Abstract
Benzothiadiazole (BTH) is a so-called plant activator and protects plants from diseases by activating the salicylic acid (SA) signaling pathway. By microarray screening, we identified BTH- and SA-inducible WRKY transcription factor (TF) genes that were upregulated within 3 h after BTH treatment. Overexpression of one of them, WRKY45, in rice (Oryza sativa) markedly enhanced resistance to rice blast fungus. RNA interference–mediated knockdown of WRKY45 compromised BTH-inducible resistance to blast disease, indicating that it is essential for BTH-induced defense responses. In a transient expression system, WRKY45 activated reporter gene transcription through W-boxes. Epistasis analysis suggested that WRKY45 acts in the SA signaling pathway independently of NH1, a rice ortholog of Arabidopsis thaliana NPR1, which distinguishes WRKY45 from known Arabidopsis WRKY TFs. Two defense-related genes, encoding a glutathione S-transferase and a cytochrome P450, were found to be regulated downstream of WRKY45 but were not regulated by NH1, consistent with the apparent independence of the WRKY45- and NH1-dependent pathways. Defense gene expression in WRKY45-overexpressed rice plants varied with growth conditions, suggesting that some environmental factor(s) acts downstream of WRKY45 transcription. We propose a role for WRKY45 in BTH-induced and SA-mediated defense signaling in rice and its potential utility in improving disease resistance of rice, an importance food resource worldwide.
INTRODUCTION
Plants respond to microbial pathogen attack by activating a variety of defense responses that are mediated through multiple signaling pathways. In many dicot plants, salicylic acid (SA) plays a crucial role in mediating one of the signaling pathways leading to defense responses, including induction of PATHOGENESIS-RELATED (PR) genes. After pathogen infection, endogenous levels of SA and its conjugates increase markedly in dicots, such as tobacco (Nicotiana tabacum), cucumber (Cucumis sativus), and Arabidopsis thaliana, preceding the induction of PR genes and the onset of disease resistance (Malamy et al., 1990; Métraux et al., 1990; Rasmussen et al., 1991). Exogenous application of SA induces PR gene expression and disease resistance in dicots. In contrast with dicots, which contain low basal levels of SA, rice has basal levels of SA two orders of magnitude higher (Raskin et al., 1990). SA levels do not increase after inoculation of rice with either the bacterial pathogen Pseudomonas syringae or the fungal pathogens Magnaporthe grisea and Rhizoctonia solani (Silverman et al., 1995). By contrast, SA levels in the leaves of 28 rice (Oryza sativa) cultivars were correlated with general blast resistance, suggesting that SA may serve as a chemical barrier against pathogen infection (Silverman et al., 1995). Yang et al. (2004) proposed that SA modulates redox balance and protects rice plants from oxidative stress such as that elicited by M. grisea.
Benzothiadiazole (BTH) is a functional analog of SA and one of the so-called plant activators that protect various plants from infectious diseases (Görlach et al., 1996; Lawton et al., 1996). In dicots, plant activators induce defense responses by activating the SA signaling pathway. When BTH is applied to plants at high dosages, it induces constitutive activation of defense responses, including several defense-related genes (direct defense; Kohler et al., 2002; van Hulten et al., 2006). By contrast, when BTH is applied at relatively low dosages, plants' defense responses do not activate immediately but become apparent only after pathogen infection (Conrath et al., 2002). This mode of action, which is common among plant activators, is termed potentiation or priming. In rice, BTH enhances resistance to R. solani (Rohilla et al., 2002) and M. grisea (Schweizer et al., 1999) and is used in the field, but the molecular mechanisms underlying its action remain to be investigated.
In dicots, several regulatory proteins have been implicated in the transcriptional regulation of defense genes under the control of the SA signaling pathway. Among those is Arabidopsis NPR1, an important positive regulator in this pathway that is required for transducing the SA signal to downstream PR gene activation (Cao et al., 1997). Chern et al. (2001) have reported that overexpression of Arabidopsis NPR1 in rice enhances resistance to Xanthomonas oryzae. In addition, Arabidopsis NPR1 interacts with the bZIP-type transcription factors (TFs) of rice, as in Arabidopsis (Chern et al., 2001). Those investigators have shown that NH1, a rice ortholog of NPR1, also interacts with a bZIP-type TF in rice and that NH1 overexpression in rice confers high levels of resistance to X. oryzae (Chern et al., 2005). These observations suggest that rice has a signaling pathway for disease resistance that is similar to the SA-dependent pathway in dicots.
The WRKY family of TFs has been suggested to play a role in controlling the transcription of defense genes through the W-box in their promoters, which is a key cis-element for defense-related transcriptional regulation (Rushton et al., 1996; Eulgem et al., 1999; Maleck et al., 2000). In Arabidopsis, the WRKY family consists of an estimated 74 members that fall into three major groups on a structural basis (Eulgem, 2005). A large number of WRKY genes are expressed in response to pathogen infection (Kalde et al., 2003), and the function of several WRKY TFs has been implicated in the defense reactions in Arabidopsis (Yu et al., 2001; Chen and Chen, 2002; Robatzek and Somssich, 2002). An Arabidopsis group III WRKY TF, WRKY70, activates SA-regulated genes downstream of NPR1 and represses jasmonic acid (JA)–responsive genes, integrating SA and JA signaling during systemic acquired resistance (Li et al., 2004). Recent studies have placed several WRKY TFs, including At WRKY70, downstream of NPR1 in the SA signaling pathway on the basis of transcriptional profiling using Arabidopsis npr1 mutant plants (Wang et al., 2006). Based on chromatin immunoprecipitation experiments in parsley (Petroselinum crispum), it has been proposed that WRKY TFs act in a network of mutually competing participants with temporal displacement occurring at defined cis-elements preoccupied by other family members in a stimulus-dependent manner (Turck et al., 2004; Ülker and Somssich, 2004). More recent works have shown various roles of WRKY TFs in defense programs of Arabidopsis (Journot-Catalino et al., 2006; Wang et al., 2006; Xu et al., 2006). The WRKY TFs form a superfamily in rice, as in dicots (Xie et al., 2005). Several WRKY genes are expressed in response to the rice blast fungal elicitor (Kim et al., 2000), infection with causal agents of bacterial blight and fungal blast diseases (Wen et al., 2003; Ryu et al., 2006), and defense signal molecules SA and JA (Ryu et al., 2006). Os WRKY03 (Liu et al., 2005) and Os WRKY71 (Liu et al., 2006) were functionally characterized, and both were placed upstream of NH1. However, our knowledge of the functions of WRKY TFs in the defense programs in rice is limited.
In an attempt to elucidate the molecular mechanisms underlying BTH-induced disease resistance, we identified WRKY45, which is transcriptionally upregulated by BTH and SA. Induction of rice blast resistance by BTH was abolished in WRKY45 knockdown rice plants, and overexpression of WRKY45 markedly enhanced resistance, indicating that this WRKY TF plays a crucial role in the BTH-inducible defense program of rice. Epistasis analysis revealed that WRKY45 is involved in a signaling pathway downstream of SA but independent of NH1. The presence of two apparently independent pathways was further supported by the fact that different genes were regulated by the two transcriptional regulators. These findings distinguish WRKY45 from any known WRKY TFs in Arabidopsis and suggest that the signaling pathways downstream of SA are evolutionally divergent between rice and Arabidopsis. To our interest, defense gene expression in these transformants varied between growth conditions. Similar growth condition dependence of PR gene expression also occurred in BTH-treated wild-type rice plants. On the basis of these results, we propose a role of WRKY45 in the BTH-induced and SA-mediated defense signaling in rice and its potential utility in improving disease resistance of rice, an importance food resource worldwide.
