Abstract
Although several potential telomere binding proteins have been identified in higher plants, their in vivo functions are still unknown at the plant level. Both knockout and antisense mutants of RICE TELOMERE BINDING PROTEIN1 (RTBP1) exhibited markedly longer telomeres relative to those of the wild type, indicating that the amount of functional RTBP1 is inversely correlated with telomere length. rtbp1 plants displayed progressive and severe developmental abnormalities in both germination and postgermination growth of vegetative organs over four generations (G1 to G4). Reproductive organ formation, including panicles, stamens, and spikelets, was also gradually and severely impaired in G1 to G4 mutants. Up to 11.4, 17.2, and 26.7% of anaphases in G2, G3, and G4 mutant pollen mother cells, respectively, exhibited one or more chromosomal fusions, and this progressively increasing aberrant morphology was correlated with an increased frequency of anaphase bridges containing telomeric repeat DNA. Furthermore, 35S:anti-RTBP1 plants expressing lower levels of RTBP1 mRNA exhibited developmental phenotypes intermediate between the wild type and mutants in all aspects examined, including telomere length, vegetative and reproductive growth, and degree of genomic anomaly. These results suggest that RTBP1 plays dual roles in rice (Oryza sativa), as both a negative regulator of telomere length and one of positive and functional components for proper architecture of telomeres.
INTRODUCTION
Telomeres, which form the extreme ends of linear eukaryotic chromosomes, consist of tandem repeats of short, G-rich sequence elements. They are responsible for preserving chromosome integrity and for protection against end-to-end fusion and recombination with other chromosomes and for exonucleolytic degradation (Blackburn, 1991; Greider, 1996). Telomeres are packaged into specialized nucleoprotein complexes, and nonhistone telomere binding proteins appear to be essential components for telomere structure and functions (Collins, 2000; Blackburn, 2001; Shore, 2001). Telomere binding proteins can be divided into two distinct classes according to their substrate specificities. Members of the first group, which bind to the single-stranded 3′ extension at telomere termini, are necessary for chromosome capping and telomerase regulation. The second class comprises double-stranded telomeric repeat binding proteins. Double-stranded telomere binding proteins, including yeast Rap1 and Taz1 and human TRF1/Pin2 and TRF2, have been extensively characterized (Shore, 1994; Chong et al., 1995; Bilaud et al., 1997; Cooper et al., 1997). These proteins are involved in the control of telomere length and stability in vivo. A negative feedback function that regulates telomere length has been identified for Rap1 and TRF1/Pin2, which inhibit the action of telomerase at the ends of telomeres (van Steensel and de Lange, 1997; Wotton and Shore, 1997; Smogorzewska et al., 2000). Recent studies have revealed that human telomere complex is composed of six proteins (de Lange, 2005). Three subunits, TRF1, TRF2, and POT1, directly interact with TTAGGG telomere repeats, while three additional subunits, TIN2, TPP1, and Rap1, are associated with a complex. This human telomere-protein complex, referred to as shelterin, exhibits DNA remodeling activity and acts to change the structure of the telomeric DNA, thereby protecting chromosome ends (de Lange, 2005).
Although the plant telomere repeat sequence TTTAGGG is highly conserved with that of human, its structural compositions and biological roles have remained much less well understood in higher plants. Recently, cDNAs encoding potential telomeric DNA binding proteins have been identified from several plant species, including rice (Oryza sativa), Arabidopsis thaliana, and tobacco (Nicotiana tabacum) (Yu et al., 2000; Chen et al., 2001; Hwang et al., 2001; Yang et al., 2003; Karamysheva et al., 2004; Kuchar and Fajkus, 2004; Schrumpfova et al., 2004; Hwang and Cho, 2007). Like human TRF1/Pin2 and TRF2, the predicted proteins possess a single DNA binding Myb-like domain, displaying sequence identity to double-stranded telomere binding proteins from yeast and human. In Arabidopsis, there are at least 12 TRF-like (TRFL) genes that fall into two different gene families (Karamysheva et al., 2004). Bacterially expressed proteins from TRFL family 1 formed homo- and heterodimers and specifically interacted with double-stranded plant telomeric repeats in vitro. These results suggest that plant telomeres are also comprised of telomere-specific protein components that contribute to their proper structure and functions. Possible in vivo functions of telomere binding protein Ng-TRF1 were shown in tobacco BY-2 suspension cells. Both 35S:Ng-TRF1 and 35S:anti-Ng-TRF1 transgenic BY-2 cells exhibited significant changes in telomere length and cell viability, indicating that Ng-TRF1 is involved in the control of telomere length and stability in tobacco cultured cells (Yang et al., 2004). However, in vivo functions of plant telomere binding proteins have yet to be determined at the plant level.
We aim to elucidate the physiological roles of telomere binding proteins with respect to telomere structure and functions in higher plants. In this report, we have used rice as a molecular genetic and cytological model system and obtained plants containing a T-DNA copy integrated into the RICE TELOMERE BINDING PROTEIN1 (RTBP1) gene and transgenic plants carrying 35S:RTBP1 and 35S:anti-RTBP1 constructs. Pulse-field gel electrophoresis showed that both knockout and antisense lines exhibited markedly longer telomeres compared with those of the wild-type plants. Homozygous rtbp1 lines displayed progressive and severe developmental abnormalities in both vegetative and reproductive organs accompanied by genome instability during four consecutive generations (G1 to G4). In G2 rtbp1 mutants, abnormal chromosome bridges were detected in 11.4% of anaphases examined, while the anaphase bridges increased to 17.2 and 26.7% in G3 and G4 mutants, respectively. These results could lead to a better understanding of RTBP1 function not only at the cellular level but also in the whole plant and suggest that RTBP1 participates in the control of telomere length and telomere stability in rice plants.
