. Biochem. J. 375 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat, B., and Davies, G. (1997). Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7 637–644. [DOI] [PubMed] [Google Scholar]
- Hoffman, M., Jia, Z.H., Peña, M.J., Cash, M., Harper, A., Blackburn, A.R., Darvill, A., and York, W.S. (2005). Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr. Res. 340 1826–1840. [DOI] [PubMed] [Google Scholar]
- Hrmova, M., Farkaš, V., Lahnstein, J., and Fincher, G.B. (2007). A barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1,3;1,4)-beta-d-glucans. J. Biol. Chem. 282 12951–12962. [DOI] [PubMed] [Google Scholar]
- Irwin, D.C., Cheng, M., Xiang, B.S., Rose, J.K.C., and Wilson, D.B. (2003). Cloning, expression and characterization of a family-74 xyloglucanase from Thermobifida fusca. Eur. J. Biochem. 270 3083–3091. [DOI] [PubMed] [Google Scholar]
- Ishimaru, M., and Kobayashi, S. (2002). Expression of a xyloglucan endo-transglycosylase gene is closely related to grape berry softening. Plant Sci. 162 621–628. [Google Scholar]
- Johansson, P., Brumer, H., Baumann, M.J., Kallas, Å.M., Henriksson, H., Denman, S.E., Teeri, T.T., and Jones, T.A. (2004). Crystal structures of a poplar xyloglucan endotransglycosylase reveal details of transglycosylation acceptor binding. Plant Cell 16 874–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallas, Å.M., Piens, K., Denman, S.E., Henriksson, H., Fäldt, J., Johansson, P., Brumer, H., and Teeri, T.T. (2005). Enzymatic properties of native and deglycosylated hybrid aspen (Populus tremula × tremuloides) xyloglucan endotransglycosylase 16A expressed in Pichia pastoris. Biochem. J. 390 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooiman, P. (1960). On the occurrence of amyloids in plant seeds. Acta Bot. Neerl. 9 208–219. [Google Scholar]
- Kraulis, P. (1991). MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26 283–291. [Google Scholar]
- Lerouxel, O., Cavalier, D.M., Liepman, A.H., and Keegstra, K. (2006). Biosynthesis of plant cell wall polysaccharides—A complex process. Curr. Opin. Plant Biol. 9 621–630. [DOI] [PubMed] [Google Scholar]
- Lorences, E.P., and Fry, S.C. (1993). Xyloglucan oligosaccharides with at least 2 alpha-d-xylose residues act as acceptor substrates for xyloglucan endotransglycosylase and promote the depolymerization of xyloglucan. Physiol. Plant. 88 105–112. [Google Scholar]
- Lu, W., Wang, Y., Jiang, Y., Li, J., Liu, H., Duan, X., and Song, L. (2006). Differential expression of Litchi XET genes in relation to fruit growth. Plant Physiol. Biochem. 44 707–713. [DOI] [PubMed] [Google Scholar]
- Martinez-Fleites, C., Guerreiro, C.I.P.D., Baumann, M.J., Taylor, E.J., Prates, J.A.M., Ferreira, L.M.A., Fontes, C.M.G.A., Brumer, H., and Davies, G.J. (2006). Crystal structures of Clostridium thermocellum xyloglucanase, XGH74A, reveal the structural basis for xyloglucan recognition and degradation. J. Biol. Chem. 281 24922–24933. [DOI] [PubMed] [Google Scholar]
- Matthews, B.W. (1968). Solvent content in protein crystals. J. Mol. Biol. 33 491–497. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A., and Murphy, M.E.P. (1994). Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50 869–873. [DOI] [PubMed] [Google Scholar]
- Michel, G., Chantalat, L., Duee, E., Barbeyron, T., Henrissat, B., Kloareg, B., and Dideberg, O. (2001). The kappa-carrageenase of P. carrageenovora features a tunnel-shaped active site: A novel insight in the evolution of clan-B glycoside hydrolases. Structure 9 513–525. [DOI] [PubMed] [Google Scholar]
- Mohand, F.A., and Farkas, V. (2006). Screening for hetero-transglycosylating activities in extracts from nasturtium (Tropaeolum majus). Carbohydr. Res. 341 577–581. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994). AmoRe: An automated package for molecular replacement. Acta Crystallogr. A 50 157–163. [Google Scholar]