RESULTS
Identification of BTH-Responsive WRKY TFs
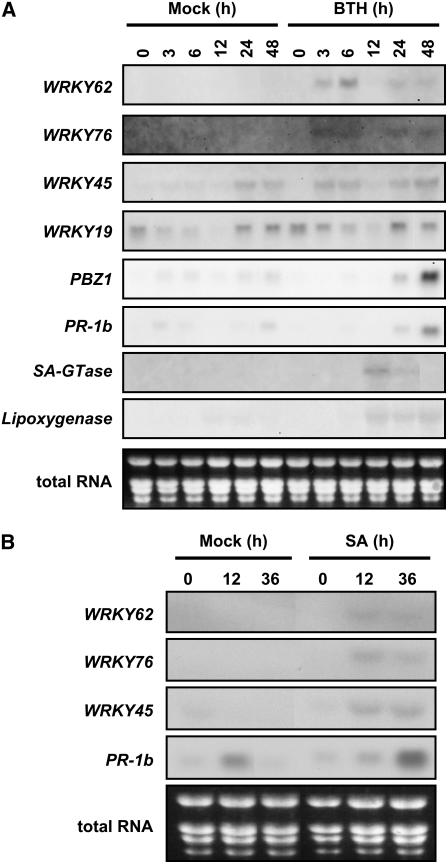
To screen for BTH-responsive genes in rice, we performed microarray analysis of ∼44,000 rice genes in BTH-treated rice plants using a 60-mer oligo DNA microarray. We treated plants with 0.5 mM BTH, extracted RNAs from leaves of BTH- and mock-treated plants at 24 h after treatment, and compared the gene expression between the two groups with four biological replicates. Statistical analysis using analysis of variance false discovery rate (q value) < 0.05 (Sharov et al., 2005) identified 326 BTH-responsive genes (see Supplemental Table 1 online), including several WRKY TF genes and many defense-related genes. Several studies have implicated WRKY TFs in plant defense responses, so we chose four WRKY genes for which full-length cDNAs were available for further characterization: Os WRKY62 and -76 (group II) and Os WRKY19 and -45 (group III) (Xie et al., 2005), as well as other defense-related genes (Table 1). We examined early-phase patterns of BTH response of the WRKY genes and some defense-related genes by RNA gel blot hybridization (Figure 1). The expression of three of the four WRKY genes (WRKY45, Os WRKY62, and Os WRKY76) was upregulated within 3 h after BTH application. Early-phase BTH response of Os WRKY19 was not clear in this experiment because of substantial signal levels in mock-treated plants, but RT-PCR confirmed the BTH inducibility of this gene (data not shown). BTH-induced upregulation was also confirmed with PR-1b and PBZ1 at 24 h after treatment and with an SA-GTase homolog and lipoxygenase at 12 h (Figure 1A). The WRKY genes were upregulated before all other defense-related genes examined.
Table 1.
Summary of Microarray Data for the BTH-Inducible Genes Characterized in This Study
| Accession Number | Locus ID | Annotation | Fold Change (BTH/Mock) | FDR(Q Value) | References |
|---|---|---|---|---|---|
| AK068337 | Os09g0417600 | Os WRKY76 | 313.0 | <0.00001 | |
| AK067834 | Os09g0417800 | Os WRKY62 | 425.0 | <0.00001 | |
| AK066255 | Os05g0322900 | WRKY45 | 35.8 | <0.00001 | |
| AK108389 | Os05g0571200 | Os WRKY19 | 14.2 | <0.00001 | |
| AK064395 | Os09g0518200 | SA-glucosyltransferase | 60.2 | <0.00001 | |
| AK072241 | Os12g0559200 | Lipoxygenase | 12.1 | <0.00001 | Schaffrath et al. (2000) |
| AK107926 | Os01g0382000 | PR-1b | 5.5 | <0.00001 | Agrawal et al. (2000b) |
| AK071613 | Os12g0555500 | PBZ1 | 4.2 | 0.04300 | Midoh and Iwata (1996) |
| AK103453 | Os10g0528300 | GST (Tau class) | 97.8 | <0.00001 | |
| AK072220 | Os07g0418500 | Cytochrome P450 | 38.6 | <0.00001 | |
| AF251277 | Os07g0129200 | PR-1a | 10.1 | 0.01200 | Agrawal et al. (2000a) |
Results of differential expression between BTH- and mock-treated rice plants 24 h after treatments are shown with fold changes and q values. A data set for all the BTH-responsive genes with q values < 0.05 can be found in Supplemental Table 1 online. A complete set of microarray data was deposited to the Gene Expression Omnibus repository under accession number GSE7567. FDR, false discovery rate.
Figure 1.
BTH- and SA-Induced Expression of WRKY and Defense-Related Genes in Rice.
(A) BTH-induced expression. Total RNA (5 μg per lane) from the third and fourth leaves of BTH- and mock-treated plants at the indicated time points after BTH application were hybridized with an RNA probe to the 3′-untranslated regions of the gene of interest, as described in Methods. The experiments were performed in two replicates with similar results.
(B) SA-induced expression. RNA samples were prepared from SA- and mock-treated rice plants at 12 and 36 h after SA application and hybridized as in (A).
It is generally thought that BTH, a functional analog of SA, triggers defense responses in plants similar to those triggered by SA. Our microarray analysis in rice supported this notion (data not shown), and we verified SA-responsive upregulation of WRKY45, -62, and -76, as shown in Figure 1B. SA response of Os WRKY19 was again unclear because this gene substantially responded to mock treatment (data not shown).
Overexpression of WRKY45 Enhances Blast Resistance in Rice
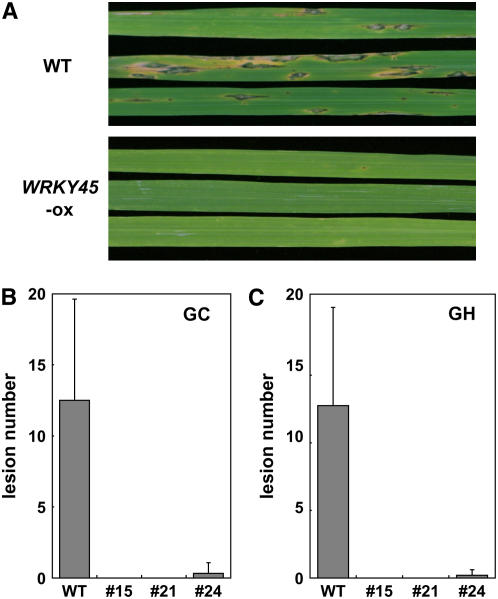
As an initial step to functionally characterize the four BTH-inducible WRKY TFs, we generated transgenic rice lines that express their cDNAs under the control of the constitutive maize (Zea mays) ubiquitin promoter and tested the transformants for resistance to a compatible race of blast fungus. The transformant rice plants were grown in a growth chamber and inoculated with blast fungus and then disease symptoms were characterized 7 d later. The transformants overexpressing Os WRKY62, -76, and -19 were as susceptible as wild-type plants under our assay conditions (data not shown). By contrast, those overexpressing WRKY45 (WRKY45-ox) showed remarkable decreases in susceptible-type lesions compared with wild-type plants (Figures 2A and 2B).
Figure 2.
Blast Resistance of WRKY45-ox Plants.
WRKY45-ox homozygote T3 (fourth generation) and control wild-type plants grown in a growth chamber ([A] and [B]) or in a greenhouse (C) were inoculated with M. grisea (race 007.0) at the five- to six-leaf stage, and disease symptoms were characterized 7 d after inoculation.
(A) Disease symptoms on the sixth leaves of wild-type and WRKY45-ox (line 15) plants grown in a growth chamber.
(B) and (C) Numbers of lesions on plants grown in a growth chamber ([B]; GC) and a greenhouse ([C]; GH). The bars represent the number of susceptible-type lesions in the 10-cm central region of the sixth leaves of individual T3 WRKY45-ox (lines #15, #21, and #24) and wild-type rice plants. Averages of 5 to 10 plants with standard deviations are shown. Greenhouse conditions were 28/23°C (day/night) with natural light in December. Essentially similar results were obtained when the plants were grown in March (data not shown).
BTH-Inducible Blast Resistance Is Compromised in WRKY45 Knockdown Rice
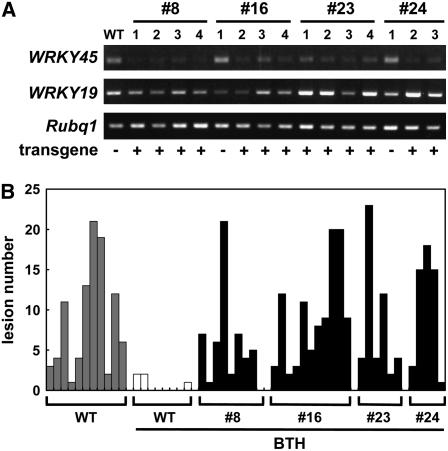
To characterize the loss-of-function phenotype conferred by WRKY45, we generated RNA interference (RNAi) transgenic rice lines by expressing an inverted-repeat sequence of the 3′ region of WRKY45 cDNA, the sequence of which is conserved only at a very low level among members of the WRKY family in rice. BTH induction of WRKY45 expression was reduced in the T1 (second generation) plants of four RNAi lines that had inherited the transgene but not in T1 segregants that had lost the transgene; these results indicate successful transgene-dependent knockdown of WRKY45 (WRKY45-kd; Figure 3A). BTH-induced upregulation of Os WRKY19, a group III BTH-inducible WRKY gene, was not affected in these transformants (Figure 3A), thus demonstrating the specificity of gene silencing. We tested the BTH-inducible blast resistance associated with WRKY45 (Figure 3B). Compared with untreated wild-type plants, BTH-treated wild-type plants showed marked reduction in the number of susceptible-type lesions. By contrast, BTH-treated WRKY45-kd plants exhibited a comparable number of blast lesions to that of untreated wild-type plants. These results indicate that WRKY45 plays an essential role in BTH-induced blast resistance in rice.