RESULTS
Isolation of a T-DNA Insertion Mutant of RTBP1 and Construction of 35S:RTBP1 and 35S:anti-RTBP1 Transgenic Rice Plants
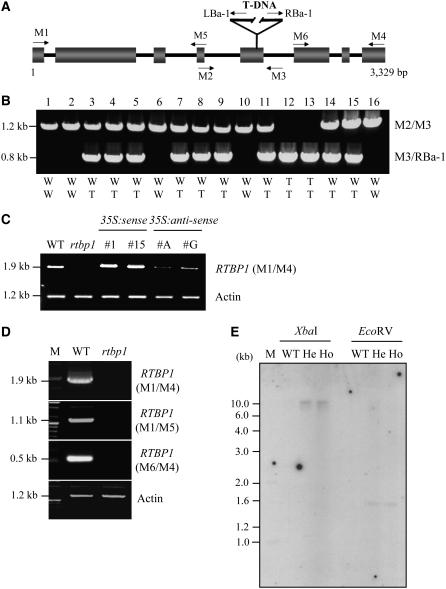
In the past few years, there has been a marked increase of interest in structure and functions of plant telomeres. Most of the work has dealt with the identification of the proteins that interact with telomere sequence. Consequently, a number of proteins that bind in vitro to oligonucleotides containing telomeric TTTAGGG repeats have been isolated from several plant species. Until now, however, only a few of these proteins have been shown to reflect a preference for a structural feature of plant telomeres in vivo. RTBP1 was previously identified as a double-stranded telomere binding protein in rice (Yu et al., 2000). However, its in vivo function was not known. It was therefore pertinent to establish if the presence or absence of RTBP1 affects the architecture of plant telomeres. To define the cellular functions of RTBP1 in rice, we employed a reverse genetic approach. The gene-specific primer M1 along with a T-DNA–specific primer LBa-1 were used to screen DNA pools from a collection of 20,500 T-DNA–transformed rice mutant lines (Jeon et al., 2000). A 2.2-kb PCR product was amplified using these primers. After PCR screening of successively smaller mutant pools, we were able to isolate a single rice line that contained a T-DNA insertion in the RTPB1 gene and referred to the mutant as rtbp1. The insertion was mapped to the fifth exon in RTBP1 located on chromosome 2 (line 2D-00626; Figure 1A). Plants homozygous for the T-DNA insertion were identified by multiplex PCR with primers M2, M3, and RBa-1 (Figure 1B). T-DNA disruption of RTBP1 was further verified by RT-PCR, demonstrating that the rice mutant seedlings contained a negligible amount of both full-length and partial RTBP1 mRNAs (Figures 1C and 1D). This indicates that rtbp1 is null for the RTBP1 gene. Genomic DNA gel blot analysis using a β-glucuronidase (GUS) cDNA probe confirmed that the rtbp1 mutant plants contained a single copy of T-DNA integrated into the RTBP1 gene (Figure 1E). We also established transgenic rice that overexpressed or suppressed RTBP1 by introducing a cauliflower mosaic virus 35S promoter-pRTBP1 construct in the sense (35S:RTBP1) or antisense (35S:anti-RTBP1) orientation. Elevated expression of RTBP1 was observed in 35S:RTBP1 transgenic lines, while a markedly lower level of mRNA was detected in the 35S:anti-RTBP1 plants (Figure 1C).
Figure 1.
Molecular Characterization of the T-DNA Insertion into the Rice RTBP1 Gene.
(A) Schematic representation of the genomic organization of RTBP1 with the position of the integrated T-DNA. Solid bars indicate exons. Solid lines represent introns. T-DNA insertion site and orientation of left and right borders are indicated. Gene-specific (M1, M2, M3, M4, M5, and M6) and T-DNA–specific (LBa-1 and RBa-1) primers used in the genotyping and RT-PCR are shown with arrows.
(B) Genotyping of the rtbp1 mutant plants. A set of gene-specific and T-DNA–specific primers used for genomic PCR are indicated. W, wild type; T, T-DNA insertion.
(C) RT-PCR analysis of RTBP1 and actin mRNAs in the wild type, homozygous G1 rtbp1 mutant line, and 35S:RTBP1 (#1 and #15), and 35S:anti-RTBP1 (#A and #G) T2 transgenic rice plants using M1 and M4 primers. The actin gene was used as a loading control.
(D) RT-PCR analysis of RTBP1 and actin mRNAs in the wild-type plant and homozygous G1 rtbp1 mutant line. In this experiment, two different primer sets (M1/M5 and M6/M4) were used for RTBP1, and no RT-PCR products were detected, indicating that rtbp1 was null for the RTBP1 gene. The actin gene was used as a loading control. M, molecular weight marker.
(E) Genomic DNA gel blot analysis in rice genomic DNA. Total leaf genomic DNAs were isolated from wild-type plants and heterozygous and homozygous G1 mutant lines. Rice genomic DNAs (10 μg per lane) were digested with various restriction enzymes as indicated, blotted onto nylon membranes, and hybridized with the 32P-labeled cDNA probe corresponding to the GUS gene. The blots were visualized by autoradiography. M, molecular weight marker; He, heterozygous G1 rtbp1 mutant; Ho; homozygous G1 rtbp1 mutant.
Knockout Mutation and Suppression of RTBP1 Resulted in Increased Telomere Length in Rice Plants
To address whether the altered expression of RTBP1 affects telomere metabolism in rice, we measured the length of telomeres in wild-type, rtbp1, 35S:RTBP1, and 35S:anti-RTBP1 plants. Total genomic DNA was isolated from each mutant or transgenic line, digested with the restriction enzyme TaqI, and resolved by pulse-field gel electrophoresis. DNA on the gel was then blotted and hybridized with the telomere repeat probe (TTTAGGG)70 (Figure 2) Samples from callus, leaf, and root of wild-type plants migrated as a typical telomeric smear, with fragments ranging from ∼5 to 10 kb. On the other hand, the rtbp1 plant showed markedly longer telomeres, whose lengths ranged between 10 and 30 kb in both heterozygous and homozygous G1 mutant populations (Figure 2A). These long telomeres were further maintained throughout the G2 to G4 plants, reaching a new stable set point (Figure 2B). Overexpression of antisense RTBP1 mRNA caused a significant enhancement of telomere elongation in T2 progeny, resulting in telomeres 8 to 25 kb long (Figure 2C). We interpret these results as evidence that there is an inverse correlation between the amount of functional RTBP1 and the length of telomeres in rice plants. On the other hand, telomeric DNA tracts were maintained at a constant size in T2 35S:RTBP1 transgenic plants (data not shown). This raises the possibility that the amount of RTBP1 might be saturated in the wild-type plants; hence, overexpression of RTBP1 did not exert any effects on the telomere length. With this in mind, we speculate that RTBP1 is involved in negative regulation of telomere length in rice plants.
Figure 2.
Telomere Length in the Wild Type, rtbp1 Mutant Line, and 35S:RTBP1 and 35S:anti-RTBP1 Transgenic Rice Plants.
(A) Telomere length in callus, leaf, and root of wild-type plants and G1 heterozygous and G1 homozygous rtbp1 mutant lines.
(B) Marked increase in telomere length in rtbp1 mutant plants over four successive generations (G1 to G4).
(C) Enhanced telomere elongation in T2 35S:anti-RTBP1 transgenic plants (#A and #G). Pulse-field gel electrophoresis was performed more than five times independently using randomly chosen individual rice plants.
