Abstract
Glycolysis is a ubiquitous pathway thought to be essential for the production of oil in developing seeds of Arabidopsis thaliana and oil crops. Compartmentation of primary metabolism in developing embryos poses a significant challenge for testing this hypothesis and for the engineering of seed biomass production. It also raises the question whether there is a preferred route of carbon from imported photosynthate to seed oil in the embryo. Plastidic pyruvate kinase catalyzes a highly regulated, ATP-producing reaction of glycolysis. The Arabidopsis genome encodes 14 putative isoforms of pyruvate kinases. Three genes encode subunits α, β1, and β2 of plastidic pyruvate kinase. The plastid enzyme prevalent in developing seeds likely has a subunit composition of 4α4β1, is most active at pH 8.0, and is inhibited by Glu. Disruption of the gene encoding the β1 subunit causes a reduction in plastidic pyruvate kinase activity and 60% reduction in seed oil content. The seed oil phenotype is fully restored by expression of the β1 subunit–encoding cDNA and partially by the β2 subunit–encoding cDNA. Therefore, the identified pyruvate kinase catalyzes a crucial step in the conversion of photosynthate into oil, suggesting a preferred plastid route from its substrate phosphoenolpyruvate to fatty acids.
INTRODUCTION
An important metabolic function of a developing Arabidopsis thaliana seed is the deposition of storage reserves: oil in the form of triacylglycerols, but also proteins and oligo- and polysaccharides (Baud et al., 2002). As sucrose is the major photosynthetic product transported in the phloem, the embryo is required to catabolize incoming sucrose and convert it into the more efficient storage compounds mentioned above. Glycolysis is central to this process as it converts sugars into precursors for protein and fatty acid synthesis while concomitantly producing ATP by substrate level phosphorylation. In fact, stable isotope labeling has been used to demonstrate that 90% of glucose fed to developing canola (Brassica napus) (a close relative of Arabidopsis and oilseed crop) embryos is converted to pyruvate by glycolysis (Schwender and Ohlrogge, 2002). Furthermore, a clear link between glycolysis and seed metabolism is apparent in the wrinkled1 (wri1) mutant of Arabidopsis, in which a general reduction in glycolytic activity results in an 80% reduction in seed oil (Focks and Benning, 1998).
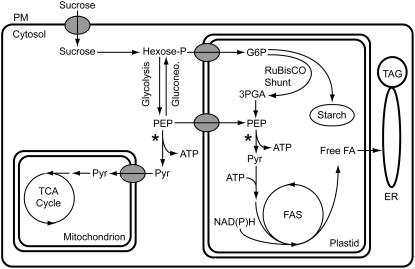
Glycolysis in plants occurs in both the cytosol and the plastid, and both compartments are connected through plastid membrane transporters (Weber, 2004; Plaxton and Podesta, 2006). The glycolytic intermediates of developing B. napus embryos appear to be in near equilibrium between the cytosol and plastid (Schwender et al., 2003), and enzymes of the full glycolytic sequence have been detected in both compartments in embryos (Eastmond and Rawsthorne, 2000). It is therefore likely that changes in glycolytic enzyme activities influence the transport of related intermediates across the plastid envelope. This compartmentation raises the question of a preferred route of glucose metabolism in developing oilseed embryos. EST analysis of developing Arabidopsis seeds indicated that mRNAs encoding cytosolic enzymes for the entire glycolytic pathway are abundant but that only mRNAs encoding plastidic enzymes for the second half of the pathway metabolizing trioses are prevalent (White et al., 2000). Microarray experiments extended these findings by detecting a shift in expression from genes encoding cytosolic glycolytic enzymes to those encoding plastidic glycolytic enzymes at the onset of storage compound accumulation. While gene expression does not necessarily reflect enzyme activity or metabolite flux, the microarray data led to the hypothesis that glucose is broken down in the cytosol to phosphoenolpyruvate (PEP), which is then imported into the plastid and metabolized by plastidic pyruvate kinase (PKp) (Figure 1; Ruuska et al., 2002). While the import of PEP by plastids of B. napus embryos has been demonstrated (Kubis et al., 2004), the chlorophyll a binding protein underexpressed1 (cue1) mutant of Arabidopsis lacking a seed-expressed PEP transporter has no reported seed reserve phenotype (Li et al., 1995). A recent steady state carbon flux analysis on cultured embryos of B. napus led to the proposal of a ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) shunt involving reactions of the reductive pentose phosphate pathway in the plastid (Schwender et al., 2004a). The proposed pathway bypasses the initial glycolytic reactions in the cytosol and generates PEP in the plastid, which could compensate for the PEP import deficiency in the cue1 mutant. Alternatively, pyruvate generated by cytosolic pyruvate kinase (PKc) could be imported and used directly for fatty acid biosynthesis. Isolated plastids from B. napus embryos are capable of incorporating 14C-labeled pyruvate into fatty acids (Kang and Rawsthorne, 1994; Eastmond and Rawsthorne, 2000), but no plastidic pyruvate transporter has been reported. A current model of primary metabolism in developing Arabidopsis seeds is shown in Figure 1. Direct molecular or genetic corroboration of this scheme is generally lacking, and the focus on PKp in developing Arabidopsis seeds should be highly informative in testing this hypothesis.
Figure 1.
Simplified Scheme of Carbon Metabolism with Emphasis on Oil Production in Cells of Green Developing Seeds.
Asterisks demark the pyruvate kinase reaction. Single arrows can indicate multiple reactions. 3PGA, 3-phosphoglycerate; ER, endoplasmic reticulum; FA, fatty acid; FAS, fatty acid synthase; G6P, glucose 6-phosphate; Gluconeo., gluconeogenesis; PM, plasma membrane; Pyr, pyruvate; TAG, triacylglycerol; TCA, tricarboxylic acid.
Pyruvate kinase (EC 2.7.1.40) occurs as both cytosolic and plastidic isoforms and catalyzes the ADP-dependent conversion of PEP to pyruvate while producing ATP. With respect to seed oil, PKp activity and concentration have been shown to correlate with the most active stage of lipid biosynthesis in developing B. napus embryos (Sangwan et al., 1992). Microarray data of developing Arabidopsis seeds show that the transcript level of a putative PKp encoding gene coincides with the most active period of triacylglycerol synthesis (Ruuska et al., 2002; Schmid et al., 2005). In addition, embryo PKp from B. napus is activated by 6-phosphogluconate, an intermediate of the oxidative pentose phosphate pathway, suggesting a coordination between the production of precursors and reducing equivalents for fatty acid synthesis (Plaxton et al., 2002).
Plant PK activities arise from the expression of multiple isozymes with different biochemical properties that depend on the tissue and plant source. Arabidopsis, for instance, has 14 annotated PK genes that likely exhibit a large degree of variation with respect to regulation of gene expression and enzyme activity (Arabidopsis Genome Initiative, 2000). PKp has been purified and characterized from castor (Ricinus communis) endosperm and B. napus suspension cell cultures (Plaxton et al., 1990, 2002; Negm et al., 1995). Both enzymes consist of α- and β-subunits and appear to exist as 3α3β heterohexamers. Both are regulated by metabolites of central carbon metabolism and have pH optima of ∼8.0. Despite existing biochemical knowledge of PKps and the proposed central role of PKp in embryos, a direct test of this enzyme's role in oil biosynthesis has not been conducted. Here, PKp from Arabidopsis was identified and biochemically characterized. Its function in seed oil biosynthesis was tested in planta.
RESULTS
Identification of Plastid-Localized and Seed-Expressed Pyruvate Kinases
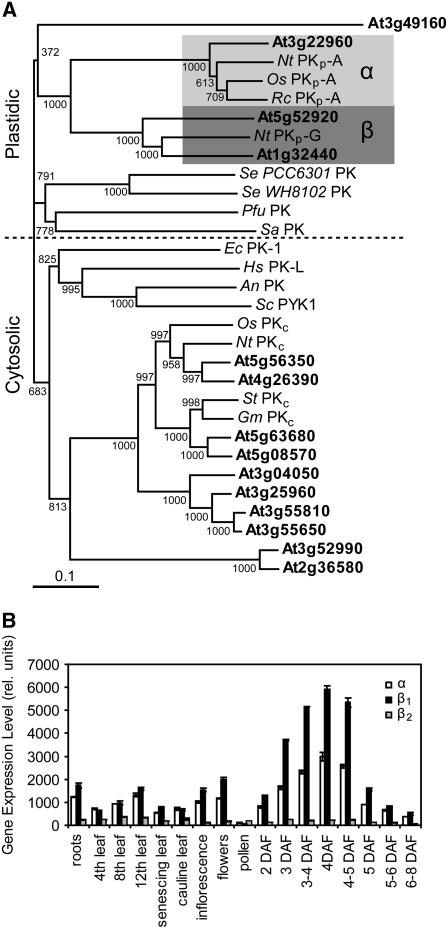
The Arabidopsis genome encodes 14 putative PK isoforms (Arabidopsis Genome Initiative, 2000). All but one (encoded by At3g49160) of the predicted PKs contain a fully conserved PK active site of [LIVAC]-x-[LIVM]-[LIVM]-[SAPCV]-K-[LIV]-E-[NKRST]-x-[DEQHS]-[GSTA]-[LIVM] (listed at the European Bioinformatics Institute; http://www.ebi.ac.uk/) and are presumably active enzymes. Several cross-kingdom PK phylogenies have been published (e.g., Hattori et al., 1995; Schramm et al., 2000; Munoz and Ponce, 2003), but only one putative PKc from Arabidopsis was included in these studies. When the 14 Arabidopsis PK amino acid sequences are aligned with bona fide PKs from other organisms, they segregate into cytosolic and plastidic clades (Figure 2A). This amino acid sequence similarity-based segregation is supported by the exclusive prediction of chloroplast transit peptides at the N termini of the four predicted PKps using ChloroP and TargetP (Emanuelsson et al., 1999, 2000). The PKps in Figure 2A are further divided between α- and β-subunits. One Arabidopsis PKp subunit (encoded by At3g22960) is most similar to described PKp-αs. Two others, which show 63% amino acid identity to each other (encoded by At5g52920, PKp-β1; At1g32440, PKp-β2), are most similar to known PKp-βs (Figure 2A).
Figure 2.
Pyruvate Kinase Similarity and Selected Gene Expression in Arabidopsis.
(A) Pyruvate kinase phylogeny. Amino acid sequences of Arabidopsis (gene loci in bold) and other bona fide pyruvate kinases were used. Bootstrap values are indicted at branches; α and β represent plastidic PK subunit families. The scale represents 10% difference. An, Aspergillus niger; Ec, Eschericia coli; Gm, Glycine max; Hs, Homo sapiens; Nt, Nicotiana tabacum; Os, Oryza sativa; Pfu, Pyrococcus furiosus; Rc, Ricinus communus; Sa, Sulfolobus acidocaldarius; Se, Synechocystis sp; Sc, Saccharomyces cerevisiae; St, Solanum tuberosum.
(B) Relative gene expression of putative Arabidopsis PKp-α (At3g22960), PKp-β1 (At5g52920), and PKp-β2 (At1g32440) encoding genes (derived from published microarray data; Schmid et al., 2005). Values are the mean ± sd (n = 3). DAF, days after flowering.
Three of the four predicted Arabidopsis PKp genes were identified as seed expressed by EST analysis of developing seeds (White et al., 2000). Two, PKp-α and PKp-β1, represent the highest level of induction of any PK gene in seeds and are coordinately expressed in all tissues, while the PKp-β2 encoding gene has very low transcript accumulation in any tissue (Figure 2B; Schmid et al., 2005).