Figure 3.
Effects of WRKY45 Knockdown on Blast Resistance.
(A) Expression of WRKY45 in response to BTH in WRKY45-kd transgenic rice plants. Wild-type rice plants and T1 WRKY45-kd transgenics were grown in growth chambers, treated with 0.5 mM BTH, and analyzed by RT-PCR for WRKY45 expression at 48 h after treatment. We also evaluated expression of Os WRKY19 and Rubq1. T1 segregants containing the transgene show reduced transcript levels of WRKY45 but not Os WRKY19 or Rubq1. Presence (+) or absence (–) of the transgene in each plant is indicated.
(B) BTH-induced blast resistance in WRKY45-kd transgenic rice plants. WRKY45-kd T1 and wild-type rice plants were grown in growth chambers, treated with 0.5 mM BTH at the four-leaf stage, and inoculated with M. grisea (race 007.0) 2 d after treatment. Disease symptoms were characterized 7 d after inoculation. Inheritance of the transgene in individual T1 segregants was examined by PCR, and the data for those containing the transgene only are shown. The bars represent the number of susceptible-type lesions in a 10-cm region of the fourth leaves of individual T1 segregants having the transgene. Average lesion numbers in the WRKY45-kd plants were comparable to those in untreated wild-type plants, whereas BTH-treated wild-type plants had very few lesions. The lesion numbers varied substantially among individual plants. This is presumably due to difficulties in evenly applying BTH and subsequently blast fungi by spraying them on rice leaves.
WRKY45 Activates Transcription through W-Boxes
WRKY TFs generally act as transcriptional regulators through W-boxes, cis-elements that are enriched in the promoters of several defense-related genes (Maleck et al., 2000). To test whether WRKY45 also modulates transcription through W-boxes and whether it activates or represses transcription, we performed transactivation experiments in rice protoplasts by cotransfecting them with an effector gene to drive WRKY45 expression and a reporter construct containing W-boxes derived from the tobacco CHN48 promoter (Yamamoto et al., 2004) located upstream of the luciferase gene. We detected increases in luciferase activity dependent on the presence of the WRKY45 effector gene (Table 2). In addition, the transcriptional activation was reduced, although not completely, when the W-boxes were mutated (Table 2). Because similar levels of reporter gene activity were observed with the mutated W-boxes in the absence of the effector gene (Table 2), the residual activity is not due to the effector gene–derived WRKY45 but is presumably due to some endogenous activity that acts through the sequence fortuitously generated in the mutated W-boxes. These results indicate that WRKY45 is a transcriptional activator and acts through W-boxes.
Table 2.
Transactivation by WRKY45 in the Rice Protoplast Transient System
| Reporter | Effector | Relative LUC Activity |
|---|---|---|
| −ELI | +W45 | 3.7 ± 1.6 |
| +ELI | −W45 | 1.8 ± 0.8 |
| +ELI | +W45 | 50.0 ± 19.2 |
| +ELIm | +W45 | 20.2 ± 3.5 |
| +ELIm | −W45 | 21.9 ± 5.6 |
Reporter plasmids (5 μg) with (+ELI) or without (–ELI) wild-type W-boxes and 3 μg of effector plasmids with (+W45) or without (–W45) WRKY45 coding sequence were cotransfected into rice protoplasts, and luciferase activities were determined. Reporter plasmids with mutant W-boxes (ELIm) were also tested. Sequences of ELI and ELIm are 5′-TTGGTCAGAAAGTCAGTCGTCGAGTTGGTCAGAAAGTCAGTC-3′ (W-box underlined) and 5′-TTGGGAAGAAAGGAAGTCGTCGAGTTGGGAAGAAAGGAAGTC-3′ (mutated nucleotides boxed), respectively. LUC activities relative to those of Renilla LUC, the internal control, are shown. Data are shown as the mean of six independent experiments with sd.
WRKY45 Acts in the SA Signaling Pathway Independently of NH1
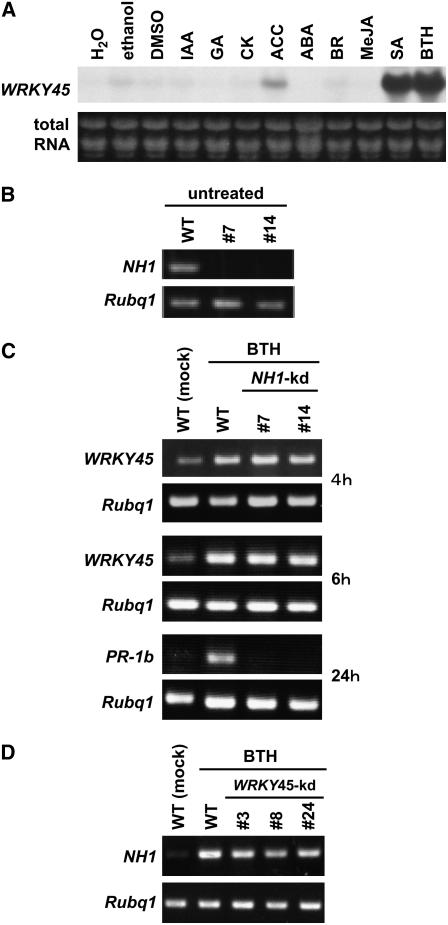
To investigate whether WRKY45 expression is responsive to only SA and BTH, we examined the effects of various signaling compounds on WRKY45 expression in leaf discs. As shown in Figure 4A, WRKY45 transcript levels responded strongly to SA and BTH. In this experimental condition, the levels of WRKY45 transcript accumulation in response to these chemicals were much higher than those observed in soil-grown plants (Figure 1), presumably because of better accessibility of the chemicals. WRKY45 transcripts responded only weakly to 1-aminocyclopropane 1-carboxylic acid and to barely detectable levels to the other chemicals tested (Figure 4A). These results indicate that the accumulation of WRKY45 transcripts is regulated mainly by the SA signaling pathway.
Figure 4.
WRKY45 Acts Downstream of SA in an Apparently NH1-Independent Pathway.
(A) Responses of WRKY45 expression to various signal compounds. Fully expanded fourth leaves of rice at the four-leaf stage were cut into pieces 2 cm long and immersed in aqueous solutions containing 0.01% Silwet L-77 and the following signal compounds: indole-3-acetic acid (IAA; Sigma-Aldrich), gibberellin G3 (GA; Wako), kinetin (CK; Sigma-Aldrich), 1-aminocyclopropane 1-carboxylic acid (ACC; Sigma-Aldrich), abscisic acid (ABA; Sigma-Aldrich), brassinolide (BR; Wako), methyl jasmonate (MeJA; Wako), 100 μM BTH, or 1 mM sodium salicylate (Nakalai) for 8 h at 30°C. Total RNAs (5 μg) were extracted, and WRKY45 transcript levels were analyzed by RNA gel blot analysis.
(B) Expression of NH1 in NH1-kd rice plants. Transcript levels of NH1 in untreated wild-type and NH1-kd plants (lines #7 and #14 and T2 homozygote) were examined by RT-PCR.
(C) Responses of WRKY45 and PR-1b expression to BTH in NH1-kd rice plants. Transcript levels of WRKY45 were analyzed by RT-PCR at 4 and 6 h, and those of PR-1b at 24 h, after BTH application to wild-type and NH1-kd (lines #7 and #14 and T2 homozygote) rice plants.
(D) Responses of NH1 expression to BTH in WRKY45-kd rice plants. Transcript levels of NH1 were analyzed 24 h after BTH application to wild-type and WRKY45-kd (lines #3, #8, and #24 and T2 homozygote) rice plants.