- Nelson, N. (1944). A photometric adaptation of the Somogyi method for the determination of glucose. J. Biol. Chem. 153 375–380. [Google Scholar]
- Nishikubo, N., Awano, T., Banasiak, A., Bourquin, V., Ibatullin, F., Funada, R., Brumer, H., Teeri, T.T., Hayashi, T., Sundberg, B., and Mellerowicz, E.J. (May 15, 2007). Xyloglucan endo-transglycosylase (XET) functions in gelatinous layers of tension wood fibers in poplar - A glimpse into the mechanism of the balancing act of trees. Plant Cell Physiol. http://dx.doi.org/10.1093/pcp/pcm055. [DOI] [PubMed]
- Nishitani, K., and Tominaga, R. (1992). Endoxyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J. Biol. Chem. 267 21058–21064. [PubMed] [Google Scholar]
- Nourizad, N., Ehn, M., Gharizadeh, B., Hober, S., and Nyren, P. (2003). Methylotrophic yeast Pichia pastoris as a host for production of ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum). Protein Expr. Purif. 27 229–237. [DOI] [PubMed] [Google Scholar]
- Pantoliano, M.W., Petrella, E.C., Kwasnoski, J.D., Lobanov, V.S., Myslik, J., Graf, E., Carver, T., Asel, E., Springer, B.A., Lane, P., and Salemme, F.R. (2001). High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 6 429–440. [DOI] [PubMed] [Google Scholar]
- Pauly, M., Albersheim, P., Darvill, A., and York, W.S. (1999. a). Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J. 20 629–639. [DOI] [PubMed] [Google Scholar]
- Pauly, M., Andersen, L.N., Kauppinen, S., Kofod, L.V., York, W.S., Albersheim, P., and Darvill, A. (1999. b). A xyloglucan-specific endo-beta-1,4-glucanase from Aspergillus aculeatus: Expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9 93–100. [DOI] [PubMed] [Google Scholar]
- Perrakis, A., Sixma, T.K., Wilson, K.S., and Lamzin, V.S. (1997). wARP: Improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr. D 53 448–455. [DOI] [PubMed] [Google Scholar]
- Planas, A. (2000). Bacterial 1,3–1,4-beta-glucanases: Structure, function and protein engineering. Biochim. Biophys. Acta 1543 361–382. [DOI] [PubMed] [Google Scholar]
- Popper, Z.A., and Fry, S.C. (2003). Primary cell wall composition of bryophytes and charophytes. Ann. Bot. (Lond.) 91 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper, Z.A., and Fry, S.C. (2004). Primary cell wall composition of pteridophytes and spermatophytes. New Phytol. 164 165–174. [DOI] [PubMed] [Google Scholar]
- Powell, H.R. (1999). The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr. D 55 1690–1695. [DOI] [PubMed] [Google Scholar]
- Rao, P.S., and Srivastava, H.C. (1973). Tamarind. In Industrial Gums—Polysaccharides and Their Derivatives, J.N. BeMiller, ed (New York: Academic Press), pp. 369–411.
- Reiter, W.D. (2002). Biosynthesis and properties of the plant cell wall. Curr. Opin. Plant Biol. 5 536–542. [DOI] [PubMed] [Google Scholar]
- Ren, Y.L., Picout, D.R., Ellis, P.R., and Ross-Murphy, S.B. (2004). Solution properties of the xyloglucan polymer from Afzelia africana. Biomacromolecules 5 2384–2391. [DOI] [PubMed] [Google Scholar]
- Rose, J.K.C., and Bennett, A.B. (1999). Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: Parallels between cell expansion and fruit ripening. Trends Plant Sci. 4 176–183. [DOI] [PubMed] [Google Scholar]
- Rose, J.K.C., Braam, J., Fry, S.C., and Nishitani, K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 43 1421–1435. [DOI] [PubMed] [Google Scholar]
- Rose, J.K.C., Brummell, D.A., and Bennett, A.B. (1996). Two divergent xyloglucan endotransglycosylases exhibit mutually exclusive patterns of expression in nasturtium. Plant Physiol. 110 493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel, A., and Cambillau, C., eds (1991). TURBO-FRODO program. In Silicon Graphics Geometry Partner Directory. (Mountain View, CA: Silicon Graphics), pp. 86–87.