The Predicted PKp Subunits Are Plastid Localized
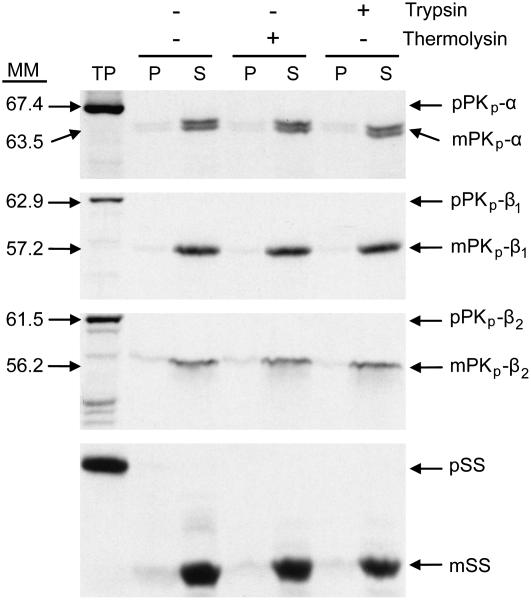
In vitro chloroplast import followed by protease protection assays were conducted to obtain evidence for plastid localization of the PKp subunits. When in vitro–produced 35S-labeled PKp subunits were incubated with isolated pea (Pisum sativum) chloroplasts, the proteins were imported and processed to their presumably mature forms (Figure 3). All three were resistant to treatment with thermolysin and trypsin and were found in the soluble fraction. Based on the data, the three putative PKp subunits are localized to chloroplasts and are imported and processed into mature, soluble, stromal proteins as predicted by the analysis of their amino acid sequence. The molecular masses indicated in Figure 3 were derived from Rf (retention factor) analysis and are in agreement with the transit peptide cleavage site predictions made by ChloroP. The full-length precursor proteins were calculated to be 65.1, 63.5, and 62.6 kD, and removal of the predicted transit peptides produces mature proteins of 59.6, 56.8, and 56.8 kD for PKp-α, PKp-β1, and PKp-β2, respectively. The mobility shift of PKp-α indicated cleavage of a potentially smaller (∼1 kD) peptide, but for PKp-β1 and PKp-β2, there were no discrepancies between the predicted and observed transit peptide cleavage sites.
Figure 3.
Subcellular Localizations of Pyruvate Kinase Subunits.
In vitro import of PKp subunits into isolated pea chloroplasts. After import, chloroplasts were subjected to either no treatment (−) or to post-treatment (+) with either thermolysin or trypsin. Intact chloroplasts were subsequently recovered by centrifugation through 40% Percoll cushion and fractionated into a total membrane (P) and a supernatant (S) fraction. All fractions were analyzed by SDS-PAGE and fluorography. MM, molecular masses of precursor and mature proteins based on Rf analysis; TP, 10% of translation reaction added; p, precursor form; m, mature form; pSS, precursor of the small subunit of Rubisco included as control.
Heteromeric Subunit Composition of Recombinant PKps
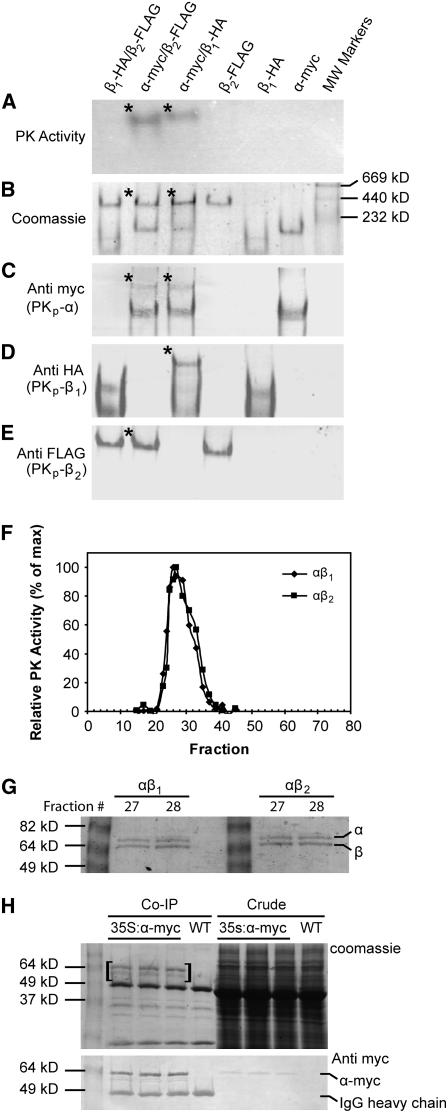
A recombinant approach was taken to study Arabidopsis seed PKp subunit composition due to the scarcity of embryo tissue for native protein purification. The cDNAs encoding the three putative PKp subunits lacking the predicted transit peptides were isolated by RT-PCR and were inserted into an Escherichia coli expression vector with an N-terminal 6X-His tag and a thrombin site for removal of the tag after purification. Gel electrophoresis (SDS-PAGE) and subsequent immunoblotting were used to confirm purification of single proteins and complete cleavage of the tag (see Supplemental Figure 1A online). Epitope-tagged versions of the proteins were also generated that in addition to the N-terminal His-tag had short C-terminal epitope tags. PKp-α was fused with c-myc (EQKLISEEDL), PKp-β1 with HA (YPYDVPDYA), and PKp-β2 with FLAG (DYKDDDDKG). The antibodies used for detection of the epitopes were shown to be lacking of any cross-reactivity. Initial tests indicated that in liquid assays, none of the subunits had PK activity on their own and that only αβ1 and αβ2 combinations were active (see Supplemental Figure 1B online). PK activity was not observed in the absence of ADP, indicating that no contaminating PEP phosphatase activity was present (see Supplemental Figure 1B online). Native-PAGE with epitope-tagged proteins was used to explore this subunit requirement in more detail. Figures 4A to 4E show five identical native-PAGE gels developed in different ways. The gel in Figure 4A was stained for PK activity. Only the αβ1 and αβ2 mixtures were active. Moreover, the activities coincided with less-mobile bands as shown by the Coomassie Brilliant Blue (CBB)–stained gel (Figure 4B). In the case of αβ1, detection of the individual α-myc and β1-HA fusion proteins with specific antibodies revealed a higher molecular mass complex coinciding in mobility with the active complex in Figure 4A, suggesting that the active complex is composed of both α- and β1-subunits (Figures 4C and 4D). The result is less clear for the αβ2 complex because the β2 subunit alone forms a higher molecular mass complex with the same mobility as the active enzyme (Figures 4A, 4B, and 4E). However, the α-myc protein is present in the αβ2 higher molecular complex (Figure 4C). Thus, it is likely that the higher molecular mass active complex in the αβ2 mixture is also composed of both subunits.
Figure 4.
In Vitro and in Vivo Interaction of PKp Subunits.
(A) to (E) Identically loaded native-PAGE gels showing in vitro interaction of PKp subunits. A total of 10 pmol (∼0.6 μg) of each subunit was used per lane. The asterisk denotes bands corresponding to the active PK complexes.
(A) PK activity stained gel.
(B) CBB-stained gel.
(C) Immunological detection of α-myc with anti c-myc.
(D) Immunological detection of β1-HA with anti HA.
(E) Immunological detection of β2-FLAG with anti FLAG.
(F) PK activity elution profile after fast protein liquid chromatography over Superdex-200.
(G) SDS-PAGE gel of most active fractions from (F). Total protein (75 ng) was loaded per lane.
(H) CBB-stained gel and anti-myc immunoblot of co-IP proteins. Total proteins were extracted from wild-type and 35S:α-myc silique tissue and were subjected to co-IP. Approximately 37.5 μg of crude protein and half of the total eluate were loaded per lane. Bands unique to the 35S:α-myc co-IP lanes (in brackets) were excised and identified. IgG heavy chain is indicated on the immunoblot for reference.
Gel filtration chromatography was used to estimate the molecular masses of the active PKp heteromers. Figure 4F shows that for both αβ1 and αβ2, PK activity eluted as a single peak. The molecular masses of these active complexes were calculated to be 463 ± 10 kD for αβ1 and 476 ± 10 kD for αβ2, which is consistent with octomeric complexes of 60 kD α-subunits and 57 kD β-subunits. Based on SDS-PAGE analysis, the most active fast protein liquid chromatography fractions contained equal amounts of α- and β-subunits (Figure 4G). Thus, the active PKs appear to be heterooctomers composed of four α- and four β-subunits.
The in vivo interaction of the PKp subunits was tested using coimmunoprecipitation (co-IP). Three constructs containing full-length, epitope-tagged cDNAs encoding the α-, β1-, and β2-subunits driven by the cauliflower mosaic virus (CaMV) 35S promoter were introduced into Arabidopsis. Only the α-myc fusion protein could be directly immunoprecipitated from plant tissue. One possible explanation is that the epitope tags in the β-subunits were not accessible in the native complex. The SDS-PAGE gels in Figure 4H show the result of co-IP experiments with silique tissue from wild-type and three 35S:α-myc plants. A small amount of α-myc protein was detected by an anti-myc immunoblot in crude extracts from 35S:α-myc plants. After immunoprecipitation, the α-myc protein was enriched and became visible on CBB-stained gels. In addition, another protein running slightly faster than α-myc was visible on the CBB-stained gel. The CBB-stained and immunoreactive bands running at 49 kD and at a slightly less molecular mass were also present in the wild-type control. These were presumably products of the degradation of the anti-myc IgG during elution from the Protein-A sepharose. All three PKp subunits are very close in size and could comigrate during SDS-PAGE. Therefore, a gel slice including a section above and below the α-myc protein from the co-IP reaction (indicated by brackets in Figure 4H) was excised, and the contained proteins were subjected to tryptic digest and liquid chromatography–tandem mass spectrometry. Analysis of the mass spectrometry data identified peptides of all three PKp subunits with significant individual ion scores (Mowse scores >31, P < 0.05): PKp-α (encoded by At3g22960), PKp-β1 (encoded by At5g52920), and PKp-β2 (encoded by At1g32440), with 29, 7, and 3 nonredundant peptides representing 67%, 21%, and 10% coverage of the predicted mature proteins, respectively (see Supplemental Figure 2 online).
Kinetic Characterization of PK Complexes
Enzyme activity analysis was performed using reconstituted PKp complexes. The maximum PK activity for the αβ1 and αβ2 heteromers was reached within 1 min of mixing the subunits (see Supplemental Figure 1C online). Reciprocal titrations in saturating substrate conditions showed that plots of PK activity versus subunit equivalents follow hyperbolic curves when one subunit is held constant and the other added in increasing amounts (see Supplemental Figures 1D and 1E online). Furthermore, when equal molar ratios of subunits were used, enzyme activity increased linearly with increasing protein concentration (see Supplemental Figure 1F online). These results suggest that the association (and thus activity) of the subunits is dependent only on protein concentration and that there is little or no cooperativity of subunit association. Additionally, a series of subunit inactivation experiments using site-directed mutagenesis and chemical inactivation were performed, and the results indicated that PK activity is dependent on both α- and β-subunits (see Supplemental Figure 3 online). Further kinetic experiments were done using equal molar ratios of αβ1 or αβ2 mixtures. The enzymes had a strict requirement for Mg2+ and K+ and were completely inactivated after 3 min of incubation at 50 to 55°C. The pH optima were found to be pH 7.8 to 8.0 (Table 1), and subsequent reactions were conducted at pH 8.0. The Vmax of αβ1 was ∼2-fold higher than that of αβ2, and the substrate concentration at half maximal initial velocity (S0.5) values for ADP and PEP were 1.5- and 3-fold lower, respectively, for αβ1 (Table 1). Both complexes displayed sigmoidal saturation kinetics for ADP and PEP, with αβ1 having greater Hill coefficients (Table 1). Both enzymes were capable of catalyzing phosphoryl group transfer from PEP to NDPs other then ADP. Activities detected using 10 mM CDP, GDP, or UDP were 85, 57, and 67% and 13, 24, and 19% for αβ1 and αβ2, respectively, compared with an ADP control. Clearly, the αβ2 enzyme preferred ADP, while αβ1 was less discriminatory.
Table 1.
PK Kinetic Constants
| Enzyme
|
||
|---|---|---|
| Constant | αβ1 | αβ2 |
| pH Optimum | 8.0 | 7.8 |
| Vmax (units/mg) | 13.5 ± 0.3 | 7.6 ± 0.4 |
| PEP S0.5 (μM) | 75.1 ± 7.0 (2.0) | 118.6 ± 11.5 (1.2) |
| ADP S0.5 (μM) | 113.9 ± 9.1 (1.9) | 303.6 ± 27.8 (1.2) |
| Kcat (s−1) | 9.5 × 104 | 5.3 × 104 |
Hill coefficients for PEP and ADP are given in parentheses. Values are the mean ± sd (n = 4).