Phenotypic Analysis of rtbp1
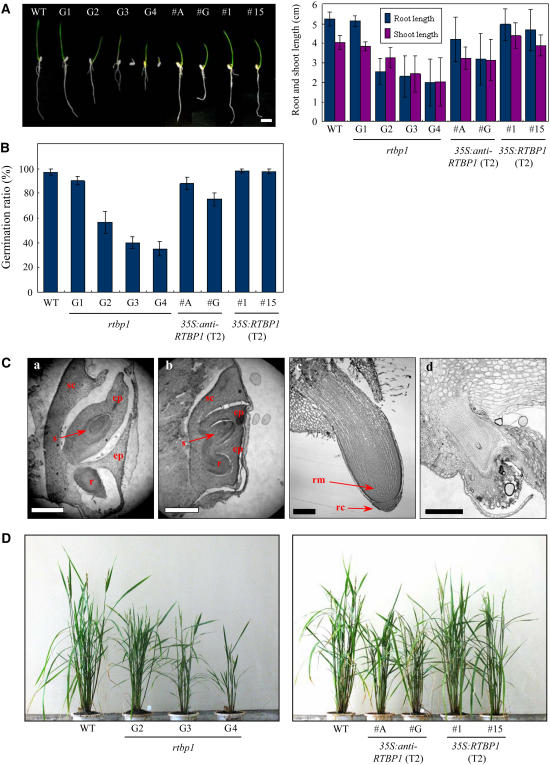
To explore the mechanistic outcome of decrease in the level of RTBP1 (Figure 1) and changes in telomere length (Figure 2), we examined the rtbp1 plants for phenotypic differences from the wild-type plants. We observed that rtbp1 displayed abnormal growth of vegetative organs. As presented in Figure 3A, rtbp1 exhibited a gradual retardation in growth of roots and shoots over four successive generations (G1 to G4). The aberrant growth of rtbp1 was generally apparent in early stages of development, resulting in severely shortened seedlings with less pronounced tap roots and increased numbers of lateral roots under the light growth conditions through G3 to G4 generations. The germination efficiency of the rtbp1 line, scored as radicle emergence from the seed coat, was also reduced progressively to 90, 55, 40, and 35% in G1, G2, G3, and G4, respectively (Figure 3B). Mutant seeds, which failed to germinate, were analyzed by microscopy. The results indicate that the G3 rtbp1 mutant seeds may not be dormant because radicle and coleoptile tissues appeared to be formed 1 d after imbibition (Figure 3C, b). At 2 d after imbibition, however, mutant radicle and coleoptile were unable to overcome the constraints of the surrounding tissues, resulting in the reduction of germination rate (Figure 3C, d). In addition, the root apical meristem and root cap were not normally developed in these G3 mutant seeds. The germinating G3 and G4 mutant seeds were able to develop intact seedlings, but their growth was significantly retarded compared with that of the wild type. Consequently, the height and number of leaves of mature 4-month-old G4 homozygous rtbp1 progeny grown in the greenhouse conditions displayed only ∼50% to those of wild-type rice (Figure 3D, left panel). Thus, both the germination and postgermination growth of the rtbp1 mutant line was markedly impaired compared with the wild-type plants.
Figure 3.
Developmental Defects of Vegetative Organs in the rtbp1 Mutant Line and 35S:anti-RTBP1 Transgenic T2 Rice Plants.
(A) Progressive and severe retardation of vegetative growth of the rtbp1 mutant during four successive generations. Wild type, rtbp1 (G1 to G4 generations), and transgenic T2 35S:anti-RTBP1 (#A and #G) and T2 35S:RTBP1 (#1 and #15) seedlings were germinated and grown in MS agar for 5 d under the light growth conditions (left panel), and the growth patterns of roots and shoots were monitored (right panel). The values are means ± sd (n = 50).
(B) Germination ratio of the wild type, rtbp1, and transgenic sense and antisense lines. The germination efficiency was scored as radicle emergence from the seed coat. The values are means ± sd (n = 50).
(C) Microscopy analysis of wild-type (a) and G3 rtbp1 mutant (b) seeds at 1 d after imbibition and of wild-type (c) and G3 mutant (d) roots at 2 d after imbibition. sc, scutellum; cp, coleoptile; ep, epiblast; s, shoot apex; r, radicle; rm, root meristem; rc, root cap. Bars = 500 μm in (a) and (b) and 200 μm in (c) and (d).
(D) Gross morphology of 4-month-old wild-type, three different generations (G2 to G4) of the rtbp1 mutant, and transgenic T2 35S:anti-RTBP1 (#A and #G) and T2 35S:RTBP1 (#1 and #15) plants grown under greenhouse conditions.
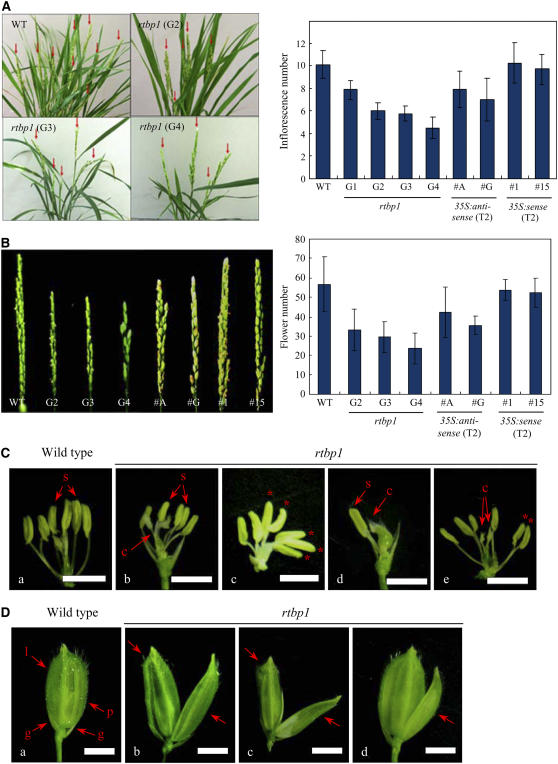
We next analyzed the development of reproductive organs. Under our experimental conditions, the wild-type rice contains 10 ± 1 panicles per plant. However, the average number of panicle in rtbp1 decreased gradually through the G1 to G4 generations, and the G4 mutant progeny contained only 4 ± 1 panicles per plant (Figure 4A). We also compared the formation of floral organs in the panicles between the wild-type and rtbp1 line. The number of flowers per panicle was significantly reduced in the rtbp1 mutant relative to that of the wild type. Normal rice has 55 ± 15 flowers per panicle on average, whereas the flower number per panicle was reduced markedly to 30 ± 8 and 24 ± 10 in the G3 and G4 mutant populations, respectively (Figure 4B). Interestingly, the morphology of rtbp1 flowers appeared to be different to that of wild-type flowers. The normal flower is typified by one pair of glumes, one lemma, one palea, and six stamens (Jeon et al., 2000). The flowers of the rtbp1 mutant, however, showed varying numbers of stamens; in the G3 mutant population, 6.8% of rtbp1 flowers contained a reduced number of stamens, while 9.5% of G4 mutant flowers had abnormal stamens (Figure 4C, Table 1). In some of the G3 and G4 flowers, there is only one stamen (Figure 4C, d), and sometimes the reduced number of stamen resulted in the combined anthers in the one filament (Figure 4C, c and e). In addition, an abnormal structure of spikelets was often detected in the G4 mutant due to the abnormal growth and structure of palea and lemma (Figure 4D, b, c and d), while we did not observe such abnormality in the wild-type flowers. The morphology of glumes, however, was similar to that of the wild type (Figure 4D). These results indicate that the development of reproductive organs occurs abnormally in the rtbp1 mutant. Overall, our phenotypic analysis implies that suppression of the RTBP1 gene yields severe defects in both vegetative and reproductive organs of rice plants.
Figure 4.
Developmental Defects of Floral Organs in the rtbp1 Mutant Line and 35S:anti-RTBP1 Transgenic T2 Rice Plants.
(A) Comparison of the wild type and rtbp1 (G2 to G4 generations) after bolting (left panel) and the number of panicle per plant in the wild type, rtbp1 (G2 to G4 generations), T2 35S:anti-RTBP1 (#A and #G), and T2 35S:RTBP1 (#1 and #15) rice (right panel). The values are means ± sd (n = 30).