In Arabidopsis, NPR1 plays a major role in the SA signaling pathway downstream of SA. A recent article by Wang et al. (2006) reported that >99% of BTH-responsive genes in Arabidopsis are under the regulation of NPR1, including several WRKY genes. Five of the NPR1-regulated WRKY genes (At WRKY70, -53, -54, -38, and -66) belong to group III, as does Os WRKY45. To comparatively characterize the signaling pathway downstream of SA in rice with that in Arabidopsis, we investigated the epistatic relationship between WRKY45 and NH1 (AK120715), a rice ortholog of NPR1 (Chern et al., 2005), using NH1 knockdown (NH1-kd) rice plants in which BTH-dependent blast resistance is impaired (S. Sugano, C.-J. Jiang, and H. Takatsuji, unpublished results). In NH1-kd plants, the levels of NH1 transcripts were markedly reduced (Figure 4B), while those of its two closest paralogs (AK065952 and AK067198) were unaffected (data not shown). BTH-induced upregulation of PR-1b was severely suppressed in these plants, indicating that PR-1b is regulated downstream of NH1 (Figure 4C). By contrast, NH1-kd plants were indistinguishable from wild-type plants with respect to BTH-induced upregulation of WRKY45 (Figure 4C). These results indicate that WRKY45 is not downstream of NH1 at the level of transcription, clearly distinguishing WRKY45 from the Arabidopsis WRKY TFs that have been placed downstream of NPR1. Yu et al. (2001) have suggested that NPR1 is transcriptionally regulated by a WRKY TF in Arabidopsis, so we also examined the expression of NH1 in response to BTH and the effects of WRKY45 knockdown on this regulation to see whether WRKY45 is upstream of NH1. As shown in Figure 4D, BTH upregulated NH1 expression in wild-type rice plants, and this upregulation was scarcely affected by WRKY45 knockdown. Thus, it appears that WRKY45 acts independently of NH1 in the SA signaling pathway.
Different Sets of Genes Are Differentially Regulated Downstream of WRKY45 and NH1
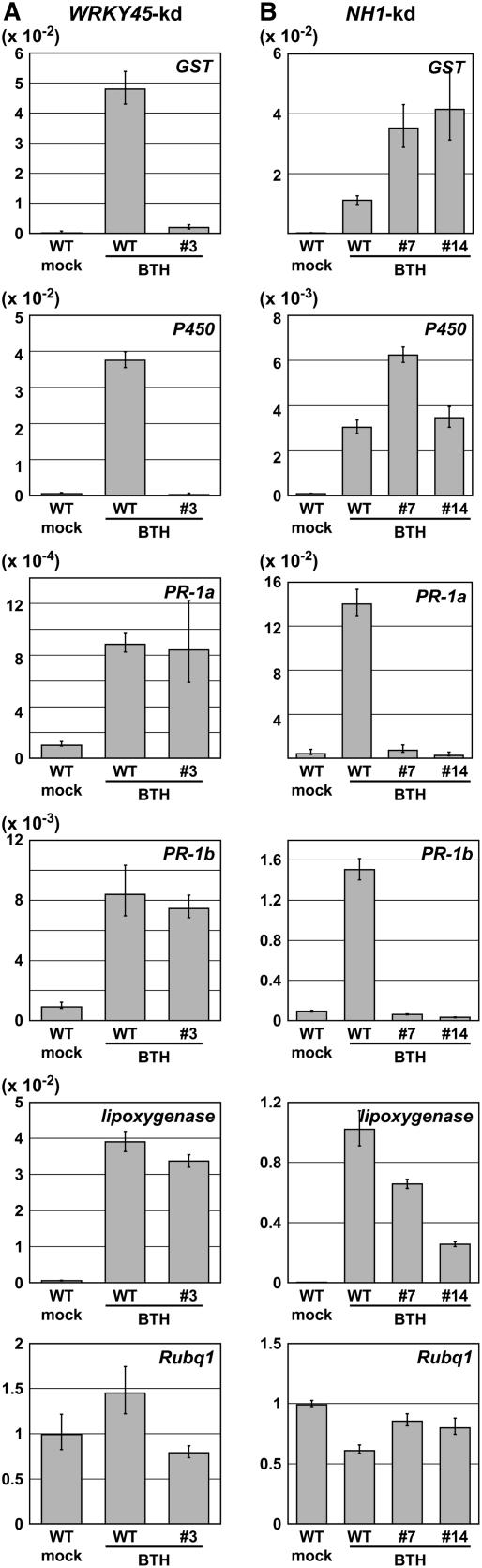
The results of epistasis analysis shown above prompted us to compare the genes regulated by WRKY45 and NH1. Among candidate WRKY45-dependent BTH-regulated genes selected by a preliminary microarray screening in WRKY45-kd rice (data not shown), we chose two genes encoding a glutathione S-transferase (GST; AK103453) and a cytochrome P450 (AK072220) (Table 1) and examined their regulatory patterns. WRKY45-kd and wild-type rice plants were BTH and mock treated, and the transcript levels of those genes were determined by quantitative RT-PCR 24 h after treatments (Figure 5A). The results demonstrated that BTH-responsive upregulation of these two genes was evidently reduced in WRKY45-kd plants compared with wild-type plants, indicating that these genes are under the regulation of WRKY45 either directly or indirectly. By contrast, the upregulation of these genes by BTH was not abolished in NH1-kd plants (Figure 5B), indicating that NH1 is not involved in the BTH-responsive regulation of these genes. These results further support the conclusion that WRKY45 and NH1 are involved in apparently independent signaling pathways. PR-1a and PR-1b showed coregulated patterns of expression that are obviously different from those of the GST and cytochrome P450 genes. BTH-responsive upregulation of PR-1a and PR-1b was consistently abolished in NH1-kd plants, indicating that these genes are under the regulation of NH1. The WRKY45 dependence of these genes varied in different experiments: the BTH responses of these genes were not affected by WRKY45 knockdown in some experiments (Figure 5A) but were affected in other experiments (see Supplemental Figure 1 online). The variation of the results was not dependent on the lines used (data not shown). These results suggest that the BTH responses of PR-1a and PR-1b are conditionally dependent on WRKY45, and some minor differences in experimental conditions can influence their apparent WRKY45 dependence. The BTH response of a lipoxygenase gene (AK072241) appeared unaffected by the knockdown of either WRKY45 or NH1 (Figures 5A and 5B; see Supplemental Figure 1 online), suggesting that the BTH-responsive regulation of this gene is dependent on neither transcriptional regulator. Collectively, these results reveal at least three classes of coregulated genes among BTH-responsive genes: WRKY45-dependent and NH1-independent ones, NH1-dependent and conditionally WRKY45-dependent ones, and those independent of either transcriptional regulator.
Figure 5.
Quantitative RT-PCR Analysis of the Dependence of the BTH-Responsive Genes on WRKY45 and NH1.
T2 homozygotes of WRKY45-kd and NH1-kd rice plants and wild-type plants were treated with 0.5 mM BTH. The fourth leaves from four plants were harvested and pooled 24 h after treatment, and the expression of selected BTH-responsive genes was analyzed by quantitative RT-PCR to examine the dependence of the expression on WRKY45 (A) and NH1 (B). Averages of three determinations relative to those of Rubq1 are shown with sd. The expression level of Rubq1 is relative to that in mock-treated wild-type plants. The experiments were done four or five times with similar results except for those with PR1a and PR1b in WRKY45-kd (see Supplemental Figure 1 online). The magnitudes of BTH responses varied substantially between the experiments in (A) and (B), presumably due to variable penetration of BTH into tissues and/or minor variations in the temporal patterns of expression between the experiments. After repeated experiments, however, we confirmed that the variations in the magnitudes of responses did not influence our conclusions regarding the signaling pathways. The genes analyzed are those listed in Table 1. WRKY45-kd line #3 and NH1-kd lines #7 and #14 were used.
A BLAST search revealed that rice PR1b is one of the closest homologs of Arabidopsis PR1 (At2g14610), which is BTH inducible and NPR1 regulated (Cao et al., 1997). As for other BTH-responsive rice genes described above (AK103453, AK072220, and AK072241), BLAST searches revealed several Arabidopsis homologs with similar levels of homology to the corresponding rice genes, some of which are BTH responsive, while others are not (Wang et al., 2006). In this situation, it is difficult to specify counterpart relationships between the rice and Arabidopsis genes without further experiments.