Numerous metabolites and signaling compounds (fully listed in Methods) were tested as effectors of Arabidopsis PKp activity at subsaturating concentrations of PEP and ADP. Those compounds that had a significant effect are listed in Table 2. Only one activator, 6-phosphogluconate, was identified, and it only activates αβ2. The rest of the effectors acted as inhibitors, with the most effective being Glu and oxalate (Table 2). Values for the constants I50 and Ka were calculated (see Methods for definitions) for effectors capable of 50% relative activation or inhibition, and they clearly show that αβ2 is the more sensitive enzyme with respect to these compounds. Neither enzyme was sensitive to treatment with DTT or sodium tetrathionate, suggesting a lack of redox regulation contrary to what was previously observed for plastidic glucose 6-phosphate dehydrogenase isoforms of Arabidopsis (Wakao and Benning, 2005).
Table 2.
Metabolite Effectors of PK Activity
| Concentration Tested (mM) | Relative Activity (%)
|
I50 (mM)
|
Ka (mM)
|
||||
|---|---|---|---|---|---|---|---|
| Metabolite | αβ1 | αβ2 | αβ1 | αβ2 | αβ1 | αβ2 | |
| 6-PG | 0.05 | 96 | 192 | – | – | – | 0.02 |
| Glutamate | 5.00 | 59 | 30 | 6.20 | 2.10 | – | – |
| Oxalate | 0.20 | 71 | 50 | 0.41 | 0.21 | – | – |
| Iso-citrate | 10.00 | 70 | 75 | ND | ND | – | – |
| AMP | 1.00 | 77 | 96 | ND | ND | – | – |
| ATP | 1.00 | 77 | 87 | ND | ND | – | – |
| Glyoxylate | 5.00 | 83 | 79 | ND | ND | – | – |
| OAA | 2.00 | 82 | 88 | ND | ND | – | – |
Values represent the mean of at least four repeats. Activity is percentage relative to a noneffector control (set at 100). 6-PG, 6-phoshpogluconate; OAA, oxaloacetate; ND, not determined.
Disrupting the PKp-β1 Encoding Gene Causes a Reduction of PKp Activity and Seed Oil Content
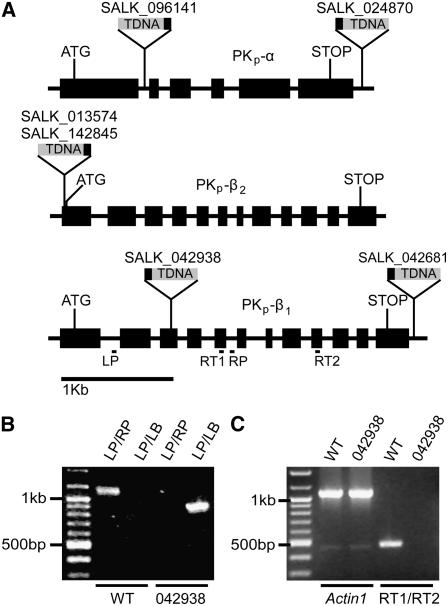
Multiple independent SALK T-DNA Insertion lines were obtained to study the in vivo function of the Arabidopsis PKps (Alonso et al., 2003). Six lines were reanalyzed for exact insertion site location (PKp-α, SALK_096141 and SALK_024870; PKp-β1, SALK_042938 and SALK_042681; PKp-β2, SALK_013574 and SALK_142845), but only one (SALK_042938 in PKp-β1) carried an insertion in a translated portion of the gene (Figure 5A). The line SALK_096141 has an insertion in an intron of the PKp-α encoding gene, but RNA levels were unaffected in these plants. Genotyping by PCR revealed homozygous SALK_042938 mutants completely lacking transcripts from the PKp-β1 encoding gene as detected by RT-PCR (Figures 5B and 5C). Transcript amounts for the PKp-α and PKp-β2 encoding genes were unchanged (see Supplemental Figure 4 online). The mutant will be referred to as pkp1.
Figure 5.
Identification of a SALK T-DNA Mutant in PKp-β1.
(A) Gene structure of the three PK subunits and locations of T-DNA insertions. Black box on T-DNA is the left border. LP and RP depict locations of primers used for genotyping in (B). RT1 and RT2 depict locations of primers used for RT-PCR in (C). ATG, start codon; STOP, stop codon.
(B) PCR-based genotyping of SALK_042938. LP and RP refer to At5g52920-specific primers shown in (A) and in Table 4. LB refers to the T-DNA left border primer in Table 5.
(C) RT-PCR to measure PKp-β1 encoding gene expression in SALK_042938. Actin1 (At2g37620) is the control. RT1 and RT2 refer to At5g52920-specific primers shown in (A) and in Table 5.
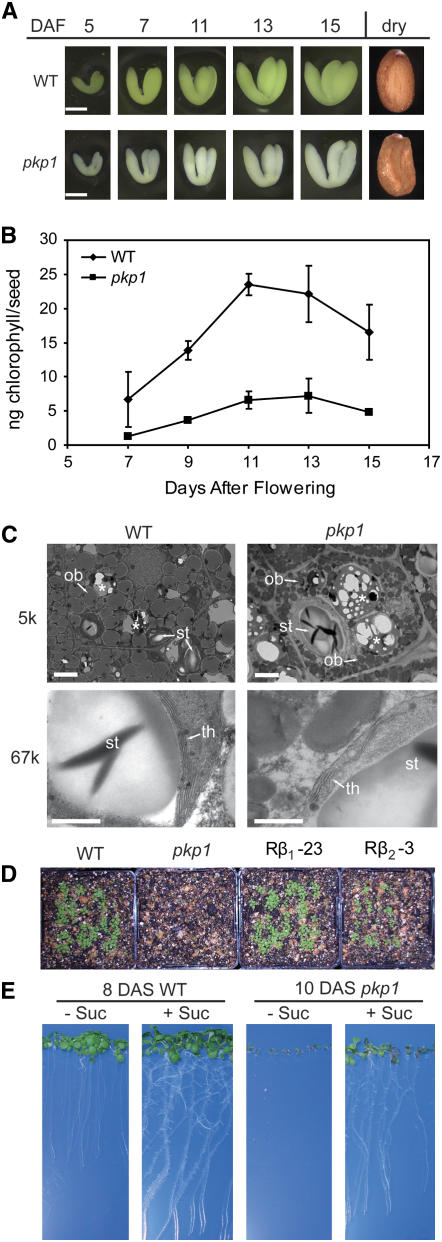
Notably, of the six analyzed mutants, only pkp1 had any visible seed phenotype. The mature seeds of pkp1 were wrinkled when observed under a dissecting microscope, and developing mutant embryos contained less chlorophyll (Figures 6A and 6B). Transmission electron microscopy was done to assess any ultrastructural perturbations in the pkp1 mutant. The 5k-magnification (Figure 6C, top panels) micrographs show representative cotyledon cells from 13 d after flowering (DAF) embryos. Wild-type cells are full of oil bodies by this time and still contain some starch in the plastids. The pkp1 mutant has much smaller oil bodies than the wild type but has much larger starch granules. The 67k-magnification images (Figure 6C, bottom panels) show in more detail the thylakoid structure of the mutant. While organized similarly, the pkp1 thylakoids are less extensive than the wild type. The smaller oil bodies, reduced thylakoid membranes, and wrinkled seeds of pkp1 suggest a reduction in lipid biosynthesis and possibly storage compound accumulation. Therefore, oil and protein were quantified in the mature pkp1 seeds (Table 3). The mutant accumulated only 40% as much oil as the wild type, yet there was only a 15% reduction in protein. Test crosses and analysis of the F1 progeny indicated that the low oil phenotype of pkp1 is a recessive trait largely dependent on the genotype of the embryo. A homozygous pkp1 sporophyte did however result in a 15% reduction in seed oil when pollinated with wild-type pollen (Table 3), indicating a small maternal effect.
Figure 6.
pkp1 Seed Phenotypes.
(A) Seed phenotypes of pkp1 and the wild type. Embryos were dissected out of developing seeds at the time indicated. Fully dessicated mature seeds are shown as well. Bars = 0.2 mm.
(B) Total chlorophyll content of pkp1 and wild-type developing seeds. Forty seeds were measured per sample. Values are the mean ± sd (n = 3).
(C) Electron micrographs of cells from 13 DAF wild-type and pkp1 cotyledons. Starch granules (st) and oil bodies (ob) are marked with arrows in the top panels. Higher magnification in the bottom panels reveals thylakoid membranes (th) inside plastids. The 5k and 67k denote magnification used. Asterisks mark protein bodies. Bars = 2 μm in top panels and 0.5 μm in bottom panels. Left panels, wild type; right panels, pkp1 mutant.
(D) Seedling establishment on soil. 100 seeds per line were sown directly on soil after sterilization. Picture was taken are 12 d after sowing. Rβ1-23, pkp1 rescued with CaMV 35S–driven PKp-β1; Rβ2-3, pkp1 rescued with CaMV 35S–driven PKp-β2.
(E) Sucrose-dependent seedling establishment of pkp1. Seeds were germinated on Murashige and Skoog medium without (−) or with (+) 2% sucrose. The wild type and the pkp1 mutant were compared at similar developmental stages.
Table 3.
Wild-Type and pkp1 Seed Storage Compound Accumulation
| WT | pkp1 | WT♂ × pkp1♀ | pkp1♂ × WT♀ | |
|---|---|---|---|---|
| Total FAME (μg/seed) | 6.77 ± 0.7 | 2.71 ± 0.2 | 5.68 ± 0.8 | 6.9 ± 0.2 |
| Protein (μg/seed) | 5.23 ± 0.6 | 4.39 ± 0.5 | ND | ND |
| Seed mass (μg/seed) | 19.20 ± 0.9 | 14.40 ± 1.1 | ND | ND |
Values are the mean of three repeats ± sd. Seed mass determined by measuring the weight of 500 seeds three times. ND, not determined; WT, wild type.
During propagation, it was observed that pkp1 did not establish when sown directly on soil, and this phenotype could be rescued by ectopic expression of either PKp-β1 or PKp-β2 (Figure 6D). A common phenotype of low oil mutants is an inability to establish in the absence of a sugar source (Lu and Hills, 2002; Cernac et al., 2006). Indeed, pkp1 will not establish unless provided with sucrose in the medium (Figure 6E). The pkp1 seedlings were photographed 2 d later than the wild type because germination is slightly delayed in the mutant, and at 10 DAF, the seedlings are at the same developmental stage as the wild type at 8 DAF.
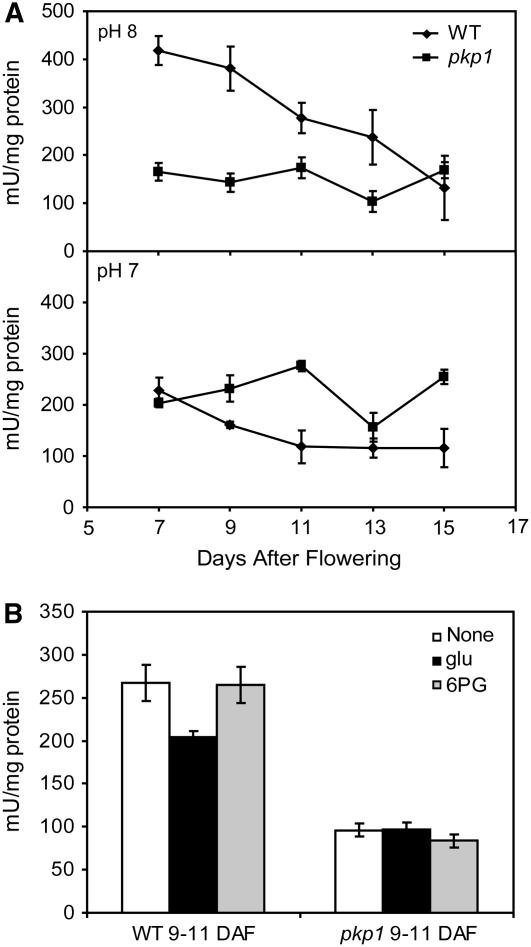
Enzyme activities in developing seeds were measured to determine the extent of the PKp defect in pkp1. The presence of a potentially large number of different cytosolic and plastidic PK isoforms complicates the interpretation of activity assays using crude extracts. However, two factors aide in validating seed PKp activity in crude extracts: (1) all 14 PK encoding genes are not highly expressed in any given tissue at the same time (Schmid et al., 2005); (2) cytosolic PKs typically have pH optima of approximately pH 7.0, whereas plastidic PKs have pH optima of approximately pH 8.0 (Hu and Plaxton, 1996; Smith et al., 2000; Plaxton et al., 2002; this work). Figure 7A shows the time course of PK activity using protein extracts of seeds dissected from staged siliques. At pH 8.0, wild-type PK activity is greatest at 7 DAF and steadily declines throughout seed development. This pattern very closely agrees with the expression profiles of PKp-α and PKp-β1 encoding genes, as shown in Figure 2. The pkp1 mutant at pH 8.0, however, has threefold reduced PK activity at 7 DAF and does not change within experimental limitations throughout the rest of the time course. Pyruvate kinase activity was not reduced in the mutant at pH 7.0 but was actually increased at 11 and 15 DAF. Mutant and wild-type protein extracts were also assayed in the presence of 5 mM Glu or 0.2 mM 6-phosphogluconate (Figure 7B). Recall that Glu at 5 mM inhibited recombinant αβ1 by ∼40%, while αβ2 was inhibited by 70% (Table 2). Native PK activity in 9 to 11 DAF wild-type seed extract was inhibited only ∼25% by 5 mM Glu, while that from pkp1 was unaffected. As shown above, 6-phosphogluconate is a potent activator of αβ2 (Table 2) yet had no effect on wild-type or pkp1 seed PK activity.