(B) Panicle structure (left panel) and the number of flowers per panicle (right panel) in the wild type, rtbp1 (G2 to G4 generations), T2 35S:anti-RTBP1 (#A and #G), and T2 35S:RTBP1 (#1 and #15) rice. The values are means ± sd (n = 70).
(C) Close-up view of the stamens of wild-type (a), G3 rtbp1 (b and c), and G4 rtbp1 (d and e) flowers. Multiple anthers in one filament are indicated by asterisks. c, carpel; s, stamen. Bars = 0.5 cm.
(D) A spikelet from the wild type (a) and the G4 rtbp1 line (b, c, and d). g, glume; l, lemma; p, palea. Bars = 0.5 cm.
Table 1.
Frequency of Abnormal Stamen and Anaphase Bridges
| Genotype | Abnormal Stamen | Anaphase Bridge |
|---|---|---|
| Wild type | 0/60 (0%) | 0/80 (0%) |
| rtbp1 G2 | 2/62 (3.2%) | 8/70 (11.4%) |
| rtbp1 G3 | 4/59 (6.8%) | 10/58 (17.2%) |
| rtbp1 G4 | 6/63 (9.5%) | 16/60 (26.7%) |
| 35S:anti-RTBP1 #A | 1/58 (1.7%) | 2/73 (2.7%) |
| 35S:anti-RTBP1 #G | 3/72 (4.2%) | 3/68 (4.4%) |
As a next experiment, we investigated the phenotypic differences of 35S:anti-RTBP1 transgenic plants. Because 35S:anti-RTBP1 lines contained significantly lower levels of RTBP1 transcript relative to the wild type (Figure 1C), we expected that the antisense plants would also display growth defects. Figures 3A, 3B, and 3D (right panel) depict the visible alterations in the 35S:anti-RTBP1 transgenic plants during the vegetative stages. As was the case for the rtbp1 line, the growth and germination ratio of these antisense lines was significantly disturbed in the T2 generation. In addition, the T2 35S:anti-RTBP1 plants displayed disordered development of reproductive organs, including abnormal number and structure of panicles, flowers, and stamens (Figures 4A and 4B, Table 1). These defects, however, were generally less pronounced than those seen for the G2 rtbp1 mutant plant; hence, 35S:anti-RTBP1 transgenic lines exhibited a phenotype intermediate between that of wild-type and rtbp1 mutant plants. Thus, it is consistent with the notion that the proper level of RTBP1 is required for the normal growth and development of rice plants.
Finally, we examined the phenotype of 35S:RTBP1 transgenic plants. In contrast with what we observed in 35S:anti-RTBP1, the RTBP1-overexpressing lines were indistinguishable from wild-type plants in both vegetative and reproductive stages in terms of development and morphology (Figures 3A, 3B, 4A, 4B, and 4D). Thus, this is in line with the result that the 35S:RTBP1 plants possess telomeres of normal length. It is still conceivable that rice cells might require a much more increased level of RTBP1 for the alteration of their phenotype, but our experimental conditions were not optimal to detect such changes. This led us to hypothesize that the basal level of RTBP1 might be already saturated or high enough to play a functional role in the wild-type plants; hence, its overexpression may result in no visible effects on the 35S:RTBP1 T2 lines. Taken together, our mutant and transgenic analyses support the notion that RTBP1 plays a critical role in the development of both vegetative and reproductive organs in rice plants.
Cytogenetic Analysis
Riha et al. (2001) isolated the telomerase-deficient Arabidopsis At-TERT−/− mutants and analyzed their phenotypes. Although Arabidopsis can survive for up to 10 generations without telomerase, the last five generations show severe and progressive developmental abnormalities in both vegetative and reproductive organs, which are associated with massive genome instability. Detailed cytogenetic studies indicated that genomic instability in G8 telomerase mutants was caused by multiple rounds of the breakage-fusion-bridge cycle due to dysfunctional telomeres (Siroky et al., 2003). In addition, perturbation of At-Pot1 or At-Pot2 gene expression, which encode Pot (protection of telomeres)-like proteins in Arabidopsis, induced serious growth aberrations and sterility with bridged chromosomes and aneuploidy (Shakirov et al., 2005). These results indicate that the developmental anomalies correlated with telomere dysfunction are intimately tied with meiosis disorders in Arabidopsis. Thus, we considered the possibility that disturbed phenotypes of the rtbp1 rice mutant line are also accompanied by cytogenetic damage due to the structural dysfunction of telomeres resulting from the absence of telomere binding protein RTBP1.
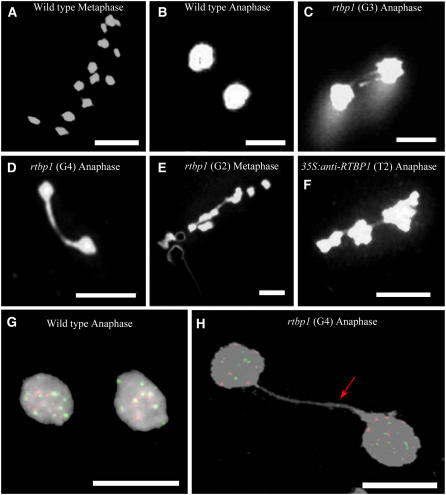
To investigate this possibility, we performed cytogenetic analysis. Meiotic progression in wild-type and rtbp1 pollen mother cells (PMCs) was monitored via 4′,6-diamidino-2-phenylindole (DAPI) staining of chromosomes. Figures 5A and 5B). show typical meiotic events in wild-type PMCs. All 12 chromosomes from wild-type cells were clearly separated throughout meiosis, resulting in the complete migration of chromosomes to the opposite pole of the cells in metaphase and anaphase, consistent with their normal morphology. On the other hand, meiotic anomalies were often detected in rtbp1 mutant cells. As shown in Table 1 and Figures 5C and 5D, we observed the presence of anaphase bridges in rtbp1 PMCs, which were not observed in wild-type cells. Among 70 anaphases examined from G2 mutant cells, eight anaphases (11.4%) displayed bridged chromosomes, while 17.2% (10/58) of G3 anaphases exhibited abnormal chromosome fusions (Figure 5C). Finally, in G4 mutant cells, the number of bridges further rose, and up to 26.7% (16/60) of anaphases contained one or more chromosome fusions (Figure 5D). Chromosome bridges were also detected in the mutant metaphase, although the frequency was lower (Figure 5E). Thus, increased frequency of anaphase bridges during four consecutive generations (G1 to G4) of rtbp1 PMCs is coupled with the gradually increased severity of the morphology of the mutant plants over these generations.
Figure 5.
Cytogenetic Analysis of Wild-Type, rtbp1 Mutant, and 35S:anti-RTBP1 T2 Transgenic PMCs.
(A) and (B) DAPI-stained metaphase and anaphase chromosomes of wild-type PMCs. Bars = 10 μM.
(C) and (D) A high incidence of anaphase chromosome fusions was detected in G3 (17.2%) and G4 (26.7%) rtbp1 mutants. Examples of anaphase chromosomes with two bridges (C) and one bridge (D) are presented. Bars = 10 μM.
(E) and (F) Abnormal chromosomal fusions were also detected during metaphase in rtbp1 mutant PMCs (E) and anaphase in 35S:anti-RTBP1 T2 transgenic PMCs (F). Bars = 10 μM.