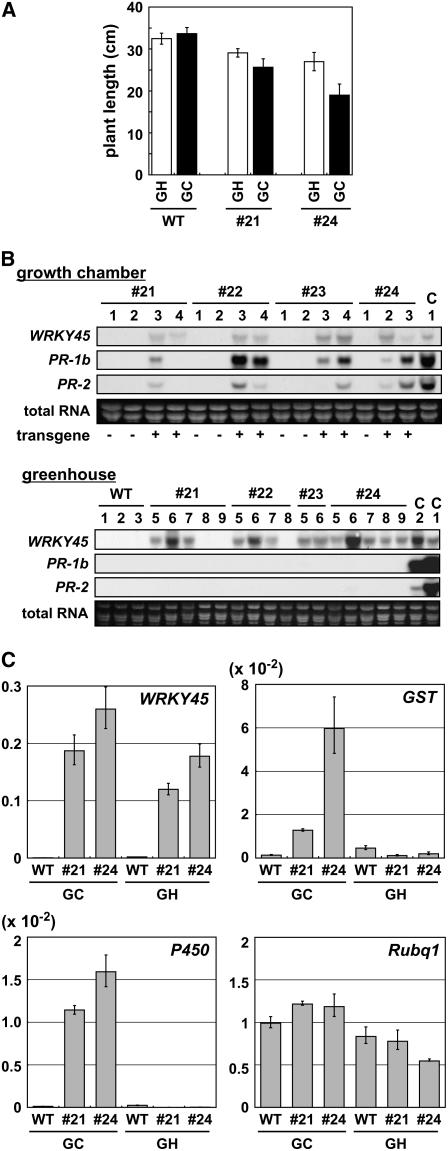
Plant Growth and PR Gene Expression in WRKY45-ox Rice Are Dependent on Growth Conditions
We fortuitously observed that the growth of WRKY45-ox rice plants was dependent on growth conditions. As shown in Figure 6A, wild-type rice plants grew even better in a growth chamber than in a greenhouse. WRKY45-ox rice plants exhibited some growth retardation compared with wild-type plants under both conditions. However, the degree of growth retardation was obviously more severe in the growth cabinet than in the greenhouse. We reasoned that this growth condition dependence might be correlated with the expression of defense-related genes, so we examined transcript levels of PR-1b and PR-2 in WRKY45-ox lines grown under the two conditions (Figure 6B). These PR genes are BTH inducible and are known to be responsive to blast fungus infection (Kim et al., 2001) and a fungal elicitor (Romero et al., 1998). When T1 WRKY45-ox plants were grown in the growth chamber, both PR genes were upregulated but not in the sibling T1 plants that lacked WRKY45 overexpression (Figure 6B). When the WRKY45-ox plants were grown in the greenhouse, however, transcripts of neither PR gene were detectable, even in the plants expressing WRKY45 at levels comparable to those in growth chamber–grown transformants (Figure 6B). Quantitative PCR revealed that the two WRKY45-regulated genes encoding GST and cytochrome P450 also exhibited similar growth condition dependence (Figure 6C). Thus, growth conditions have a profound influence on the expression of PR genes during concurrent WRKY45 overexpression, which may in turn affect the growth of WRKY45-ox plants.
Figure 6.
Effects of Growth Conditions on Plant Growth and Gene Expression in WRKY45-ox Plants.
(A) Effects of growth conditions on growth of WRKY45-ox rice plants. Lengths of wild-type and T2 homozygote WRKY45-ox rice plants grown in a growth chamber and a greenhouse were measured 15 d after sowing.
(B) Effects of growth conditions on BTH-induced expression of PR genes in WRKY45-ox plants. Individual T1 plants of each WRKY45-ox line with or without WRKY45 overexpression were analyzed for the expression of PR-1b and PR-2 (Gns5) by RNA gel blot hybridization. Rice plants were grown in growth chambers (top panels) or a greenhouse (bottom panels); greenhouse conditions were 28/23°C (day/night) with natural light in March. A mutant rice plant homozygous for Tos17-inserted Os SSI2, a rice homolog of Arabidopsis SSI2 (Kachroo et al., 2001), that constitutively expresses WRKY45 and PR (our unpublished results) was used as a positive control (C1). In the bottom panels, an RNA sample from growth chamber–grown WRKY45-ox rice plants was also run as a control (C2). Numbers at the top of lanes indicate serial numbers of the individual T1 transformants examined. The presence (+) or absence (–) of the transgene in each transformant as determined by PCR is indicated in the top panels. PR genes were expressed depending on WRKY45 expression in the growth chamber but not in the greenhouse.
(C) Quantitative PCR analysis of the effects of growth conditions on the expression of WRKY45-regulated genes in WRKY45-ox plants. The same RNA samples as used in (B) were analyzed for the expression of WRKY45, GST (AK103453), and cytochrome P450 (AK072220). Averages of three determinations relative to those of Rubq1 are shown with sd. The expression level of Rubq1 is relative to that in mock-treated wild-type plants.
This finding led us to test blast resistance of the WRKY45-ox plants that had been grown in a greenhouse to see whether the lack of the defense gene expression before fungus infection influences the resistance of these plants. As shown in Figure 2C, however, greenhouse-grown WRKY45-ox plants showed high blast resistance comparable to that of growth chamber–grown WRKY45-ox plants (Figure 2B). These results indicate that constitutive upregulation of those defense genes before fungus infection is not a prerequisite for the blast resistance of WRKY45-ox plants.
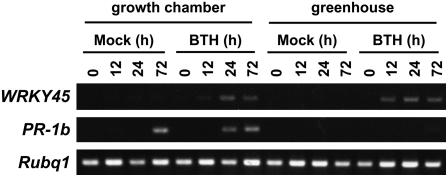
BTH Induction of PR Gene Expression Is Also Dependent on Growth Conditions
The growth condition dependence of the PR gene expression in WRKY45-ox rice prompted us to examine whether the growth conditions also affect BTH-induced regulation of defense genes. Wild-type rice plants were grown under the two conditions and were BTH and mock treated. RT-PCR analysis showed that WRKY45 was upregulated by BTH regardless of the growth condition (Figure 7). By contrast, PR-1b was upregulated in the growth chamber–grown plants but not in the greenhouse-grown plants (Figure 7). This growth condition dependence of PR gene expression is strikingly similar to that observed in the WRKY45-ox plants. Presumably, the same environmental factor(s) is responsible for the similar phenomena observed in BTH-treated wild-type plants and WRKY45-ox plants.
Figure 7.
Effects of Growth Conditions on BTH Induction of PR Genes.
Wild-type rice plants grown in a growth chamber or greenhouse were BTH or mock treated, and then RT-PCR analysis was used to evaluate the temporal patterns of expression of the WRKY45 and PR-1b genes. WRKY45 was induced by BTH under both growth conditions, whereas induction of PR-1b occurred only in growth chamber–grown plants.
DISCUSSION
WRKY45 Plays an Essential Role in BTH-Induced Disease Resistance in Rice
BTH induces resistance to various pathogens in plants. In an attempt to elucidate the molecular mechanism underlying the BTH-induced disease resistance, we identified BTH-responsive WRKY TF genes and found that one of them, WRKY45, plays a crucial role in BTH-induced defense reactions in rice. Several studies have highlighted the importance of WRKY TFs in disease resistance in dicots. For example, enhanced resistance to bacterial pathogens has been observed in Arabidopsis in which WRKY TFs, such as At WRKY29 (Asai et al., 2002) and At WRKY18 (Chen and Chen, 2002), are overexpressed. At WRKY70 exhibits opposite functions against different types of pathogens consistent with its proposed role in mediating SA-dependent signaling that antagonizes JA-mediated signaling (Li et al., 2004, 2006). Evidence of the loss-of-function phenotype for WRKY genes has been limited because functional redundancy obscures such phenotypes. Recently, however, characterization of double and triple mutants has revealed various functions of different WRKY TFs in defense reactions (Wang et al., 2006; Xu et al., 2006). We have shown in this study that constitutive overexpression of rice WRKY45 markedly enhanced blast resistance, whereas overexpression of three other BTH-inducible WRKY genes did not. BTH induction of blast resistance was markedly compromised in WRKY45-kd plants, being exempted from major functional redundancy. On the basis of these results, we conclude that rice WRKY45 plays a crucial and predominant role in BTH-inducible defense responses that strongly enhances resistance to blast infection.
WRKY45 Acts in an SA Signaling Pathway Apparently Independent of NH1
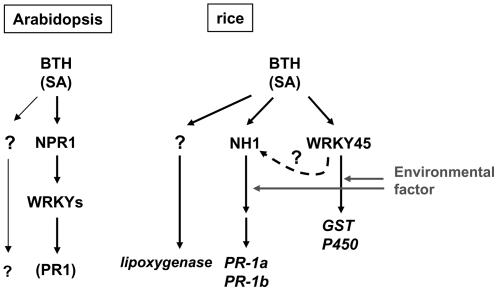
Our results have shown that WRKY45 is neither downstream nor upstream of NH1, a rice homolog of NPR1; therefore, WRKY45 appears to be involved in a pathway independent of NH1 downstream of SA (Figure 8). Expression analysis of BTH-responsive defense-related genes in WRKY45-kd and NH1-kd plants supports this notion: two BTH-responsive genes coding for a GST and a cytochrome P450 were found to be regulated by WRKY45, either directly or indirectly, but were not regulated by NH1. The dependence of PR-1a and PR-1b on the two regulators is not straightforward. BTH-responsive upregulation of these genes was consistently dependent on NH1. However, its dependence on WRKY45 in WRKY45-kd plants varied in different experiments. Moreover, PR-1b was upregulated in WRKY45-ox plants in a growth condition–dependent manner. Thus, this class of genes is dependent on both WRKY45 and NH1 under certain conditions. These observations suggest that the WRKY45-dependent and NH1-dependent pathways are not completely independent. As illustrated in Figure 8, a conditional signal flow from WRKY45 to NH1, which presumably does not involve transcriptional regulation of NH1, would account for these observations, but this hypothesis remains to be experimentally examined. Our results also suggest the presence of a pathway that involves neither WRKY45 nor NH1 leading to the regulation of lipoxygenase.