- Saladié, M., Rose, J.K.C., Cosgrove, D.J., and Catala, C. (2006). Characterization of a new xyloglucan endotransglucosylase/hydrolase (XTH) from ripening tomato fruit and implications for the diverse modes of enzymic action. Plant J. 47 282–295. [DOI] [PubMed] [Google Scholar]
- Saura-Valls, M., Fauré, R., Ragas, S., Piens, K., Brumer, H., Teeri, T.T., Cottaz, S., Driguez, H., and Planas, A. (2006). Kinetic analysis using low-molecular mass xyloglucan oligosaccharides defines the catalytic mechanism of a Populus xyloglucan endotransglycosylase. Biochem. J. 395 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims, I.M., Munro, S.L.A., Currie, G., Craik, D., and Bacic, A. (1996). Structural characterisation of xyloglucan secreted by suspension-cultured cells of Nicotiana plumbaginifolia. Carbohydr. Res. 293 147–172. [DOI] [PubMed] [Google Scholar]
- Sinnott, M.L. (1990). Catalytic mechanisms of enzymatic glycosyl transfer. Chem. Rev. 90 1171–1202. [Google Scholar]
- Steele, N.M., Sulová, Z., Campbell, P., Braam, J., Farkaš, V., and Fry, S.C. (2001). Ten isoenzymes of xyloglucan endotransglycosylase from plant cell walls select and cleave the donor substrate stochastically. Biochem. J. 355 671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulova, Z., Lednicka, M., and Farkas, V. (1995). A colorimetric assay for xyloglucan-endotransglycosylase from germinating seeds. Anal. Biochem. 229 80–85. [DOI] [PubMed] [Google Scholar]
- Sundqvist, G., Stenvall, M., Berglund, H., Ottosson, J., and Brumer, H. (2007). A general, robust method for the quality control of intact proteins using LC-ESI-MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 10.1016/j.jchromb.2007.01.011. [DOI] [PubMed]
- Thompson, D.S. (2005). How do cell walls regulate plant growth? J. Exp. Bot. 56 2275–2285. [DOI] [PubMed] [Google Scholar]
- Tine, M.A.S., Silva, C.O., de Lima, D.U., Carpita, N.C., and Buckeridge, M.S. (2006). Fine structure of a mixed-oligomer storage xyloglucan from seeds of Hymenaea courbaril. Carbohydr. Polym. 66 444–454. [Google Scholar]
- Tuskan, G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604. [DOI] [PubMed] [Google Scholar]
- Vincken, J.P., York, W.S., Beldman, G., and Voragen, A.G.J. (1997). Two general branching patterns of xyloglucan, XXXG and XXGG. Plant Physiol. 114 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocadlo, D.J., Davies, G.J., Laine, R., and Withers, S.G. (2001). Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature 412 835–838. [DOI] [PubMed] [Google Scholar]
- Wang, Q., Ellis, P.R., Ross-Murphy, S.B., and Reid, J.S.G. (1996). A new polysaccharide from a traditional Nigerian plant food: Detarium senegalense Gmelin. Carbohydr. Res. 284 229–239. [DOI] [PubMed] [Google Scholar]
- Yaoi, K., Kondo, H., Noro, N., Suzuki, M., Tsuda, S., and Mitsuishi, Y. (2004). Tandem repeat of a seven-bladed beta-propeller domain in oligoxyloglucan reducing-end-specific cellobiohydrolase. Structure 12 1209–1217. [DOI] [PubMed] [Google Scholar]
- Yokoyama, R., and Nishitani, K. (2001). A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 42 1025–1033. [DOI] [PubMed] [Google Scholar]
- Yokoyama, R., Rose, J.K.C., and Nishitani, K. (2004). A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol. 134 1088–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, W.S., and Hawkins, R. (2000). Preparation of oligomeric beta-glycosides from cellulose and hemicellulosic polysaccharides via the glycosyl transferase activity of a Trichoderma reesei cellulase. Glycobiology 10 193–201. [DOI] [PubMed] [Google Scholar]
- York, W.S., Vanhalbeek, H., Darvill, A.G., and Albersheim, P. (1990). The structure of plant cell walls. 29. Structural analysis of xyloglucan oligosaccharides by H-1-NMR spectroscopy and fast atom bombardment mass spectrometry. Carbohydr. Res. 200 9–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.