Figure 7.
PK Activity in pkp1 and Wild-Type Seeds.
(A) Total PK activity measured at pH 8.0 and 7.0 in saturating substrate conditions. One mU is defined as 1 nmol pyruvate formed per minute. Values are the mean ± sd (n = 4).
(B) Native seed PK activity at 9 to 11 DAF in response to metabolite effectors. Activity was measured at pH 8.0 with subsaturating substrate concentrations. None, no effectors; glu, 5 mM Glu; 6-PG, 0.2 mM 6-phosphogluconate. One mU is defined as 1 nmol pyruvate formed per minute. Values are the mean ± sd (n = 4).
Restoration of Seed Oil Content in pkp1 Expressing β-Subunit–Encoding cDNAs
Dealing with a single T-DNA insertion line, which could potentially harbor secondary mutations that cannot be detected by genotyping, required additional precautions in unambiguously linking the target genotype with the observed phenotype, in this case the reduction in oil content. To address this issue, we constructed transgenic lines in the pkp1 background expressing cDNAs that encode either PKp-β1 or PKp-β2 subunits and included these transgenic lines in the analysis. Expression of these cDNAs was expected to restore the oil content if this phenotype was due to the disruption in the PKp-β1 encoding gene in the pkp1 line, thereby confirming the link between genotype and phenotype.
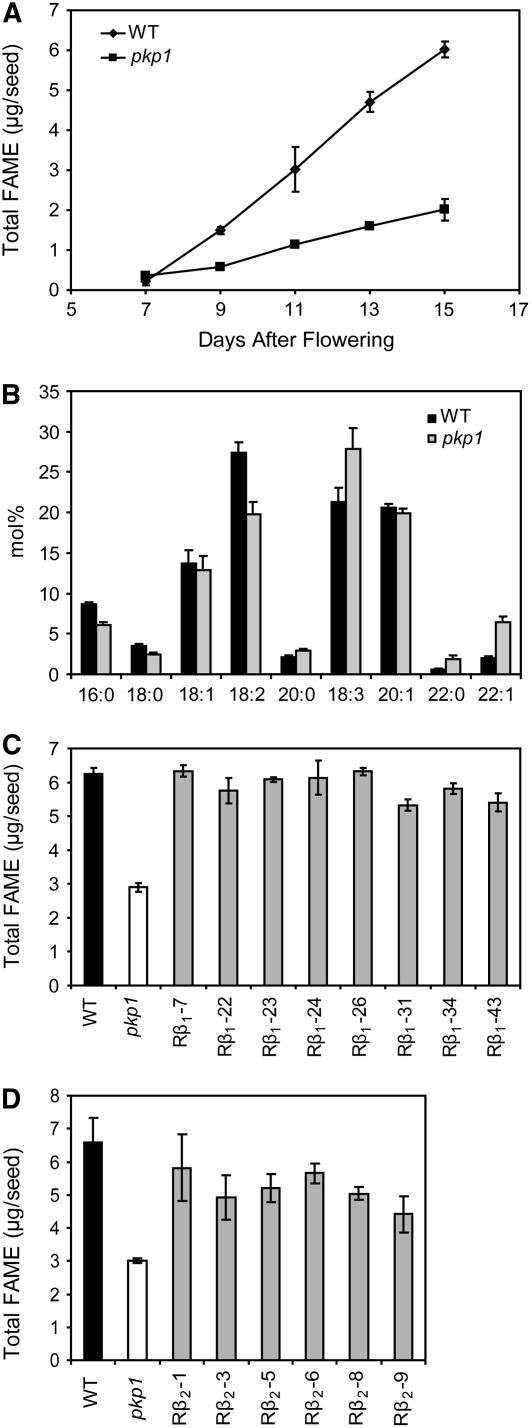
Fatty acid methylester (FAME) analysis of developing seeds revealed that pkp1 accumulates oil in the same temporal pattern as the wild type but at a much lower rate (Figure 8A). The fatty acid composition of the oil in mature seeds was also analyzed. The pkp1 mutant had a decrease in stearic (18:0), oleic (18:1), and linoleic (18:2) acids along with an increase in linolenic (18:3) acid and the proportion of very-long-chain (20 and 22 carbons) to long-chain (16 and 18 carbons) fatty acids (Figure 8B). A T-DNA construct containing a CaMV 35S driven cDNA encoding PKp-β1 was used to rescue the lipid phenotype of pkp1. Antibiotic resistance was used to select transformants, and rescued lines were identified by scoring for visual rescue of the wrinkled seed phenotype. Of 43 independent transformants, eight appeared to be rescued based on seed morphology. Homozygous T3 seeds from individual rescued lines were subjected to FAME analysis (Figure 8C). Overexpression of the PKp-β1 encoding cDNA rescues the lipid phenotype of the mutant but does not result in an increase in oil amount in excess of wild-type seed oil content in any of the lines. A PKp-β2 encoding cDNA was similarly overexpressed in pkp1, and transformants were selected. Twenty independent transformants were identified, and six of these had rescued seed morphology. FAME analysis was performed on homozygous T3 seeds from these lines and showed rescue of the low-oil phenotype of pkp1 (Figure 8D). However, the restoration of oil content was not as complete as was achieved by overexpressing the PKp-β1 encoding cDNA.
Figure 8.
Lipid Phenotype of pkp1 Seeds.
(A) Fatty acid accumulation in developing seeds. Values are the mean ± sd (n = 6).
(B) Fatty acid profile of desiccated mature seeds. Values obtained from FAME analysis of dry seeds. Values are the mean ± sd (n = 6).
(C) Rescue of low oil phenotype in mature seeds of pkp1 by overexpression of PKp-β1 encoding cDNA. The eight individual rescued lines are denoted with Rβ1 and a number. Values are the mean ± sd (n = 3).
(D) Rescue of low oil phenotype in mature seeds of pkp1 by overexpression of PKp-β2 encoding cDNA. The six individual rescued lines are denoted with Rβ2 and a number. Values are the mean ± sd (n = 3).
The complementation data strongly suggest that the seed oil phenotype of pkp1 is caused by the T-DNA insertion located in the PKp-β1 encoding gene. To further exclude the possibility of second site mutations causing the phenotype, an antisense construct was generated to reduce the transcript amount of the PKp-β1 encoding gene specifically in seeds. The Arabidopsis 12S seed storage protein promoter was used for this purpose. Of 20 wild-type transformants identified, three had seeds with phenotypes reminiscent of the pkp1 mutant. None of the empty vector control lines displayed altered seed morphology (wrinkledness). Analysis of FAMEs was performed on the wrinkled seeds, and the three lines contained 3.92 ± 0.6, 2.73 ± 0.3, and 2.22 ± 0.2 μg of oil per seed. The recapitulation of the pkp1 seed phenotype using antisense repression independently corroborates that the low oil phenotype can be caused by reduction or abolishment of expression of the PKP1 gene encoding PKp-β1.
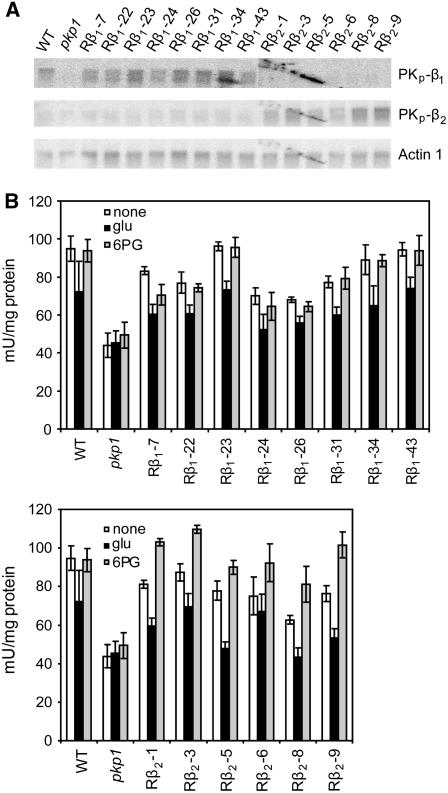
Rescued Lines Have Subunit-Specific Restoration of PK Activity
Pyruvate kinase gene expression and enzyme activity in the rescued lines expressing β-subunit–encoding cDNA in pkp1 were measured. Figure 9A shows RNA gel blots of 9 to 11 DAF silique tissue probed with PKp gene-specific probes. The top panel shows no accumulation of PKp-β1 transcript in pkp1 (as demonstrated by RT-PCR in Figure 5C) and restoration only in the Rβ1 lines overexpressing the PKp-β1 encoding cDNA. In the middle panel, very little PKp-β2 transcript is present in wild-type, pkp1, or the Rβ1 lines. However, the Rβ2 lines clearly have increased amounts of the transcript. Based on gene expression, the Rβ1 and Rβ2 lines appear to have αβ1 or αβ2, respectively, as the dominant PK in silique tissue and thus provided an opportunity to further test the metabolite regulation observed for the recombinant αβ1 and αβ2 complexes. PK activity was first measured at pH 8.0 from 9 to 11 DAF silique material in the absence of effectors. The top and bottom panels of Figure 9B show that PK activity was restored in the Rβ1 and Rβ2 lines. The inclusion of 5 mM Glu in the assay mixture resulted in a moderate inhibition of native PK activity in the wild type and all of the rescued lines, as was observed for the wild type in Figure 7B. The PK activity from the Rβ2 lines, however, responded differently from the wild type and the Rβ1 lines to the presence of 0.2 mM 6-phosphogluconate. PK was activated by 6-phosphogluconate in the Rβ2 lines, which provides needed correlation between the observed metabolite regulation of recombinant and native PK activity.
Figure 9.
PKp Gene Expression and Enzyme Activity in pkp1 Lines Rescued with Ectopic Overexpression of PKp-β1 or PKp-β2.
(A) Expression of PKp encoding genes in siliques of wild-type, pkp1, and rescued lines. Rβ1 represents overexpression of PKp-β1, and Rβ2 denotes overexpression of PKp-β2. An Actin1 probe was used for loading control.
(B) Native PK activity of 9 to 11 DAF siliques in response to metabolite effectors. Top panel includes wild-type, pkp1, and pkp1 lines rescued with overexpression of PKp-β1 (Rβ1). Bottom panel includes wild-type, pkp1, and pkp1 lines rescued with overexpression of PKp-β2 (Rβ2). Activity was measured at pH 8.0 with subsaturating substrate concentrations. None, no effectors; glu, 5 mM Glu; 6-PG, 0.2 mM 6-phosphogluconate. One mU is defined as 1 nmol pyruvate formed per minute. Values are the mean ± sd (n = 4).