(G) and (H) FISH analysis of anaphase chromosomes in the wild-type (left panel) and G4 rtbp1 (right panel) PMCs with the telomeric repeat (TTTAGGG)70 (green) and rice centromere-specific CentO satellite DNA (red) fluorescent probes. Telomeric repeat signal detected in the anaphase bridge is indicated by an arrow. No CentO signal was detected in the anaphase bridge. Bars = 5 μM.
Using the fluorescence in situ hybridization (FISH) technique, Vannier et al. (2006) recently observed anaphase bridges containing telomeric repeats in Arabidopsis tert and atrad50/tert mutant cells, which indicates that the abnormal chromosome-end fusions include telomeric repeats at the fusion points. To address whether the anaphase bridges detected in rtbp1 PMCs were the result of telomere dysfunction, we next performed a FISH experiment using two different kinds of probes: a telomeric DNA repeat probe containing (TTTAGGG)70 (the same probe as that used for pulse-field gel electrophoresis) and rice centromere-specific 165-bp CentO satellite DNA (Zhang et al., 2005). As shown in Figure 5H, we detected hybridization signal of the telomeric probe (green) in anaphase bridge of G4 rtbp1 PMCs, while we were not able to detect such hybridization of CentO probe (red), suggesting that the abnormal anaphase bridges in rtbp1 PMCs contain telomeric DNAs. Overall, these results support the notion that deficiency of RTBP1 resulted in telomere dysfunction, which in turn caused impaired growth of rtbp1 plants. In addition, chromosome bridges were also observed in the T2 generation of 35S:anti-RTBP1 lines (2.7 to 4.4%) (Figure 4F, Table 1). As was the case for development of vegetative and reproductive organs, the frequency of anaphase bridges in antisense lines was intermediate between the wild-type and G2 rtbp1 plants. These cytogenetic results argue that RTBP1 is crucially involved in the positive control of telomere stability in rice plants.
DISCUSSION
Telomeres have highly conserved architecture composed of tandemly arrayed G-rich sequences of DNA that are synthesized and maintained by the action of telomerase at the extremity of eukaryotic linear chromosomes. Telomeres of higher plants are made up of typical TTTAGGG arrays, although a few plants exceptionally contain TTAGGG vertebrate-type repeats (Weiss-Schneeweiss et al., 2004) or no detectable telomeric repeats (Pich et al., 1996). In humans, telomere structure and telomerase activity are closely coupled with the program of cellular proliferation, differentiation, senescence, and tumor growth. Despite limited insight into the plant telomeres compared with mammalian systems, plant telomeres have recently attracted much interest, since it is becoming increasingly apparent that the function and architectural features of telomeres as well as the patterns of telomerase regulation are largely conserved in higher plants and animals (reviewed in Riha and Shippen, 2003a; McKnight and Shippen, 2004; Gallego and White, 2005; Lamb et al., 2007; Zellinger and Riha, 2007). As in human and other animal species, the stable maintenance of telomeres is essential for growth and development in Arabidopsis, and a number of nuclear proteins, such as RAD50, KU70/80, MRE11, Pot1/2, ATM, and ATR, have been shown to be involved in telomere regulation (Gallego and White, 2001; Bundock et al., 2002; Bundock and Hooykaas, 2002; Riha and Shippen, 2003b; Gallego et al., 2003; Puizina et al., 2004; Shakirov et al., 2005; Vespa et al., 2005; Vannier et al., 2006). Disruption of the genes encoding these telomere-related proteins causes strikingly different developmental phenotypes accompanied by an abrupt onset of genome instability. Thus, protection and homeostasis of telomeres are crucial for the normal life cycle of Arabidopsis.
In addition to the above-mentioned telomere-related proteins, genes for putative double-stranded telomeric repeat binding factors have been isolated in several plant species. The majority of the work, however, focused on the in vitro characterization of the encoded proteins, which results in a lack of information about in vivo functions of the proteins (Yu et al., 2000; Chen et al., 2001; Hwang et al., 2001; Yang et al., 2003; Karamysheva et al., 2004; Kuchar and Fajkus, 2004; Schrumpfova et al., 2004). Recently, it was reported that telomere length and cell viability were markedly changed when expression of double-stranded telomere binding factor Ng-TRF1 was altered in transgenic tobacco BY-2 suspension culture cells, providing the first evidence for in vivo role of telomere binding protein in plants at the cellular level (Yang et al., 2004). However, functions of telomere binding proteins have yet to be determined at the whole-plant level. Rice is regarded as a model monocot cereal because of its economical value, small genome size, and ease of transformation (Jeon et al., 2000). Recent genomic analysis revealed that rice telomeric regions are significantly heterogeneous in both sequence and structure, suggesting the existence of dynamic mechanisms of controlling telomere length (Mizuno et al., 2006). In addition, most telomere studies in plants have been done with the dicot Arabidopsis. These factors prompted us to investigate the possible function of RTBP1 in rice.
In this study, we examined the effects of inactivating, silencing, or overexpressing RTBP1 on rice telomere, morphology, and development. We detected typical telomeric smears consisting of fragments 5 to 10 kb long in wild-type rice plants (Figure 2A), which agrees with the results determined by both fiber-FISH and terminal restriction fragment assay (Mizuno et al., 2006). However, we found that the rtbp1 mutant lines had markedly longer telomeres (10 to 30 kb), whereas the 35S:anti-RTBP1 constructs contained 8- to 25-kb telomeric tracts. Thus, the activity of the RTBP1 gene seems to be inversely related to telomere length, suggesting that RTBP1 is involved in a negative control of telomere length homeostasis in rice plants. This raises the tantalizing possibility that RTBP1 inhibits the action of telomerase at the extreme ends of telomeres in vivo, as is the case for human TRF1/PIN2 (van Steensel and de Lange, 1997; Wotton and Shore, 1997; Smogorzewska et al., 2000). It is worth noting that markedly lengthened telomeric DNA tracts were also found in heterozygous G1 progeny (Figure 2A). This suggests that control of telomere length was sensitive to cellular amount of RTBP1. In addition, although the severity of the phenotype of rtbp1 increased gradually during each of four successive generations, long telomeres in G1 plants were maintained at almost constant size in subsequent G2 to G4 generations of rtbp1 mutant (Figure 2B). Thus, an as yet unidentified RTBP1-independent factor(s) may also be in place that controls telomere homeostasis in rice plants. Isolation and characterization of telomere-related mutants other than rtbp1 will be necessary to examine this possibility.