Figure 8.
Proposed Model for BTH/SA Signaling Pathway in Rice.
In Arabidopsis, most BTH-responsive genes are regulated downstream of NPR1, and this regulation is mediated by several WRKY TFs acting downstream of NPR1. In rice, WRKY45 and NH1 constitute apparently independent signaling pathways. GST and cytochrome P450 genes are regulated by WRKY45 but not by NH1. PR-1a and PR-1b are dependent on NH1 and are conditionally dependent also on WRKY45. Posttranslational signal flow from WRKY45 to NH1 (dashed arrow) would account for the dependence of these genes on both NH1 and WRKY45. The lipoxygenase gene does not appear to depend on either NH1 or WRKY45, so it might be regulated by another regulator. An environmental factor acts downstream of WRKY45 transcription and might also act on the NH1-dependent pathway (gray arrows).
It has previously been reported that rice has high SA levels and that those levels do not increase after pathogen infection (Raskin et al., 1990; Silverman et al., 1995), making the role of the SA signaling pathway in rice disputable. Our results, however, demonstrate that the SA signaling pathway mediated by WRKY45 or NH1 plays an active role in defense responses in rice. What is the biological significance of having (at least) two signaling pathways for defense responses downstream of SA/BTH, unlike in Arabidopsis, in which the NPR1-dependent pathway is predominant and regulates nearly all BTH-responsive genes? One possibility is that the NH1-dependent pathway has more limited functions in rice than the NPR1-dependent pathway has in Arabidopsis, with the WRKY45-dependent pathway playing a crucial role in rice. Alternatively, the WRKY45-dependent pathway may play a role in a defense program unique to rice or monocots. Comprehensive comparative analysis of downstream genes regulated by the two regulators would be of particular use in elucidating the roles of respective regulators in the defense program in rice.
Does Arabidopsis Have a WRKY45 Counterpart?
WRKY TFs are classified into three groups in light of their structures (Eulgem et al., 2000); WRKY45 falls into group III. Systematic characterization of expressional responses of group III WRKY genes in Arabidopsis has shown that four subsets of genes are present and show different response patterns to different signaling cues during compatible, incompatible, and nonhost interactions and that these subsets do not reflect phylogenetic relationships (Kalde et al., 2003). According to the phylogenetic analysis by Wu et al. (2005), At WRKY70, -54 -53, -41, -30, and -46 are more closely related to WRKY45 (denoted as Os WRKY50 in Wu et al. [2005]) than other group III WRKY TFs in Arabidopsis. Of these genes, At WRKY30 is not SA inducible, whereas the other five are (Kalde et al., 2003). At WRKY70 and -53 play a positive role in SA-mediated resistance to P. syringae. These WRKY genes have been placed downstream of NPR1 (Wang et al., 2006), distinguishing them from WRKY45. At WRKY54, which is functionally redundant with At WRKY70 with respect to negative regulation of SA synthesis, has also been placed downstream of NPR1 (Wang et al., 2006). Therefore, these four group III WRKY TFs are not orthologs of WRKY45. At WRKY41 and -46, which have not been well characterized so far, remain candidate WRKY45 orthologs. However, it is plausible that Arabidopsis does not have a counterpart to WRKY45, considering that >99% of BTH-responsive genes are under the regulation of NPR1.
An Environmental Factor(s) Acts on Defense Gene Expression Downstream of WRKY45 Transcription
The WRKY45-regulated defense genes were upregulated in WRKY45-ox plants when the plants were grown in a growth chamber, but transcripts were barely detectable in plants grown in a greenhouse. PR-1b expression in BTH-treated wild-type rice plants also showed similar growth condition dependence, whereas WRKY45 was upregulated regardless of the growth conditions. These observations strongly suggest that WRKY45 mediates BTH-induced transcriptional regulation of the defense genes and that an environmental factor(s) acts downstream of the transcription of WRKY45. The growth of WRKY45-ox rice plants was also growth condition dependent, which most likely reflects the expression of WRKY45 downstream genes. Similar growth condition–dependent phenotypes have been seen in transgenic rice plants overexpressing Arabidopsis NPR1 (Fitzgerald et al., 2004) and rice NH1 (Chern et al., 2005). These plants show lesion mimic and cell death phenotypes and dwarfed growth when they are grown in a growth chamber but not when grown in a greenhouse. Fitzgerald et al. (2004) suggested that the abundance and quality of the light that plants perceive may affect the phenotype of the NPR1-ox and NH1-ox rice plants. The same could hold for the growth condition–dependent phenotype of WRKY45-ox plants. Identification of the environmental factor and elucidation of the signaling pathway mediating it are of particular interest in understanding the molecular mechanism underlying this growth condition dependence of the phenotypes.
Possible Role of WRKY45 in BTH-Primed Potentiation
When plants are potentiated by BTH, induction of cellular defense responses upon pathogen infection occurs more rapidly or to a greater degree than in untreated plants, thereby accounting for the enhanced disease resistance in potentiated plants. Under potentiated conditions, most defense genes remain silent and become upregulated only after pathogen infection (Conrath et al., 2006). Greenhouse-grown WRKY45-ox plants show strong resistance to blast disease in spite of the lack of constitutive expression of the defense-related genes before fungus infection (Figures 2C and 6B). On the basis of these observations, it seems most likely that WRKY45-ox plants grown in a greenhouse are under a potentiated state similar to BTH-primed plants. On the other hand, the defense reactions against blast fungus we observed in growth chamber–grown WRKY45-ox plants are most likely equivalent to direct defense. To our interest, Kohler et al. (2002) reported that BTH-primed Arabidopsis plants exhibit defense responses in response to wounding and water infiltration, which indicates that the manifestation of defense responses occurs in response to abiotic stresses as well as to pathogen infection. These observations seem to parallel ours that an as yet unidentified environmental factor(s) triggers defense gene expression in WRKY45-ox and BTH-treated plants. We speculate that the environmental factor(s) could activate the same signaling pathway that pathogen infection does, leading to synergistic interaction of this signaling pathway with the SA signaling pathway downstream of WRKY45 transcription. This possible synergistic interaction of the two signaling pathways could be a key feature underlying BTH-primed potentiation of plant defense reactions.
In many cases, enhanced disease resistance conferred by overexpression of TF genes accompanies growth defects resulting in reduced productivity. This is presumably due to constitutive activation of defense responses under the regulation of introduced TFs. The overexpression of WRKY45 imposes relatively small adverse effects on plant growth, although they are dependent on the growth conditions. This is presumably due to the potentiated state established in these plants as discussed above. Our preliminary results have shown that the growth defects can be further reduced without major loss of blast resistance by optimizing the level of WRKY45 expression. After appropriate optimization, WRKY45 would serve as an effective tool to improve disease resistance of rice, an important food resource worldwide.
METHODS
Plant Materials, Chemicals, and Pathogen Treatments
All the experiments were performed with rice (Oryza sativa cv Nipponbare). Unless otherwise noted, plants typically were grown in growth chambers in soil at 28/23°C (light/dark), 50% humidity, 200 μmol m−2 s−1 light intensity, and 14-h photoperiods. BTH (in 0.5% [v/v] acetone + 0.05% [v/v] Tween 20) and SA (in 0.01% [v/v] Tween 20) were sprayed onto leaves (1 mL/plant). Mock treatments were done by spraying the solvents only. Conidia of blast fungus (Magnaporthe grisea Cavara, race 007.0) were suspended in 0.01% Tween 20 at a density of 105/mL and sprayed onto rice plants as described (Watanabe et al., 1977).
Microarray Analysis
Rice plants were BTH or mock treated at the four-leaf stage and then fourth leaves were harvested from four plants at 24 h after treatments and pooled. Total RNA was isolated from each pool using an RNeasy plant mini kit (Qiagen). The RNAs (400-ng aliquots) were labeled with a Low RNA Input Linear Amplification/Labeling Kit (Agilent Technologies) according to the manufacturer's instructions. Aliquots of Cy3-labeled cRNAs (1 μg each) of the BTH- or mock-treated samples were used for hybridization in an Agilent Rice Oligo Microarray (44K, custom-made; Agilent Technologies). Four biological replicate samples sets (of both BTH and mock) were analyzed. After hybridization, microarray slides were scanned (scanner model G2505B and software G2565BA; Agilent), and data were analyzed using Feature Extraction software (version 9.1; Agilent Technologies) at the default settings. All microarray procedures and data analyses were performed according to the manufacturer's manual (http://www.chem.agilent.com/scripts/generic.asp?lpage=11617andindcol=Nandprodcol=Y; G4140-90040). The data were normalized using GeneSpring GX 7.3 software (Agilent) and statistically analyzed using the National Institutes of Aging array analysis tool (http://lgsun.grc.nia.nih.gov/ANOVA/) (Sharov et al., 2005). We used the error model “maximum of averaged and Bayesian error variances” to reduce false positives. The false positives were controlled by measuring the false discovery rate (Benjamini and Hochberg, 1995).