Altered Substrate/Product Ratios and Accumulation of Glycolytic Precursors in the pkp1 Mutant
The effects of the pkp1 mutation on the pools of the substrates (PEP and ADP) and products (Pyr and ATP) of PK in developing seeds were analyzed. Metabolite measurements were made at 5 to 7 DAF and at 11 to 13 DAF, when PK activity and chlorophyll content, respectively, are most affected in the mutant seeds. Table 4 shows the results of these analyses. At 5 to 7 DAF, the amount of Pyr in pkp1 is decreased, resulting in a 40% reduction in the Pyr/PEP ratio. At 11 to 13 DAF, there is no difference in the ratio of Pyr/PEP between the wild type and pkp1. Instead, in pkp1, the absolute amounts of Pyr and PEP are proportionally increased. The PEP and Pyr data show that PK activity positively correlates with the Pyr/PEP ratio due mostly to effects on the steady state levels of Pyr. The ATP/ADP ratio is increased almost twofold in pkp1 when both PK activity (5 to 7 DAF) and chlorophyll content (11 to 13 DAF) are the most reduced. It seems that the metabolic perturbations in pkp1 actually result in increased energy status in the developing seeds. This is somewhat surprising as PK activity and photosynthesis are expected to contribute to the ATP pool.
Table 4.
Metabolite Levels in Wild-Type and pkp1 Developing Seeds
| DAF | PEP nmol/g FW | Pyr nmol/g FW | Pyr/PEP | ADP nmol/g FW | ATP nmol/g FW | ATP/ADP | |
|---|---|---|---|---|---|---|---|
| WT | 5 to 7 | 17.5 ± 2.5 | 89.1 ± 7.9 | 5.1 | 15.8 ± 1.6 | 53.7 ± 3.7 | 3.4 |
| 11 to 13 | 13.3 ± 1.6 | 36.5 ± 2.7 | 2.7 | 12.8 ± 1.2 | 38.8 ± 3.7 | 3.0 | |
| pkp1 | 5 to 7 | 18.6 ± 1.5 | 58.1 ± 6.1 | 3.1 | 13.0 ± 1.3 | 66.4 ± 3.4 | 5.1 |
| 11 to 13 | 22.2 ± 4.1 | 59.1 ± 10.4 | 2.7 | 10.9 ± 2.6 | 57.3 ± 7.0 | 5.3 |
Values represent the mean ± sd of at least three repeats. Pyr, pyruvate; FW, fresh weight.
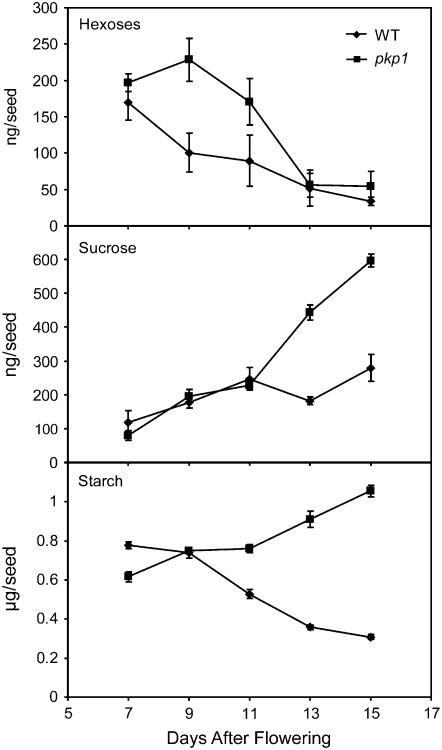
Pyruvate kinase is a control point for glycolysis because its activity has a direct impact on ATP and PEP, the latter of which is an inhibitor of phosphofructokinase in anoxic tissues, such as seeds (Plaxton and Podesta, 2006). It is reasonable that a reduction in PK activity would result in an inhibition of glycolytic flux and an accumulation of carbohydrate precursors. Thus, hexose, sucrose, and starch were also measured in developing wild-type and pkp1 seeds. Figure 10 shows that in the wild type, hexoses accumulated early during development and steadily decreased, while sucrose followed the opposite trend. The pkp1 mutant seeds followed the same trends but contained twice as much of both compounds during peak accumulation times. Starch in the wild type showed the same transient accumulation pattern as previously documented (Focks and Benning, 1998; Baud et al., 2002). However, the pkp1 mutant continued to store starch throughout embryo development (Figure 10). The data suggest that a reduction in PKp activity impairs the catabolism of carbohydrates in developing seeds, resulting in the accumulation of starch.
Figure 10.
Carbohydrate Accumulation in pkp1 and Wild-Type Seeds.
Hexoses (glucose plus fructose), sucrose, and starch levels in developing seeds. Values are the mean ± sd (n = 4).
DISCUSSION
Arabidopsis Has Two Heterooctomeric PKps
Previous kinetic analyses of plant PKs have been limited to enzymes that can be purified in the native state from dissected tissue samples (e.g., Hu and Plaxton, 1996; Smith et al., 2000; Plaxton et al., 2002; Turner et al., 2005). The results of such studies have revealed that PK isoforms vary depending on the tissue and subcellular compartment in question. Arabidopsis tissues, and especially developing seeds, represent a challenge to the traditional enzyme purification approach due to the difficulty in amassing the quantities of tissue required. However, this work on glycolysis is facilitated by the available resources and model characteristics of Arabidopsis, and the findings are crucial if we want to achieve a complete understanding of the biology of this plant. A bioinformatics approach was taken to identify the candidate seed expressed in PKp encoding genes in the Arabidopsis genome sequence. Three potential plastid-targeted, seed-resident PKs were identified based on phylogenetics and gene expression data (Figure 2). The prediction of plastid localization of these proteins was subsequently confirmed using pea chloroplast in vivo import assays (Figure 3). Indeed, PKp-α and PKp-β1 have been found in the stromal fraction of chloroplasts using a proteomics approach (Friso et al., 2004). PKp-β1 was also previously identified in a mitochondrial proteomics study, but plastid contamination could not be ruled out (Giege et al., 2003). Previous research on PKp-α orthologs from B. napus and R. communis detailed the proteins' transit peptide cleavage sites (Wan et al., 1995; Plaxton et al., 2002). The N-terminal sequence of the mature B. napus protein aligns with a region ∼40 amino acids C-terminal to the predicted Arabidopsis cleavage site, while the R. communis protein has a transit peptide of 83 amino acids. These data suggest that the transit peptide of PKp-α may be longer than the predicted 55 amino acids. However, analysis of the observed molecular masses of the precursor and mature proteins after pea chloroplast import support the predictions made by TargetP and ChloroP (Figure 3). Moreover, the Arabidopsis PKp-α protein contains a domain (79% amino acid identity to R. communis PKp-A) that is responsible for altered import characteristics of R. communis PKp-A (Wan et al., 1995). Our results indicate that the presence of this sequence does not affect the ability of Arabidopsis PKp-α to be imported into and processed in pea chloroplasts.
It was determined that the Arabidopsis PKp enzymes are most likely ∼460-kD heterooctomers of 60 and 57 kD subunits with 4α4β stoichiometry (Figure 4). The reconstitution of active PKps from individually purified inactive subunits in this study and the in vivo co-IP results leave little ambiguity as to the heteromeric structure of the Arabidopsis PKps. Some studies of recombinant PKp polypeptides from developing R. communis endosperm concluded that the PKp-α and PKp-β homologs are distinct enzymes (Blakeley and Dennis, 1993; Blakeley et al., 1995; Wan et al., 1995), while others have determined that the same proteins are actually subunits of a single heteromeric PKp complex (Plaxton et al., 1990; Plaxton, 1991; Negm et al., 1995). The data presented in this article support the conclusion that PKp is a complex composed of two different subunits.
Arabidopsis PKp Activity Is Determined by Two β-Subunits
Kinetic analysis revealed that the reconstituted Arabidopsis enzymes behave much like previously documented PKps (pH optima of ∼8.0, Mg2+ and K+ requirement, and S0.5 values for PEP and ADP in the 100 to 300 μM range). A pH optimum of 8.0 is potentially important for these enzymes. Such a pH is generated in the plastid stroma in response to light. As the plastids of Arabidopsis seeds are green and presumably photosynthetic, it is possible that in vivo PKp is light activated via alkalinization of the stroma. Such regulation of PKp could contribute to the light-induced stimulation of fatty acid synthesis in green seeds (Goffman et al., 2005). Distinct differences between the two isoforms in Arabidopsis were also discovered. The αβ1 form is a more efficient enzyme with a higher specific activity and lower S0.5 values for both PEP and ADP (Table 1). In addition, αβ1 is 3 to 5 times more efficient at using alternative nucleoside diphosphates. The αβ2 enzyme, on the other hand, is more responsive to the strong metabolite effectors Glu, oxalate, and 6-phosphogluconate (Table 2). In fact, αβ1 is completely insensitive to the activating effect of 6-phosphogluconate. This is interesting since what has been described as the major PKp from B. napus is activated by 6-phosphogluconate (Plaxton et al., 2002). Apparently, Arabidopsis and B. napus have diverged in this regard as the gene for the β2 subunit present in the 6-phosphogluconate–regulated PKp (αβ2 enzyme) from Arabidopsis is hardly expressed in any tissue, and 6-phosphogluconate regulation is not detectable in seed extracts (Figures 2B and 7B). A distinct feature of the Arabidopsis PKps is their inhibition by Glu (Table 2). Regulation by this effector has been reported for PKcs from other plants but not PKps (Hu and Plaxton, 1996; Smith et al., 2000). It should be noted that while the presence of either β1 or β2 in the Arabidopsis PKp complex results in different regulatory properties, these subunits should not be considered purely regulatory. They both contain fully conserved PK active sites in which chemical or genetic modification results in an inactive PK complex (see Supplemental Figure 3 online). These experiments, in combination with the fact that the α-subunit has no activity when assayed alone (see Supplemental Figure 1B online), support a model in which both α and either β1 or β2 subunits, respectively, are required for enzyme activity, and the specific interactions between the subunits result in differential kinetic properties.
The primary structures of the recombinant PKps may not be the same as the native enzymes; thus, the kinetic data must be considered with caution. However, as the transit peptide cleavage site predictions agree well with the observed molecular masses of precursor and mature PK subunits (Figure 3), the differences are likely to be minimal between the native and recombinant proteins. Moreover, native PK activity from developing seeds responds to metabolite effectors as predicted by analysis of the recombinant proteins (Figures 7B and 9B). These in vivo data are in agreement with the kinetic analysis of in vitro enzyme activity. It should also be noted that specific posttranslational modifications could result in changes to the enzyme structure and or function, and these possibilities were not explored. Soybean (Glycine max) PKc, for instance, can be partially degraded at the C terminus in vivo, which results in altered regulatory properties (Tang et al., 2003).
The αβ1 Form of PK Is Dominant in Seeds
Perhaps most significant is the realization that PKp from Arabidopsis exists as differentially regulated heteromers resulting from the interaction of the α-subunit with one or the other β-subunit. The two PKp-β subunits of Arabidopsis likely arose through gene duplication as indicated by the similarity of their amino acid sequence and gene structure (Figure 5A) and subsequently evolved unique regulatory features. The αβ1 enzyme is likely to be dominant in most tissues based on gene expression, but it is possible that under certain stress conditions the αβ2 enzyme is produced at higher rates. The regulatory properties of the αβ1 enzyme, notably the lower S0.5 for ADP and PEP, higher Vmax, lower sensitivity to metabolic regulation, and ability to use other NDPs more efficiently, make it a better enzyme for processing carbon at high rates, independent of the metabolic status of the tissue. The αβ2 enzyme, however, is less active and more susceptible to metabolite-based regulation and may play more specialized roles. It will be interesting to study the in vivo relationship of these subunits and whether or not one enzyme complex can contain a mixture of β-subunits that would further fine-tune PK activity for specific metabolic demands. Some of these regulatory differences become obvious in the pkp1 lines overproducing the β2 units, as it is not sufficient for maximal oil accumulation in seeds.