Another striking phenotype of rtbp1 is severely disturbed development of vegetative and reproductive organs. Closer inspection revealed that both germination and postgermination growth of shoots and roots as well as generation of floral tissues, including stamens, spikelets, lemma, palea, and anthers, of homozygous rtbp1 lines were gradually and severely impaired over four successive generations (Figures 3 and 4). Furthermore, cytogenetic analysis showed that up to 26.7% of anaphases of the rtbp1 PMCs suffered one or more chromosomal fusions, which might cause a significant degree of genome instability (Figure 5, Table 1). With the aid of FISH analysis, we could detect the telomeric repeat sequence in the abnormal anaphase bridges of mutant PMCs (Figure 5H). Accumulating evidence in mammalian systems indicates that bridged anaphase chromosomes and unequal segregation of chromosomes are typical results from the fusion of unprotected chromosome termini due to structural defects of telomeres (Hande et al., 1999; Artandi et al., 2000). Arabidopsis mutants in which telomere-related genes are disrupted suffer genomic instability due to anaphase chromosomal fusions, as do the later generation progeny of telomerase-deficient mutants (Riha et al., 2001; Riha and Shippen, 2003b; Siroky et al., 2003; Puizina et al., 2004; Shakirov et al., 2005; Vespa et al., 2005; Vannier et al., 2006). In this regard, it should be noted that increased frequency of anaphase bridges in rtbp1 PMCs occurs in parallel with progressively increased aberrant morphology of the mutant rice plants (Table 1). In addition, although telomere length is reset to a new, longer size after just one generation of RTBP1 loss and the length of telomeres does not change in subsequent generations (Figure 2), the rate of chromosome fusion continues to increase. Thus, abnormal phenotypes of rtbp1 are not due to the increased telomere length per se but because of genome instability caused by uncapped telomeres. Based on these results, we propose that RTBP1 may play dual functions as a negative regulator of telomere length and as one of the required functional components for the proper architecture of telomeres in rice plants.
Because our extensive screening of the T-DNA–transformed 20,500 rice mutant collection yielded only one mutant line for RTBP1, we established transgenic 35S:anti-RTBP1 rice plants for comparison (Figure 1). It is interesting to note that the 35S:anti-RTBP1 lines exhibited a phenotype intermediate between the wild-type and rtbp1 knockout plants in all of the criteria examined, including telomere length (Figure 2), both vegetative and reproductive growth (Figures 3 and 4), and degree of genomic anomaly (Figure 5, Table 1). Because the 35S:anti-RTBP1 progeny contained lower but detectable levels of RTBP1 mRNA (Figure 1), these results, along with those obtained from rtbp1, provide evidence that the amount of functional RTBP1 is critically associated with the program of normal life cycle of rice plants.
Human telomeres are protected by sheltrin, a complex consisting of six telomere-associated proteins (de Lange, 2005). Sheltrin has a DNA remodeling activity and provides for the dynamic structural flexibility of the chromosome ends, which is necessary for the protection of telomeres. Electron microscopy examination of mammalian telomeric DNA revealed large duplex loops with tails, which were termed t-loops (Griffith et al., 1999). This structure was proposed to be a solution to the problem of telomere protection. Subsequent studies showed that human TRF2 is involved in the formation of t-loops (Zhu et al., 2000; Stansel et al., 2001). Telomeric DNA of higher plants is also arranged into t-loops. Recent electron microscopy studies showed frequent t-loops with highly varying sizes, ranging from 2 to 75 kb, in Pisum sativum telomeres and suggest that t-loops will be found in plant and animal species that contain detectable telomeric repeat sequences (Cesare et al., 2003). In the case of the P. sativum t-loops, the largest loops detected were ∼85 kb. It seems likely that there is some active mechanism to generate such large structures. Thus, it would be of interest to investigate whether RTBP1 participates in t-loop formation of rice telomeres.
It was reported that, although telomere tracts are fairly uniformly distributed in 14 wild accessions of Arabidopsis, the Wassilewskija ecotype exhibited a bimodal size distribution, with different individuals harboring telomeres with 2 to 5 kb or 4 to 9 kb, indicating that telomere length is not modulated by a single genetic factor (Shakirov and Shippen, 2004). In addition, analysis of the subtelomeric region of 35 wild-type accessions revealed higher structural variation relative to centromere region, which implies the variety of genomic events that drive the fluidity of chromosome termini (Kuo et al., 2006). Maillet et al. (2006) analyzed telomere lengths in 16 different interecotype crosses between Arabidopsis plants with different telomere sizes and observed that the interecotype hybrid plants presented a new telomere-length set point, intermediate between that of the two parent plants. These recent studies support the notion that plant telomeres are structurally and functionally dynamic. Therefore, further functional characterizations of RTBP1 and identification of the proteins it interacts with will be critical to a more complete understanding of such dynamic structures and functions of telomeres in rice, a model monocot cereal plant.
METHODS
Plant Growth
Dry rice (Oryza sativa var japonica cv Dongjin) seeds were soaked once with 70% ethanol and then rinsed extensively with sterilized water. Seeds of the wild type, rtbp1 mutants, 35S:RTBP1, and 35S:anti-RTBP1 were germinated on Murashige and Skoog medium containing MS basal salt (Wako Pure Chemical), 3% sucrose, 0.2% phytogel, and 0.55 mM myo-inositol. The seedlings were grown for 1 week at 25°C under continuous light and then transplanted to soil in the greenhouse and raised to maturity.
Total RNA Isolation and RT-PCR
Total RNAs of wild-type, rtbp1, 35S:RTBP1, and 35S:anti-RTBP1 rice leaves were obtained by a method described previously (Hong et al., 2005). The first-strand cDNA, synthesized from 10 μg of total RNA, was PCR amplified with RTBP1-specific primers using high-fidelity Ex-Tag polymerase (Takara) as described (Yang et al., 2002).
Genomic DNA Isolation and Pulse-Field Gel Electrophoresis
Total genomic DNA was isolated from mature rice leaves as indicated previously (Hong et al., 2005) and digested with TaqI restriction enzyme. The DNA fragments (10 μg per lane) were separated by pulse-field gel electrophoresis using a CHEF-DRIII system (Bio-Rad) according to Yang et al. (2004). The gel was blotted onto a nylon membrane filter (Bio-Rad) and then hybridized to a 32P-labeled (TTTAGGG)70 fragment under high stringency conditions (Yang et al., 2004).
PCR Screening for the rtbp1 Mutant Line
The primer M1 (5′-ATGGTGTTGCAGAAGAGGTTGGAC-3′) was used in combination with a T-DNA left border primer LBa-1 (5′-TCCGAAACTATCAGTGTCTAGCTAGA-3′) in a PCR screen on pooled rice DNA (Jeon et al., 2000) to identify rice lines containing a T-DNA copy integrated into the RTBP1 gene. Using primers M1 and LBa-1, a 2.2-kb PCR fragment was specifically amplified from the largest DNA pool and detected in consecutive smaller DNA pools until a single positive rice plant was identified. Plants homozygous for the T-DNA insertion were identified by multiplex PCR with primers M2 (5′-AAAACCCTTAGATGACCACATACA-3′), M3 (5′-AGTTAACCATCAAATCTTTCAACA-3′), and RBa-1 (5′-GGGTTGGGGTTTCTACAGGACGTAAC-3′) (Figure 2B).
Genomic DNA Gel Blot Analysis
Rice leaf genomic DNA (10 μg per lane) was digested with appropriate enzymes, separated by electrophoresis on a 0.7% agarose gel, and blotted onto a nylon membrane filter (Amersham). The filter was hybridized with 32P-labeled GUS cDNA probe under high stringency conditions as described by Yang et al. (2003). The GUS probe was prepared from 1.8-kb BamH1-EcoRI fragment (Jeon et al., 2000). The blot was visualized by autoradiography at −80°C using Kodak XAR-5 film and an intensifying screen.