RNA Gel Blot Hybridization
Total RNA was isolated using Trizol reagent (Invitrogen) and purified with an RNeasy mini kit (Qiagen). The RNAs were separated in agarose gels and transferred onto GeneScreen membranes (NEN Life Science Products) as described by Kapoor et al. (2002). Digoxigenin (DIG)-labeled antisense RNAs prepared with a DIG RNA labeling kit (SP6/T7; Roche Diagnostics) were used as probes. The blotted membranes were hybridized with the DIG-labeled probes using SSC-based buffers according to manufacturer's protocol, and signals were detected by chemiluminescence reaction using a DIG nucleic acid detection kit (Roche Diagnostics) and CDP-STAR (Roche Diagnostics). Chemiluminescence signals were detected by a LumiVision PRO densitometer (Aisin Seiki). Sequences for the probes (according to nucleotide numbers) are as follows: Os WRKY62, nucleotides 932 to 1161 (AK067834); Os WRKY76, nucleotides 1126 to 1327 (AK068337); WRKY45, nucleotides 1126 to 1426 (AK066255); Os WRKY19, nucleotides 658 to 1016 (AK108389); PBZ1, nucleotides 560 to 822 (D38170); PR-1b, nucleotides 577 to 793 (AK107926); SA-GTase homolog, nucleotides 1297 to 1710 (AK064395); lipoxygenase, nucleotides 2567 to 2747 (AK066737); and PR-2 (Gns5), nucleotides 1095 to 1295 (AK070677).
RT-PCR Analysis
RT-PCR was performed with 2 μg of total RNA treated with DNase I (Invitrogen). Reverse transcription was done using SuperScript II RNase H− (Invitrogen) and oligo(dT)23 primers (Sigma-Aldrich). PCR was performed using cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 7 min. Quantitative RT-PCR was run on a Thermal Cycler Dice TP800 system (Takara Bio) using SYBR premix Ex Taq mixture (Takara) with cycles of 95°C for 5 s and 60°C for 30 s. Rice ubiquitin 1 (Rubq1; AK121590) was used as an internal standard. Primers for PCR used in this study are listed in Supplemental Table 2 online. These primer sets were tested by dissociation curve analysis and verified for the absence of nonspecific amplification.
Plasmid Construction and Plant Transformation
To construct a plasmid for WRKY45 RNAi, part of the WRKY45 cDNA (nucleotides 1047 to 1535) was amplified by PCR and inserted into the pANDA vector with the antisense sequence upstream of the sense one as described (Miki and Shimamoto, 2004; Miki et al., 2005), except that the pDONR207 vector (Invitrogen) was used instead of pENTR as an intermediate vector. To construct a plasmid for constitutive expression of WRKY45, pUCAP/Ubi-NT was generated by replacing the HindIII-BamHI fragment containing the cauliflower mosaic virus (CaMV) 35S promoter in pUCAP/35S-NT (Kapoor et al., 2002) with the fragment containing the Zea mays polyubiquitin promoter (Ubi-1) from pAHC27 (Toki et al., 1992; Christensen and Quail, 1996). SfiI and BamHI sites were generated between the Ubi-1 promoter and the NOS terminator in pUCAP/Ubi-NT by digesting the plasmid with BamHI and SacI and inserting a double-stranded linker (5′-GATCTGGCCAAATCGGCCGGTACCGGATCCGCGGCCGCGAGTC-3′ annealed to 5′-CGCGGCCGCGGATCCGGTACCGGCCGATTTGGCCA-3′) into this site. A fragment for full-length WRKY45 cDNA was inserted between the SfiI and BamHI sites of pUCAP/Ubi-NT. Then, a fragment encoding the Ubi-1 promoter, the WRKY45 cDNA, and the Nos terminator was excised from this plasmid using HindIII and PacI and inserted between corresponding sites in the binary vector pZH1, a derivative of pPZP202 (Hajdukiewicz et al., 1994), which contains the CaMV 35S promoter, the hygromycin phosphotransferase gene, and the NOS terminator. Rice was transformed by an Agrobacterium tumefaciens (strain EHA105)–mediated procedure as described (Toki et al., 2006), except that rice seeds precultured for 3 d on medium containing 2,4-D were used for inoculation with Agrobacterium rather than 2-week-old rice callus derived from scutellum tissue of mature seeds (Toki et al., 2006).
Transient Gene Expression in Rice Protoplasts
Luciferase (LUC) reporter plasmids were constructed by inserting a CHN48-derived W-box (Yamamoto et al., 2004) or its mutant sequences upstream of the CaMV 35S minimal promoter (–46) (Odell et al., 1985) and a LUC+:Nos-terminator fragment in pUC. The effector plasmid 35S:WRKY45 was constructed by replacing the XbaI fragment of the LUC+ sequence in 35S:LUC+:NosT/pUC with a PCR-generated fragment of WRKY45 coding sequence. Protoplasts from the rice suspension–cultured cell line Oc were used for electroporation (Hattori et al., 1994). Electroporation was performed using an Electro Cell Manipulator (ECM 600; BTX) at 875 V cm−1 with a 550-μF capacitor. After incubation overnight at 30°C, cells were collected, and proteins were extracted as described (Kyozuka et al., 1987). Luciferase activities were assayed using the dual luciferase reporter assay system (Promega) according to the manufacturer's instructions. The Renilla LUC gene (Promega) under the control of the CaMV 35S promoter (0.1 μg) was cotransfected as an internal control, and the ratio of LUC activities (firefly LUC/Renilla LUC) was calculated to normalize each assay.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Table 1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Quantitative RT-PCR Analysis of the Dependence of the BTH-Responsive Genes on WRKY45.
Supplemental Table 1. BTH-Responsive Genes in Microarray Experiments.
Supplemental Table 2. Primers for PCR Used in This Study.