Mutants in the PKp-α gene, which should result in complete inactivation of both PKps, could not be isolated. Only one mutant, in the PKp-β1 encoding gene, was identified that had no transcript accumulation and a seed phenotype (Figure 5, Table 3). Methods to separate the native Arabidopsis PK isoforms (isoelectric focusing, zymograms, and native-PAGE) were unsuccessful and so PKp activity was assayed at pH 8.0 to minimize the background from cytosolic enzymes. The PK activity profile at pH 8.0 in the wild type very closely matches the expression of the α and β1 subunit encoding genes (Figure 2B), while in pkp1, the activity is greatly reduced and does not change (Figure 7A). In addition, seed PK activity was insensitive to activation by a 10-fold higher concentration of 6PG than the Ka of the αβ2 complex (Figure 7B). All these results agree with the in vitro data and corroborate a specific inactivation of the αβ1 enzyme in the pkp1 mutant. Furthermore, the mutant supports the initial hypothesis that the influx of photosynthate into embryo tissue and the high demand for lipid and amino acid precursors requires high PKp activity. The regulatory properties of the αβ1 enzyme mentioned above make it a prime candidate for this role. Taken together with the lack of transcript accumulation for the gene encoding PKp-β2 (Figure 2B), these data indicate that the αβ1 enzyme is the major PKp isoform present in developing Arabidopsis seeds. The increase in PK activity at pH 7.0 in pkp1 suggests a compensatory mechanism in the mutant. It is possible that more pyruvate is being generated in the cytosol and imported into plastids. A preliminary flux map of carbon metabolism in B. napus embryos shows that 30% of the pyruvate used for fatty acid synthesis is cytosolic in origin (Schwender and Ohlrogge, 2002; Schwender et al., 2003, 2004b). If the same is true for Arabidopsis, the increase in PK activity at pH 7.0 only marginally compensates, as pkp1 still has a 60% reduction in seed oil. Based on gene expression data (Figure 2B; see Supplemental Figure 4 online), a small amount of the PKp-β2 subunit could also be present in developing seed. The presence of this subunit could explain the incomplete loss of PK activity and seed oil in the pkp1 mutant.
Seed Metabolism Is Dependent on Proper PKp Function
The pkp1 mutant seeds are smaller and less green than wild-type seeds (Figures 6A and 6B). The reduction in chlorophyll in pkp1 developing seeds could be the result of sugar accumulation (Figure 10), as sugars are known to repress chlorophyll accumulation and photosynthetic gene expression (Jang and Sheen, 1994; Jang et al., 1997). It is also possible that a pleiotropic effect of the pkp1 mutation is reduced chlorophyll biosynthesis. Dark treatment has been shown to decrease fatty acid synthesis by 23% in B. napus embryos (Ruuska et al., 2004). However, all light-activated processes are affected by this treatment, and the reduced biosynthetic capability was linked to a lack of light-induced activation of certain enzymes. In the case of pkp1, light still penetrates the seed and is capable of inducing enzyme activities and other processes. Thus, the primary metabolic defect brought on by a reduction in PKp activity is likely the major factor contributing to the low oil phenotype of pkp1.
In both wild-type and pkp1 seeds, fatty acids accumulate in a linear time course from ∼5 DAF until at least 15 DAF (Figure 8A). The rate of accumulation, though, is reduced by ∼60% in the mutant, which correlates with the reduction in seed oil and the reduction in total PK activity at pH 8.0. The fatty acid profile of mature pkp1 seeds is very similar to that of the wri1 mutant and plants with altered biotin carboxyl carrier protein gene expression (Focks and Benning, 1998; Thelen and Ohlrogge, 2002). These mutations impair fatty acid synthesis either by reducing the supply of precursors or by inhibiting the acetyl-CoA carboxylase reaction, respectively. It is likely that in the pkp1 mutant a reduction in precursors for fatty acid synthesis is the cause of the altered fatty acid profile. Indeed, the steady state level of pyruvate is reduced in pkp1 seeds compared with the wild type at 5 to 7 DAF, which correlates with the onset of fatty acid biosynthesis (Table 4). Interestingly, the amounts of PEP and pyruvate, but not the ratio of the two, are increased in pkp1 at 11 to 13 DAF (Table 4). The idea of a reduction in the supply of precursors for fatty acid biosynthesis is further supported by the accumulation of carbohydrates in the mutant seed, a phenotype similar to that of wri1 (Figure 10). It is also possible that the reduction in the rate of fatty acid synthesis is brought on by a lack of ATP. Seeds of B. napus (and likely Arabidopsis) represent a low oxygen environment (Vigeolas et al., 2003), and even with photosynthesis PK could play an important role in the production of ATP. A mutant defective in a plastidic ATP/ADP transporter with reduced ATP import capacity into plastids has reduced oil content in its seeds (Reiser et al., 2004). Moreover, this mutant was shown to compensate for reduced ATP import by increasing the expression of the genes encoding PKp-β1 and PKp-β2. A similar increase in transcript level for any of the PKp subunits is not seen in pkp1 seeds (see Supplemental Figure 4 online). Additionally, steady state levels of ATP in the mutant seeds were actually increased relative to the wild type (Table 4), possibly as a result of increased PKc activity in the mutant (Figure 7A). Elevated PKc activity in pkp1 might be an indicator of enhanced cytosolic glycolytic flux into mitochondrial respiration, which could also elevate the ATP/ADP ratio. Either way, it is unlikely that ATP production is a major function of PKp in developing Arabidopsis seeds. The pkp1 mutant is rescued by ectopic overexpression of the PKp-β1 and PKp-β2 encoding cDNAs except that oil accumulation is recovered less fully in the Rβ2 lines (Figures 8C and 8D). No transgenics were observed that had an increase in seed oil content or in PK activity, despite higher than wild-type expression levels of the PKp-β1 or PKp-β2 encoding genes (Figure 9A). This might indicate that in these lines the amount of PKp-α is limiting as it is required for activity.
Concomitant with the reduction in seed oil in pkp1 is the accumulation of increased amounts of hexoses, sucrose, and starch (Figures 6C and 10). It seems that there is a redirection of carbon partitioning in pkp1 in which less hexose and sucrose are catabolized via glycolysis but are instead incorporated into starch. However, the starch accumulated in pkp1 seeds at 15 DAF only accounts for ∼20 to 25% of the carbon not incorporated into fatty acids (Table 3, Figure 10). One potential mechanism for the observed accumulation of carbohydrates is that elevated PEP (Table 4), which is a potent inhibitor of plant phosphofructokinases (Plaxton and Podesta, 2006), could slow the entry of hexose-phosphates into glycolysis, resulting in a misregulation of metabolism. It is interesting that in pkp1 excess carbon is not redirected into storage protein synthesis. Instead, protein levels are slightly decreased (Table 3), which could be a result of decreased glycolytic flux. The results of this study are in support of the metabolic model depicted in Figure 1 in which PEP metabolized by PKp in the plastid is the main source of precursors for fatty acid synthesis. The compartmentation of metabolism apparently serves to isolate metabolic pathways such that specific products can be generated from distinct pools of substrates. In the case of seed oil metabolism, pyruvate generated in the plastid is used mainly for fatty acid synthesis and cannot be fully replaced by cytosolic pools. It seems that Arabidopsis seeds are programmed to make oil from plastidic pyruvate and if that pathway is perturbed, as is the case in the pkp1 mutant, the carbon is stored in a different form, such as starch. Similar redirection of carbon flux from oil biosynthesis to starch accumulation has been noted in the leafy cotyledon1 (Meinke et al., 1994) and trehalose-6-phosphate synthase1 (Gomez et al., 2006) mutants of Arabidopsis. As such, the pkp1 mutant provides another example for a plant with altered carbon partitioning in developing seeds.
METHODS
Bioinformatics
Pyruvate kinase genes highly expressed in seeds were initially identified in the seed EST database (White et al., 2000). The 14 putative Arabidopsis thaliana PK-annotated sequences from The Arabidopsis Information Resource website (www.arabidopsis.org) were used for this work. Other annotated PK sequences were downloaded from the National Center for Biotechnology Information. Predicted full-length protein sequences were aligned using ClustalW (Li, 2003) available from the Biology Workbench (San Diego Supercomputer Center, University of California, San Diego; http://workbench.sdsc.edu/). A phylogenetic tree was generated in Phylip format and bootstrapped using a random number generator seed of 111 and 1000 bootstrap trials. The phylogram was visualized using the TreeView program (version 1.6.6; Page, 1996). No manual adjustments were made to the initial alignment. Global gene expression data were mined from the AtGenExpress developmental database (Schmid et al., 2005).
cDNA Cloning and Recombinant Protein Production
The cDNAs corresponding to At3g22960, At5g52920, and At1g32440 were generated from total silique RNA isolated as previously described (Verwoerd et al., 1989). Reverse transcription was done with the Qiagen Omniscript RT kit and 600 ng of total RNA. Primers listed in Table 5 were used for PCR amplification of cDNAs to generate products with and without predicted chloroplast transit peptides and with or without epitope tags. All cDNAs were inserted into pBluescript II (Stratagene) and sequenced at the Michigan State University (MSU) Research Technology Support Facility. The vector pET-15b (Novagen) was used for recombinant protein expression in Escherichia coli strain BL21 (DE3) pLysS (Novagen). The PKp-α encoding fragment was inserted into the BamHI and Klenow-filled NdeI sites of pET-15b by digesting with KasI, filling in with Klenow, and then by digesting with BamHI and ligating. The open reading frame for PKp-β1 was inserted into the BamHI and Klenow-filled NdeI sites of pET-15b by digesting with BglII, filling in with Klenow, and then by digesting with BamHI and ligating. The PKp-β2 encoding fragment was inserted into the XhoI and Klenow-filled NdeI sites of pET-15b by first digesting with SpeI, filling in with Klenow, and then by digesting with XhoI and ligating. Proteins were expressed at 28°C by inducing at an OD600 of 0.6 with 0.5 mM isopropylthio-β-galactoside and allowing the cultures to grow for an additional 4 h. His-tagged proteins were recovered over Ni-NTA resin using standard protocols. Purified proteins were exchanged into a buffer of 50 mM NaxHxPO4, pH 7.9, 150 mM NaCl, 5 mM MgCl2, and 10% glycerol. The 6X-His tags were cleaved using a thrombin cleavage capture kit available from Novagen. Proteins were quantified with the Bradford method using reagent from Sigma-Aldrich.
Table 5.