Generation of the 35S:RTBP1 and 35S:anti-RTBP1 Constructs and Transformation of Rice
The full-length pRTBP1 cDNA was inserted into the corresponding sites of the binary vector pCambia in the sense or antisense orientation. The fusion gene construct was transferred to Agrobacterium tumefaciens strain AGL1 by electroporation as described by Cho et al. (2006). Rice transformation was accomplished by Agrobacterium-mediated cocultivation methods as previously described (Jeon et al., 2000). All transgenic rice plants were generated on a medium containing 40 mg L−1 hygromycin B. The regenerated plants were grown in a greenhouse of typically 30°C during the day and 20°C at night. The light/dark cycle in the greenhouse was 14/10 h.
Cytogenetic Studies
Anaphase and metaphase spreads were obtained from PMCs of the wild type and the rtbp1 mutant and stained with DAPI as described (Martinez-Zapater and Salinas, 1998) with some modifications. Briefly, anthers were fixed in 1:3 (v/v) acetic acid:ethanol for 2 h and soaked in distilled water, followed by digestion with 2% cellulose, 1.5% macerozyme, 0.3% pectolyase, and 1 mM EDTA, pH 4.2, in distilled water for 50 to 60 min. After being squashed on slide glass, samples were stained with DAPI and observed by fluorescence microscopy (Axio Imager; Zeiss).
FISH
The telomeric probe (TTTAGGG)70 and rice centromere-specific 165-bp CentO satellite DNA were labeled with biotin-16-dUTP or digoxigenin-11-dUTP using a nick translation system (Invitrogen). CentO DNA was obtained by PCR using genomic DNA as a template (forward primer, 5′-CACGTGGGTGCGATGTTTTTG-3′; reverse primer, 5′-TGCCAATATTTGCATTAATTG-3′). FISH analysis was performed with slides obtained as described above for DAPI staining according to Koo et al. (2004). Chromosomal DNA on the slides was denatured with 70% formamide at 70°C for 2.5 min, followed by dehydration in a 70, 85, 95, and 100% ethanol series at −20°C for 3 min each. The probe mixture was applied to the denatured chromosomal DNA and covered with a glass cover slip. Slides were placed in a humid chamber at 37°C for 18 h. Probes were detected with avidin-FITC or anti-digoxigenin-Cy3 (Roche). The chromosomes were counterstained with DAPI.
Accession Numbers
The GenBank accession numbers for the Arabidopsis TERT, tobacco Ng-TRF1, and RTBP1 are NM121691, AF543195, and AF242298, respectively.
Acknowledgments
We thank June M. Kwak (University of Maryland, College Park) and the members of W.T.K.'s laboratory for their help and discussion. This work was supported by grants from the Plant Diversity Research Center (21st Century Frontier Research Program funded by the Ministry of Science and Technology of Korea) and the BioGreen 21 Program (funded by the Rural Development Administration of the Republic of Korea) to W.T.K. and from the Crop Functional Genomics Program (CG1111) to G.A. M.Y.B. was a recipient of a BK21 graduate student scholarship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Woo Taek Kim (wtkim@yonsei.ac.kr).
References
- Artandi, S.E., Chang, S., Lee, S.L., Alson, S., Gottlieb, G.J., Chin, L., and Depinho, R.A. (2000). Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406 641–645. [DOI] [PubMed] [Google Scholar]
- Bilaud, T., Brun, C., Ancelin, K., Koering, C.E., Laroche, T., and Gilson, E. (1997). Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17 236–239. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. (1991). Structure and function of telomeres. Nature 350 569–573. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. (2001). Switching and signaling at the telomere. Cell 106 661–673. [DOI] [PubMed] [Google Scholar]
- Bundock, P., and Hooykaas, P. (2002). Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock, P., van Attikum, H., and Hooykaas, P. (2002). Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res. 30 3395–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare, A.J., Quinney, N., Willcox, S., Subramanian, D., and Griffith, J.D. (2003). Telomere looping in P. sativum (common garden pea). Plant J. 36 271–279. [DOI] [PubMed] [Google Scholar]
- Chen, C.M., Wang, C.T., and Ho, C.H. (2001). A plant gene encoding a myb-like protein that binds telomeric GGTTTAG repeats in vitro. J. Biol. Chem. 276 16511–16519. [DOI] [PubMed] [Google Scholar]
- Cho, S.K., Chung, H.S., Ryu, M.Y., Park, M.J., Lee, M.M., Bahk, Y.-Y., Kim, J., Pai, H.S., and Kim, W.T. (2006). Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-box E3 ubiquitin ligase homolog. Plant Physiol. 142 1664–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, L., van Steensel, B., Broccoli, D., Erdjument-Bromage, H., Hanish, J., Tempst, P., and de Lange, T. (1995). A human telomeric protein. Science 270 1663–1667. [DOI] [PubMed] [Google Scholar]
- Collins, K. (2000). Mammalian telomeres and telomerase. Curr. Opin. Cell Biol. 12 378–383. [DOI] [PubMed] [Google Scholar]
- Cooper, J.P., Nimmo, E.R., Allshire, R.C., and Cech, T.R. (1997). Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385 744–747. [DOI] [PubMed] [Google Scholar]
- de Lange, T. (2005). Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 19 2100–2110. [DOI] [PubMed] [Google Scholar]
- Gallego, M.E., Jalut, N., and White, C.I. (2003). Telomerase dependence of telomere lengthening in Ku80 mutant Arabidopsis. Plant Cell 15 782–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego, M.E., and White, C.I. (2001). RAD50 function is essential for telomere maintenance in Arabidopsis. Proc. Natl. Acad. Sci. USA 98 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego, M.E., and White, C.I. (2005). DNA repair and recombination functions in Arabidopsis telomere maintenance. Chromosome Res. 13 481–491. [DOI] [PubMed] [Google Scholar]
- Greider, C.W. (1996). Telomere length regulation. Annu. Rev. Biochem. 65 337–365. [DOI] [PubMed] [Google Scholar]
- Griffith, J.D., Comeau, L., Rosenfield, S., Stansel, R.M., Bianchi, A., Moss, H., and de Lange, T. (1999). Mammalian telomeres end in a large duplex loop. Cell 97 503–514. [DOI] [PubMed] [Google Scholar]
- Hande, M.P., Samper, E., Lansdorp, P., and Blasco, M.A. (1999). Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J. Cell Biol. 144 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, J.-P., Kim, S.M., Ryu, M.Y., Choe, S., Park, P.B., An, G., and Kim, W.T. (2005). Structure and expression of OsMRE11 in rice. J. Plant Biol. 48 229–236. [Google Scholar]
- Hwang, M.G., and Cho, M.H. (2007). Arabidopsis thaliana telomeric DNA-binding protein 1 is requited for telomere length homeostasis and its Myb-extension domain stabilizes plant telomeric DNA binding. Nucleic Acids Res. 35 1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, M.G., Chung, I.K., Kang, B.G., and Cho, M.H. (2001). Sequence-specific binding property of Arabidopsis thaliana telomeric DNA binding protein 1 (AtTBP1). FEBS Lett. 503 35–40. [DOI] [PubMed] [Google Scholar]
- Jeon, J.S., et al. (2000). T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22 561–570. [DOI] [PubMed] [Google Scholar]
- Karamysheva, Z.N., Surovtseva, Y.V., Vespa, L., Shakirov, E.V., and Shippen, D.E. (2004). A C-terminal Myb-extension domain defines a novel family of double-strand telomeric DNA binding proteins in Arabidopsis. J. Biol. Chem. 279 47799–47807. [DOI] [PubMed] [Google Scholar]
- Koo, D.-H., Plaha, P., Lim, Y.P., Hur, Y., and Bang, J.-W. (2004). A high-resolution karyotype of Brassica rapa ssp. pekinensis revealed by pachytene analysis and multicolor fluorescence in situ hybridization. Theor. Appl. Genet. 109 1346–1352. [DOI] [PubMed] [Google Scholar]
- Kuchar, M., and Fajkus, J. (2004). Interactions of putative telomere-binding proteins in Arabidopsis thaliana: Identification of functional TRF2 homolog in plants. FEBS Lett. 578 311–315. [DOI] [PubMed] [Google Scholar]
- Kuo, H.-F., Olsen, K.M., and Richards, E.J. (2006). Natural variation in a subtelomeric region of Arabidopsis: Implications for the genomic dynamics of a chromosome end. Genetics 173 401–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J.C., Yu, W., Han, F., and Birchler, J.A. (2007). Plant chromosomes from end to end: Telomeres, heterochromatin and centromeres. Curr. Opin. Plant Biol. 10 116–122. [DOI] [PubMed] [Google Scholar]
- Maillet, G., White, C.I., and Gallego, M.E. (2006). Telomere-length regulation in inter-ecotype crosses of Arabidopsis. Plant Mol. Biol. 62 859–866. [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater, J.M., and Salinas, J. (1998). Arabidopsis Protocol. (Totowa, NJ: Human Press).