Supplementary Material
Acknowledgments
We thank M. Kuroda (National Agricultural Research Center, Hokuriku Research Center, Niigata, Japan) for providing the binary vector pZH1, K. Shimamoto (Nara Institute of Science and Technology, Nara, Japan) for providing the RNAi vector pANDA, N. Hayashi (National Institute of Agrobiological Sciences [NIAS]) for providing the blast strain, Y. Nishizawa (NIAS) for advice on the rice blast resistance assay, H. Fukayama and M. Miyao (NIAS) for advice regarding rice cultivation, H. Nakagawa and T. Izawa (NIAS) for providing plasmids for transient expression experiments and advice, T. Taya and K. Ando (Agilent, Tokyo, Japan) for advice regarding presentation of microarray data, and Y. Ohashi and S. Seo (NIAS) for valuable suggestions on the manuscript. We also thank the Rice Genome Resource Center at NIAS for the use of the rice microarray analysis system and Y. Nagamura and R. Motoyama for technical support. This work was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Green Technology Project, IP-4006).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hiroshi Takatsuji (takatsuh@nias.affrc.go.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agrawal, G.K., Jwa, N.S., and Rakwal, R. (2000. a). A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut, phytohormones, and protein phosphatase inhibitors. Biochem. Biophys. Res. Commun. 274 157–165. [DOI] [PubMed] [Google Scholar]
- Agrawal, G.K., Rakwal, R., and Jwa, N.S. (2000. b). Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signals. Biochem. Biophys. Res. Commun. 278 290–298. [DOI] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. [Ser A] B57 289–300 (Ser A). [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63. [DOI] [PubMed] [Google Scholar]
- Chen, C., and Chen, Z. (2002). Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern, M., Fitzgerald, H.A., Canlas, P.E., Navarre, D.A., and Ronald, P.C. (2005). Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant Microbe Interact. 18 511–520. [DOI] [PubMed] [Google Scholar]
- Chern, M.S., Fitzgerald, H.A., Yadav, R.C., Canlas, P.E., Dong, X., and Ronald, P.C. (2001). Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 27 101–113. [DOI] [PubMed] [Google Scholar]
- Christensen, A.H., and Quail, P.H. (1996). Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5 213–218. [DOI] [PubMed] [Google Scholar]
- Conrath, U., et al. (2006). Priming: Getting ready for battle. Mol. Plant Microbe Interact. 19 1062–1071. [DOI] [PubMed] [Google Scholar]
- Conrath, U., Pieterse, C.M., and Mauch-Mani, B. (2002). Priming in plant-pathogen interactions. Trends Plant Sci. 7 210–216. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. (2005). Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10 71–78. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 199–206. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Schmelzer, E., Hahlbrock, K., and Somssich, I.E. (1999). Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 18 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, H.A., Chern, M.S., Navarre, R., and Ronald, P.C. (2004). Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol. Plant Microbe Interact. 17 140–151. [DOI] [PubMed] [Google Scholar]
- Görlach, J., Volrath, S., Knauf-Beiter, G., Hengy, G., Beckhove, U., Kogel, K.H., Oostendorp, M., Staub, T., Ward, E., Kessmann, H., and Ryals, J. (1996). Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Hattori, T., Terada, T., and Hamasuna, S.T. (1994). Sequence and functional analyses of the rice gene homologous to the maize Vp1. Plant Mol. Biol. 24 805–810. [DOI] [PubMed] [Google Scholar]
- Journot-Catalino, N., Somssich, I.E., Roby, D., and Kroj, T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18 3289–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, P., Shanklin, J., Shah, J., Whittle, E.J., and Klessig, D.F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 98 9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalde, M., Barth, M., Somssich, I.E., and Lippok, B. (2003). Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol. Plant Microbe Interact. 16 295–305. [DOI] [PubMed] [Google Scholar]
- Kapoor, S., Kobayashi, A., and Takatsuji, H. (2002). Silencing of the tapetum-specific zinc finger gene TAZ1 causes premature degeneration of tapetum and pollen abortion in petunia. Plant Cell 14 2353–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.Y., Lee, S.H., Park, H.C., Bae, C.G., Cheong, Y.H., Choi, Y.J., Han, C., Lee, S.Y., Lim, C.O., and Cho, M.J. (2000). Identification of rice blast fungal elicitor-responsive genes by differential display analysis. Mol. Plant Microbe Interact. 13 470–474. [DOI] [PubMed] [Google Scholar]
- Kim, S., Ahn, I.-P., and Lee, Y.-H. (2001). Analysis of genes expressed during rice-Magnaporthe grisea interactions. Mol. Plant Microbe Interact. 14 1340–1346. [DOI] [PubMed] [Google Scholar]
- Kohler, A., Schwindling, S., and Conrath, U. (2002). Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 128 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka, J., Hayashi, Y., and Shimamoto, K. (1987). High frequency plant regeneration from rice protoplasts by novel nurse culture methods. Mol. Gen. Genet. 206 408–413. [Google Scholar]
- Lawton, K.A., Friedrich, L., Hunt, M., Weymann, K., Delaney, T., Ryals, J., Kessmann, H., and Staub, T. (1996). Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10 71–82. [DOI] [PubMed] [Google Scholar]
- Li, J., Brader, G., Kariola, T., and Palva, E.T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46 477–491. [DOI] [PubMed] [Google Scholar]
- Li, J., Brader, G., and Palva, E.T. (2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Bai, X., Wang, X., and Chu, C. (August 17, 2006). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. http://dx.doi.org/10.1016/j.jplph.2006.07.006. [DOI] [PubMed]
- Liu, X.Q., Bai, X.Q., Qian, Q., Wang, X.J., Chen, M.S., and Chu, C.C. (2005). OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res. 15 593–603. [DOI] [PubMed] [Google Scholar]
- Malamy, J., Carr, J.P., Klessig, D.F., and Raskin, I. (1990). Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 250 1002–1004. [DOI] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26 403–410. [DOI] [PubMed] [Google Scholar]
- Métraux, J.P., Signer, H., Wyss-Benz, M., Gaudin, J., Ryals, J., Ward, E., Raschdorf, K., Schmid, E., Blum, W., and Inverardi, B. (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250 1004–1006. [DOI] [PubMed] [Google Scholar]
- Midoh, N., and Iwata, M. (1996). Cloning and characterization of a probenazole-inducible gene for an intracellular pathogenesis-related protein in rice. Plant Cell Physiol. 37 9–18. [DOI] [PubMed] [Google Scholar]
- Miki, D., Itoh, R., and Shimamoto, K. (2005). RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 138 1903–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, D., and Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45 490–495. [DOI] [PubMed] [Google Scholar]
- Odell, J.T., Nagy, F., and Chua, N.H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313 810–812. [DOI] [PubMed] [Google Scholar]
- Raskin, I., Skubatz, H., Tang, W., and Meeuse, B.J.D. (1990). Salicylic acid levels in thermogenic and non-thermogenic plants. Ann. Bot. (Lond.) 66 369–373. [Google Scholar]
- Rasmussen, J.B., Hammerschmidt, R., and Zook, M.N. (1991). Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv syringae. Plant Physiol. 97 1342–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohilla, R., Singh, U.S., and Singh, R.L. (2002). Mode of action of acibenzolar-S-methyl against sheath blight of rice, caused by Rhizoctonia solani Kühn. Pest Manag. Sci. 58 63–69. [DOI] [PubMed] [Google Scholar]
- Romero, G.O., Simmons, C., Yaneshita, M., Doan, M., Thomas, B.R., and Rodriguez, R.L. (1998). Characterization of rice endo-[beta]-glucanase genes (Gns2-Gns14) defines a new subgroup within the gene family. Gene 223 311–320. [DOI] [PubMed] [Google Scholar]
- Rushton, P.J., Torres, J.T., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I.E. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Ryu, H.-S., Han, M., Lee, S.-K., Cho, J.-I., Ryoo, N., Heu, S., Lee, Y.-H., Bhoo, S., Wang, G.-L., Hahn, T.-R., and Jeon, J.-S. (2006). A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 25 836–847. [DOI] [PubMed] [Google Scholar]
- Schaffrath, U., Zabbai, F., and Dudler, R. (2000). Characterization of RCI-1, a chloroplastic rice lipoxygenase whose synthesis is induced by chemical plant resistance activators. Eur. J. Biochem. 267 5935–5942. [DOI] [PubMed] [Google Scholar]
- Schweizer, P., Schlagenhauf, E., Schaffrath, U., and Dudler, R. (1999). Different patterns of host genes are induced in rice by Pseudomonas syringae, a biological inducer of resistance, and the chemical inducer benzothiadiazole (BTH). Eur. J. Plant Pathol. 105 659–665. [Google Scholar]
- Sharov, A.A., Dudekula, D.B., and Ko, M.S.H. (2005). A web-based tool for principal component and significance analysis of microarray data. Bioinformatics 21 2548–2549. [DOI] [PubMed] [Google Scholar]
- Silverman, P., Seskar, M., Kanter, D., Schweizer, P., Métraux, J.P., and Raskin, I. (1995). Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiol. 108 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki, S., Hara, N., Ono, K., Onodera, H., Tagiri, A., Oka, S., and Tanaka, H. (2006). Early infection of scutellum tissue with Agrobacterium allows high speed transformation of rice. Plant J. 47 969–976. [DOI] [PubMed] [Google Scholar]
- Toki, S., Takamatsu, S., Nojiri, C., Ooba, S., Anzai, H., Iwata, M., Christensen, A.H., Quail, P.H., and Uchimiya, H. (1992). Expression of a maize ubiquitin gene promoter-bar chimeric gene in transgenic rice plants. Plant Physiol. 100 1503–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck, F., Zhou, A., and Somssich, I.E. (2004). Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene PcPR1-1 in parsley. Plant Cell 16 2573–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker, B., and Somssich, I.E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7 491–498. [DOI] [PubMed] [Google Scholar]
- van Hulten, M., Pelser, M., van Loon, L.C., Pieterse, C.M., and Ton, J. (2006). Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 5602–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T., Igarashi, H., Matsumoto, K., Seki, S., Mase, S., and Sekizawa, Y. (1977). The characteristics of probenazole (Oryzemate) for the control of rice blast. J. Sci. 2 291–296. [Google Scholar]
- Wen, N., Chu, Z., and Wang, S. (2003). Three types of defense-responsive genes are involved in resistance to bacterial blight and fungal blast diseases in rice. Mol. Genet. Genomics 269 331–339. [DOI] [PubMed] [Google Scholar]
- Wu, K.L., Guo, Z.J., Wang, H.H., and Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12 9–26. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Zhang, Z.L., Zou, X., Huang, J., Ruas, P., Thompson, D., and Shen, Q.J. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18 1310–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S., Nakano, T., Suzuki, K., and Shinshi, H. (2004). Elicitor-induced activation of transcription via W box-related cis-acting elements from a basic chitinase gene by WRKY transcription factors in tobacco. Biochim. Biophys. Acta 1679 279–287. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Qi, M., and Mei, C. (2004). Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 40 909–919. [DOI] [PubMed] [Google Scholar]
- Yu, D., Chen, C., and Chen, Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13 1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.