Primer Sequences
| Gene or SALK Line | Primer |
|---|---|
| PKp-β2 | P,PE(f) 5′-ACTAGTATTAAAATCTCCGAAGATAG-3′ |
| P(r) 5′-CTCGAGTCATCCACCTATCTTTATCT-3′ | |
| PE(r) 5′-CTCGAGTCATCCCTTGTCATCGTCATCCTTATAATCTCCACCTATCTTTATCTT-3′ | |
| O,OE(f) 5′-GGTACCCCTCAGGTTTCTCTGCTCAT-3′ | |
| O(r) 5′-GGTACCACTGTGAGTGATTCAAAAAA-3′ | |
| OE(r) 5′-GGTACCTCATCCCTTGTCATCGTCATCCTTATAATCTCCACCTATCTTTATCTT-3′ | |
| RT(f) 5′-GGGGATGTACCGCAGCCGATA-3′ | |
| RT(r) 5′-GGATGCCGAGGTTCTGACAGG-3′ | |
| SALK_013574 | RP 5′-TTTCACACAACAAATTCGTTCATT-3′ |
| LP 5′-CAGCTTCCGCGAGTTTCCAAATCA-3′ | |
| SALK_142845 | Same as SALK_013574 |
| PKp-α | P,PE(f) 5′-GGCGCCTCCTCGTCATCATCTCC-3′ |
| P(r)-5′-GGATCCTTACGGGACGTTCATTACCT-3′ | |
| PE(r) 5′-GGATCCTTACAAATCCTCCTCACTAATCAACTTTTGCTCCGGGACGTTCATTACCTG-3′ | |
| OE(f) 5′-GGTACCAGCCAACTGTCCTGAGATTT-3′ | |
| OE(r) 5′-GGTACCTTACAAATCCTCCTCACTAATCAACTTTTGCTCCGGGACGTTCATTACCTG-3′ | |
| SALK_096141 | LP 5′-CCAAATTCAACACTCTCACACTTCG-3′ |
| RP 5′-CCATCCCACCATCAACCAAAA-3′ | |
| RT(f) 5′-GCTGCTCGTTCCCGTGGAGG-3′ | |
| RT(r) 5′-TTGAAGCGGTACAGACTCAT-3′ | |
| SALK_024870 | LP 5′-TCTCGGACATGCTGCAATCAA-3′ |
| RP 5′-TTCGCATCAGTCATCTTCGTCTTC-3′ | |
| PKp-β1 | P,PE(f) 5′-AGATCTGCTCGTGTTGAGACTGA-3′ |
| P(r) 5′-GGATCCTTAAACCTTGCGGACTTGGA-3′ | |
| PE(r) 5′-GGATCCTTATGCATAATCGGGAACATCATAGGGATAAACCTTGCGGACTTGGAT-3′ | |
| O,OE(f) 5′-GGTACCCTTCACTACTCTGTCTCAGC-3′ | |
| O(r) 5′-GGTACCCAAAAACGAGGTTCTACATA-3′ | |
| OE(r) 5′-GGTACCTTATGCATAATCGGGAACATCATAGGGATAAACCTTGCGGACTTGGAT-3′ | |
| SALK_042938 | LP 5′-TGAGATAGCATTTCAATTTGATGCG-3′ |
| RP 5′-GGCAAATCATTCACTTAGGATGGA-3′ | |
| RT1(f) 5′-CTGGGATGAATGTTGCTAGG-3′ | |
| RT2(r) 5′-GTCAACTTTGTTCTCCACTCC-3′ | |
| SALK_042681 | LP 5′-ACAGCCAATCTGGCGATCTCA-3′ |
| RP 5′-TATTACAGGTCTATTTCTTTCGG-3′ | |
| T-DNA LB | 5′-GTTCACGTAGTGGGCCATCG-3′ |
| ACTIN-1 | RT(f) 5′-AACAATCGATGGACCTGACTCG-3′ |
| RT(r) 5′-TGCGACAATGGAACTGGAATGG-3′ |
Primers used for cloning of recombinant proteins without predicted chloroplast transit peptide with (PE) or without (P) epitope tags, for cloning of full-length proteins for overexpression in plants with (OE) or without (O) epitope tags, for genotyping of SALK_KO lines (LP and RP), and for expression analysis (RT).
Pea Chloroplast Import Assays
The cDNAs encoding PKp-α, PKp-β1, and PKp-β2 inserted into pBluescript II (Stratagene) were used in this study. These genes were transcribed/translated, and proteins were subsequently labeled with [35S]-Met using the TNT-coupled wheat germ extract system according to the manufacturer's recommendations (Promega). The PKp plasmids were linearized prior to translation with the T3 or T7 RNA polymerase TNT-coupled wheat germ extract system. The plasmid containing the gene encoding the Rubisco small subunit used for control purposes has been described (Olsen and Keegstra, 1992). Pea plants (Pisum sativum var Little Marvel; Olds Seed) were grown under natural light in the greenhouse at 18 to 20°C. Chloroplasts were isolated from 8- to 12-d-old plants as described previously (Bruce et al., 1994). Binding or import reactions were performed according to published protocols (Tranel et al., 1995). Post-treatments of import reactions with either thermolysin or trypsin were performed as described previously (Jackson et al., 1998). All fractions were analyzed by SDS-PAGE (Laemmli, 1970) and fluorography (Tranel et al., 1995).
Native-PAGE Analysis and Gel Filtration Chromatography
Native-PAGE was performed using 7.5% acrylamide Ready-Gels from Bio-Rad. Freshly prepared protein was used, and 5 pmols of each PK subunit were loaded per well. The gels were run at 4°C at 140 V. PK activity staining was done as previously described, except that 50 mM HEPES-KOH, pH 8.0, buffer was used (Rivoal et al., 2002). Immunoblotting was done using standard protocols and monoclonal anti-c-myc, anti-FLAG, and anti-HA antibodies from Sigma-Aldrich. Antibodies were tested for specificity against epitope-tagged and untagged versions of all three PK subunits. Gel filtration was accomplished using a Superdex 200 HR10/30 column with a flow rate of 0.4 mL min−1 and a buffer of 50 mM HEPES-KOH, pH 8.0, 10% glycerol, 50 mM KCl, 5 mM MgCl2, 1 mM EDTA, and 0.04% NaN3. A standard curve was generated with the HMW gel filtration calibration kit from GE Healthcare. For the samples, 300 μL of 0.5 mg mL−1 (per subunit) solution was injected, and 0.35 mL fractions were collected.
Co-IP Analysis
Full-length epitope-tagged versions of PKp-α, PKp-β1, and PKp-β2 were produced by PCR with primers listed in Table 5. The respective DNA fragments were then inserted into the KpnI site of a modified pCAMBIA1300 vector (CAMBIA), which contained the EcoRI/HindIII expression cassette from pBIN121 (Clontech). Arabidopsis was stably transformed with these constructs, and protein expression was monitored by immunoblotting. Silique tissue was ground in 3 volumes (w/v) of extraction buffer containing 50 mM HEPES-KOH, pH 8.0, 5 mM MgCl2, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.1% Triton X-100, 10% glycerol, 2 mM benzamidine, 2 mM amino-n-caproic acid, 1 mM PMSF, and 1 mM PEP. Debris was removed by centrifugation at 16,000g for 10 min, and the supernatant was used for SDS-PAGE or co-IP. For co-IP, 250 μL of supernatant was precleared by incubation for 1 h at 4°C with 30 μL of a 50% slurry of Protein-A sepharose. The supernatant was kept and mixed with 5 μg of the appropriate antibody and was then rocked at 4°C. After 1 h, 30 μL of Protein-A sepharose slurry was added and the mixture rocked for an additional hour. After this time, the Protein-A sepharose with bound antibody and proteins was washed four times in extraction buffer and then mixed with SDS-PAGE sample buffer, heated, and the supernatant loaded on gels. Excised bands were submitted to the MSU Research Technology Support Facility for tryptic digest and liquid chromatography–tandem mass spectrometry. The generated data were then compared against the Arabidopsis proteome using MASCOT software (Matrix Science).
PK Enzyme Assays and Kinetic Analysis
All chemicals were from Sigma-Aldrich. Pyruvate kinase activity was detected by coupling the production of pyruvate to the conversion of NADH to NAD+ by lactate dehydrogenase unless otherwise noted. Reactions were kept at 25°C, were started by the addition of enzyme mix, and were linear for at least 5 min. Absorbance at 340 nm was monitored using a FLUOstar Optima 96-well plate reader (BMG Labtech). Standard PKp reaction mixtures contained 50 mM HEPES-KOH, pH 8.0, 5% polyethylene glycol 8000, 50 mM KCl, 15 mM MgCl2, 1 mM DTT, 2 mM PEP, 1 mM ADP, 0.2 mM NADH, and 2 units mL−1 desalted rabbit muscle lactate dehydrogenase. PEP phosphatase activity was corrected for by omitting ADP from the reaction. Reactions at pH 7.0 were done using 50 mM MOPS, pH 7.0, instead of HEPES.
For kinetic analysis, 2.5 pmol of each subunit were mixed and used per reaction. S0.5 and Vmax values were determined by fitting the Hill equation to plots of initial velocity versus substrate concentration using origin 7.0 (OriginLab). pH optimum curves were generated using a 25 mM MES and 25 mM Bis-Tris-propane buffer over a range of pH levels. For inhibitor/activator studies, metabolite stocks were made equimolar with MgCl2 and were pH adjusted to 8.0. Metabolites were tested at pH 8.0 with 100 μM PEP and 150 μM ADP for αβ1 and 150 μM PEP and 300 μM ADP for αβ2. The metabolites tested were as follows: glucose, fructose, 6GP, G1P, F6P, F16bP, DHAP, G3P, 2PG, acetate, OAA, citrate, iso-citrate, 2-oxoglutarate, succinate, fumerate, malate, gro3P, 2P-glycolate, glycolate, Ala, Arg, Asn, Asp, Cys, Gln, Glu, Gly, His, Leu, Lys, Met, Ser, AMP, ADP, ATP, ADP-Glc, UDP-Glc, UDP-Gal, R5P, 6PG, KPi, NaNO3, and NH4Cl, all at 10 mM; MgPPi, NADH, NADPH, NAD+, NADP+, oxalate, Ile, Phe, Pro, Thr, Trp, Tyr, Val, CoA, Mal-CoA, Ac-CoA, and shikimate, all at 0.5 mM; F26bP, oleoyl-CoA, oleate, and LPA, all at 0.05 mM. Oxalate, glyoxylate, and OAA were found to inhibit the LDH reaction so PK activity was measured by coupling to the ATP-dependent conversion of glucose to glucose-6-phosphate by hexokinase followed by the NAD+-dependent conversion of glucose-6-phosphate to 6-phosphogluconate by glucose-6-phosphate dehydrogenase. The reaction mix was adjusted to contain no NADH or LDH, but instead to have 1 mM NAD+, 5 mM glucose, 2 units mL−1 hexokinase, and 2 units mL−1 G6PDH. I50 and Ka values are the concentration of a metabolite required for 50% maximum inhibition or activation, respectively. They were calculated by fitting a modified Hill equation to plots of initial velocity versus effector concentration as previously described (Ballicora et al., 2005).
Site-directed mutagenesis of PK subunits was done using the QuikChange-XL site-directed mutagenesis kit using primers designed to the manufacturer's specifications (Qiagen). An absolutely conserved Lys residue in the PK active site was mutated to Leu for each subunit (PKp-α, K344L; PKp-β1, K325L; PKp-β2, K314L). This mutation has previously been shown to abolish PK activity (Sakai, 2005). Chemical inactivation was achieved by incubating 5 μM solutions of purified subunits with either water or 100-fold molar excess of 2,4,6-trinitrobenzenesulfonic acid in the dark at room temperature for 1 h. This treatment has been shown to inactivate PK subunits by covalent modification of Lys residues without abolishing protein interactions (Hollenberg et al., 1971). Excess 2,4,6-trinitrobenzenesulfonic was quenched with an equal volume of 100 mM Tris-Cl, pH 7.5, for 20 min on ice, and the proteins were used directly in enzyme assays.
Plant Growth and Transformation
All Arabidopsis plants were of the Columbia (Col-2) ecotype, except for the SALK T-DNA lines, which were Col-0. All seeds were first sterilized in 20% bleach and 0.05% Triton X-100 for 20 min and were then rinsed five times with water and plated on half-strength Murashige and Skoog medium, pH 5.9, 0.9% agar, and 2% sucrose. When appropriate, kanamycin or hygromycin B were included in the medium at 50 μg mL−1 and 25 μg mL−1, respectively. Seeds were stratified at 4°C for 3 d prior to being germinated in an incubator (AR-75; Percival Scientific) at a photon flux density of 60 to 80 μmol m −2 s−1 with a light period of 16 h (22°C) and a dark period of 8 h (18°C). Seedlings were transferred to 3.5-inch square pots and were grown in a soil mix as described previously (Xu et al., 2002) with a 16-h photoperiod, a day temperature of 22°C, and a night temperature of 20°C at a photon flux density of 100 to 120 μmol m −2 s −1. The plants were fertilized with half-strength Miracle-Gro plant food (Scotts) every 15 d. Wild-type and mutant Arabidopsis plants were prepared for transformation as previously described (Cernac and Benning, 2004). When ready, plants were transformed using the floral dip method (Clough and Bent, 1998). Competent cells of Agrobacterium tumefaciens strain C58C1 GV3101 pMP90 (Koncz and Schell, 1986) were prepared and transformed as previously described (Shen and Forde, 1989).