- McKnight, T.D., and Shippen, D.E. (2004). Plant telomere biology. Plant Cell 16 794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, H., Wu, J., Kanamori, H., Fujisawa, M., Namiki, N., Saji, S., Katagiri, S., Katayose, Y., Sasaki, T., and Matsumoto, T. (2006). Sequencing and characterization of telomere and subtelomere regions on rice chromosomes 1S, 2S, 2L, 6L, 7S, 7L and 8S. Plant J. 46 206–217. [DOI] [PubMed] [Google Scholar]
- Pich, U., Fuchs, J., and Schubert, I. (1996). How do Alliaceae stabilize their chromosome ends in the absence of TTTAGGG sequences? Chromosome Res. 4 207–213. [DOI] [PubMed] [Google Scholar]
- Puizina, J., Siroky, J., Mokros, P., Schweizer, D., and Riha, K. (2004). Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha, K., McKnight, T.D., Griffing, L.R., and Shippen, D.E. (2001). Living with genome instability: Plant response to telomere dysfunction. Science 291 1797–1800. [DOI] [PubMed] [Google Scholar]
- Riha, K., and Shippen, D.E. (2003. a). Telomere structure, function and maintenance in Arabidopsis. Chromosome Res. 11 263–275. [DOI] [PubMed] [Google Scholar]
- Riha, K., and Shippen, D.E. (2003. b). Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. USA 100 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrumpfova, P., Kuchar, M., Mikova, G., Skrisovska, L., Kubicarova, T., and Fajkus, J. (2004). Characterization of two Arabidopsis thaliana myb-like proteins showing affinity to telomeric DNA sequence. Genome 47 316–324. [DOI] [PubMed] [Google Scholar]
- Shakirov, E., and Shippen, D.E. (2004). Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 16 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov, E.V., Surovtseva, Y.V., Osbun, N., and Shippen, D.E. (2005). The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell. Biol. 25 7725–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore, D. (1994). Rap1: A protean regulator in yeast. Trends Genet. 10 408–412. [DOI] [PubMed] [Google Scholar]
- Shore, D. (2001). Telomeric chromatin: Replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev. 11 189–198. [DOI] [PubMed] [Google Scholar]
- Siroky, J., Zluvova, J., Riha, K., Shippen, D.E., and Vyskot, B. (2003). Rearrangement of ribosomal DNA clusters in lare generation telomerase-deficient Arabidopsis. Chromosoma 112 116–123. [DOI] [PubMed] [Google Scholar]
- Smogorzewska, A., van Steensel, B., Bianchi, A., Oelmann, S., Schaefer, M.R., Schnapp, G., and de Lange, T. (2000). Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansel, R.M., de Lange, T., and Griffith, J.D. (2001). T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20 5532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier, J.-B., Depeiges, A., White, C., and Gallego, M.E. (2006). Two roles for Rad50 in telomere maintenance. EMBO J. 25 4577–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel, B., and de Lange, T. (1997). Control of telomere length by the human telomeric protein TRF1. Nature 385 740–743. [DOI] [PubMed] [Google Scholar]
- Vespa, L., Couvillion, M., Spangler, E., and Shippen, D.E. (2005). ATM and ATR make distinct contributions to chromosome end protection and the maintenance of telomeric DNA in Arabidopsis. Genes Dev. 19 2111–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss-Schneeweiss, H., Riha, K., Jang, C.G., Puizina, J., Scherthan, H., and Schweizer, D. (2004). Chromosome termini of the monocot plant Othocallis siberica are maintained by telomerase, which specifically synthesises vertebrate-type telomere sequences. Plant J. 37 484–493. [DOI] [PubMed] [Google Scholar]
- Wotton, D., and Shore, D. (1997). A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11 748–760. [DOI] [PubMed] [Google Scholar]
- Yang, S.W., Jin, E.S., Chung, I.K., and Kim, W.T. (2002). Cell cycle-dependent regulation of telomerase activity by auxin, ABA and protein phosphorylation in tobacco BY-2 suspension culture cells. Plant J. 29 617–626. [DOI] [PubMed] [Google Scholar]
- Yang, S.W., Kim, D.H., Lee, J.J., Chun, Y.J., Lee, J.-H., Kim, Y.J., Chung, I.K., and Kim, W.T. (2003). Expression of the telomeric repeat binding factor gene NgTRF1 is closely coordinated with the cell division program in tobacco BY-2 suspension culture cells. J. Biol. Chem. 278 21395–21407. [DOI] [PubMed] [Google Scholar]
- Yang, S.W., Kim, S.K., and Kim, W.T. (2004). Perturbation of NgTRF1 expression induces apoptosis-like cell death in tobacco BY-2 cells and implicates NgTRF1 in the control of telomere length and stability. Plant Cell 16 3370–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, E.Y., Kim, S.E., Kim, J.H., Ko, J.H., Cho, M.H., and Chung, I.K. (2000). Sequence-specific DNA recognition by the Myb-like domain of plant telomeric protein RTBP1. J. Biol. Chem. 275 24208–24214. [DOI] [PubMed] [Google Scholar]
- Zellinger, B., and Riha, K. (2007). Composition of plant telomeres. Biochim. Biophys. Acta 1769 399–409. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Yi, C., Bao, W., Liu, B., Cui, J., Yu, H., Cao, X., Gu, M., Liu, M., and Cheng, Z. (2005). The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata. Plant Physiol. 139 306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X.-D., Kuster, B., Mann, M., Petrini, J.H.J., and de Lange, T. (2000). Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25 347–352. [DOI] [PubMed] [Google Scholar]