T-DNA Mutant Isolation and Characterization
T-DNA insertion lines were obtained from the SALK T-DNA insertion population (Alonso et al., 2003). Mutants were selected on growth medium containing kanamycin, and T-DNA insertions were confirmed using PCR primers specific for gene sequences and the T-DNA left border. Primers were designed using the i-sect tool (http://signal.salk.edu/tdnaprimers.html). Insertion sites were confirmed by sequencing the PCR product of the left border primer and a gene-specific primer. Expression of the target gene was analyzed using RT-PCR. Total RNA was isolated from seedlings using the Qiagen RNeasy kit. Total RNA (600 ng) was used for reverse transcription using the Qiagen Omniscript RT kit. PCR was done using 5% of the RT product. Actin1 (At2g37620) was used for control purposes. The PCR consisted of 25 cycles of 95°C for 30 s, 60°C for 45 s, and 72°C for 1 min, followed by a 10 min 72°C extension. Complementation of the mutant was done using full-length cDNAs inserted into the KpnI site of CaMV 35S containing the pCAMBIA1300 derivative mentioned above. An antisense construct was generated using the same full-length cDNA for PKp-β1 inserted in the antisense orientation into the KpnI site of the binary vector pBinAR-Hyg (Dörmann and Benning, 1998) containing the 12S seed storage protein promoter from Arabidopsis (Ohlrogge et al., 2006). Analysis of gene expression in the rescued lines was done using RNA gel blots. Total silique RNA was extracted using a previously described protocol (Verwoerd et al., 1989) followed by DNase treatment and cleanup using the Qiagen RNeasy kit. RNA gel blot analysis (5 μg total RNA) was performed as previously described (Dörmann and Benning, 1998). The blots were analyzed using a phosphor imager (Molecular Dynamics).
Chlorophyll was quantified in leaves and seeds as previously described (Lichtenthaler, 1987). Seeds were imaged using a Leica MZ 12.5 dissecting microscope (Leica Microsystems) equipped with a Spot Insight color camera (Diagnostic Instruments). For electron microscopy, seeds were dissected out of staged silques and soaked in water for 1 h. Embryos were dissected from seed coats and were embedded in 2% agarose. The embryos were fixed for 2 h at room temperature with 2.5% glutaraldehyde and 2.5% paraformaldehyde in 0.1 M cacodylate buffer and then post-fixed in 1% (w/v) osmium tetrachloride in 0.1 M cacodylate buffer. The samples were then dehydrated in a graded series of acetone, embedded in Poly BD 812 resin, and sectioned. The thin sections (∼70 to 100 nm) were stained with uranyl acetate and lead citrate prior to examination in a JEOL 100CX electron microscope.
Enzyme activities were measured from proteins extracted in a buffer of 50 mM Tris-Cl, pH 7.5, 5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.1% Triton X-100, 10% glycerol, 2 mM benzamidine, 2 mM amino-n-caproic acid, and 1 mM PMSF. PK activity was measured as described above.
Seed Metabolite Analysis
Seed oil quantification by fatty acid methyl ester analysis was done as previously described (Focks and Benning, 1998). Seed storage proteins were extracted from 50 mg of dry seed by first grinding in a mortar and pestle followed by two extractions of the tissue with 30 volumes of hexane. The delipidated seed material was pelleted by centrifugation at 13,000g for 10 min. The pellet was then dried in a speed-vac and extracted twice for 15 min with 0.5 volumes of 50 mM Tris-HCl, pH 8, 200 mM NaCl, 5 mM EDTA, 0.1% Tween 20, 2 mM benzamidine, 2 mM amino-n-caproic acid, and 1 mM PMSF. Water bath sonication was used to resuspend the pellets. The supernatants from the extractions were combined, and a 1:5 dilution was used to quantify protein using the Bio-Rad DC protein assay kit. Glucose, fructose, sucrose, and starch were extracted from developing seeds and quantified as previously described (Focks and Benning, 1998). PEP and pyruvate were extracted from developing seeds with perchloric acid and quantified using an NADH fluorescence assay as previously described (Hausler et al., 2000). ADP and ATP were measured in the same extracts with an ATP bioluminescence assay kit (Sigma-Aldrich) using a previously described protocol (Ruuska et al., 2000).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers (www.arabidopsis.org) used in this study are as follows: At3g22960 (encoding PKp-α), At5g52920 (encoding PKp-β1), At1g32440 (encoding PKp-β2), At2g36580, At3g04050, At3g25960, At3g49160, At3g52990, At3g55650, At3g55810, At4g26390, At5g08570, At5g56350, and At5g63680. GenBank accession numbers for non-Arabidopsis protein sequences used are as follows: Nt PKp-A, Q40545; Os PKp-A, NP_001059042; Rc PKp-A, Q43117; Nt PKp-G, Q40546; Se PCC6301 PK, YP_172116; Se WH8102 PK, NP_897391; Pfu PK, NP_578917; Sa PK, YP_256251; Ec PK-1, AAA24392; Hs PK-L, BAA02515; An PK, Q12669; Sc PYK1, NP_009362; Os PKc, BAD81116; Nt PKc, Q42954; St PKc, P22200; and Gm PKc, Q42806.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. PKp Protein Purification and Subunit Association Biochemistry.
Supplemental Figure 2. Protein Sequences, Predicted Transit Peptides, and Proteomics Coverage of PKp-α, PKp-β1, and PKp-β2.
Supplemental Figure 3. Analysis of Subunit Requirement for PK Activity by Site-Directed Mutagenesis and Chemical Inactivation.
Supplemental Figure 4. Semiquantitative RT-PCR Analysis of PKp-α and PKp-β2 Gene Expression in pkp1 Seeds.
Supplementary Material
Acknowledgments
We thank Alex Cernac for initial input in getting this project started, for thoughtful discussions along the way, and for providing pBinAR-Hyg containing the 12S seed storage protein promoter. We also thank Ken Keegstra and Jack Preiss, who allowed portions of this research to be performed in their laboratories at the MSU Department of Energy, Plant Research Laboratory (Ken Keegstra) and at the MSU Department of Biochemistry and Molecular Biology (Jack Preiss). This work was supported in parts by grants to C.B. from the Michigan Agricultural Experiment Station, BASF Plant Science, and the U.S. Department of Energy.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christoph Benning (benning@msu.edu).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Ballicora, M.A., Dubay, J.R., Devillers, C.H., and Preiss, J. (2005). Resurrecting the ancestral enzymatic role of a modulatory subunit. J. Biol. Chem. 280 10189–10195. [DOI] [PubMed] [Google Scholar]
- Baud, S., Boutin, J., Miquel, M., Lepiniec, L., and Rochat, C. (2002). An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 40 151–160. [Google Scholar]
- Blakeley, S., Gottlob-McHugh, S., Wan, J., Crews, L., Miki, B., Ko, K., and Dennis, D.T. (1995). Molecular characterization of plastid pyruvate kinase from castor and tobacco. Plant Mol. Biol. 27 79–89. [DOI] [PubMed] [Google Scholar]
- Blakeley, S.D., and Dennis, D.T. (1993). Molecular approaches to the manipulation of carbon allocation in plants. Can. J. Bot. 71 765–778. [Google Scholar]
- Bruce, B.D., Perry, S., Froehlich, J., and Keegstra, K. (1994). In vitro import of protein into chloroplasts. In Plant Molecular Biology Manual, S.B. Gelvin and R.B. Schilperoort, eds (Boston: Kluwer Academic Publishers), pp. 1–15.
- Cernac, A., Andre, C., Hoffman-Benning, S., and Benning, C. (2006). WRI1 is required for seed germination and seedling establishment. Plant Physiol. 141 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac, A., and Benning, C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40 575–585. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Dörmann, P., and Benning, C. (1998). The role of UDP-glucose epimerase in carbohydrate metabolism of Arabidopsis. Plant J. 13 641–652. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J., and Rawsthorne, S. (2000). Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos. Plant Physiol. 122 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and Von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and Von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks, N., and Benning, C. (1998). wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso, G., Giacomelli, L., Ytterberg, A.J., Peltier, J.B., Rudella, A., Sun, Q., and van Wijk, K.J. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16 478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege, P., Heazlewood, J.L., Roessner-Tunali, U., Millar, A.H., Fernie, A.R., Leaver, C.J., and Sweetlove, L.J. (2003). Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffman, F.D., Alonso, A.P., Schwender, J., Shachar-Hill, Y., and Ohlrogge, J.B. (2005). Light enables a very high efficiency of carbon storage in developing embryos of rapeseed. Plant Physiol. 138 2267–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, L.D., Baud, S., Gilday, A., Li, Y., and Graham, I.A. (2006). Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J. 46 69–84. [DOI] [PubMed] [Google Scholar]
- Hattori, J., Baum, B.R., Mchugh, S.G., Blakeley, S.D., Dennis, D.T., and Miki, B.L. (1995). Pyruvate kinase isozymes: Ancient diversity retained in modern plant cells. Biochem. Syst. Ecol. 23 773. [Google Scholar]
- Hausler, R.E., Fischer, K.L., and Flugge, U.I. (2000). Determination of low-abundant metabolites in plant extracts by NAD(P)H fluorescence with a microtiter plate reader. Anal. Biochem. 281 1–8. [DOI] [PubMed] [Google Scholar]
- Hollenberg, P.F., Flashner, M., and Coon, M.J. (1971). Role of lysyl ɛ-amino groups in adenosine diphosphate binding and catalytic activity of pyruvate kinase. J. Biol. Chem. 246 946–953. [PubMed] [Google Scholar]
- Hu, Z.Y., and Plaxton, W.C. (1996). Purification and characterization of cytosolic pyruvate kinase from leaves of the castor oil plant. Arch. Biochem. Biophys. 333 298–307. [DOI] [PubMed] [Google Scholar]
- Jackson, D.T., Froehlich, J.E., and Keegstra, K. (1998). The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 273 16583–16588. [DOI] [PubMed] [Google Scholar]
- Jang, J.C., Leon, P., Zhou, L., and Sheen, J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell 9 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, J.C., and Sheen, J. (1994). Sugar sensing in higher plants. Plant Cell 6 1665–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, F., and Rawsthorne, S. (1994). Starch and fatty acid biosynthesis in plastids from developing embryos of oil seed rape. Plant J. 6 795–805. [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of Tl-Dna gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Kubis, S.E., Pike, M.J., Everett, C.J., Hill, L.M., and Rawsthorne, S. (2004). The import of phosphoenolpyruvate by plastids from developing embryos of oilseed rape, Brassica napus (L.), and its potential as a substrate for fatty acid synthesis. J. Exp. Bot. 55 1455–1462. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Li, H.M., Culligan, K., Dixon, R.A., and Chory, J. (1995). Cue1: A mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell 7 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K.B. (2003). ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics 19 1585–1586. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic membranes. Methods Enzymol. 148 350–382. [Google Scholar]
- Lu, C., and Hills, M.J. (2002). Arabidopsis mutants deficient in diacylglycerol acyltransferase display increased sensitivity to abscisic acid, sugars, and osmotic stress during germination and seedling development. Plant Physiol. 129 1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke, D.W., Franzmann, L.H., Nickle, T.C., and Yeung, E.C. (1994). Leafy cotyledon mutants of Arabidopsis. Plant Cell 6 1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz, M.E., and Ponce, E. (2003). Pyruvate kinase: Current status of regulatory and functional properties. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 135 197–218. [DOI] [PubMed] [Google Scholar]
- Negm, F.B., Cornel, F.A., and Plaxton, W.C. (1995). Suborganellar localization and molecular characterization of nonproteolytic degraded leukoplastid pyruvate kinase from developing castor-oil seeds. Plant Physiol. 109 1461–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge, J., Benning, C., Gao, H., Girke, T., and White, J. (2006). Plant seed specific promoters. US patent 7081565.
- Olsen, L.J., and Keegstra, K. (1992). The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J. Biol. Chem. 267 433–439. [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Plaxton, W.C. (1991). Leucoplast pyruvate kinase from developing castor oil seeds: Characterization of the enzyme's degradation by a cysteine endopeptidase. Plant Physiol. 97 1334–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton, W.C., Dennis, D.T., and Knowles, V.L. (1990). Purification of leucoplast pyruvate kinase from developing castor bean endosperm. Plant Physiol. 94 1528–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton, W.C., and Podesta, F.E. (2006). The functional organization and control of plant respiration. CRC Crit. Rev. Plant Sci. 25 159–198. [Google Scholar]
- Plaxton, W.C., Smith, C.R., and Knowles, V.L. (2002). Molecular and regulatory properties of leucoplast pyruvate kinase from Brassica napus (rapeseed) suspension cells. Arch. Biochem. Biophys. 400 54–62. [DOI] [PubMed] [Google Scholar]
- Reiser, J., Linka, N., Lemke, L., Jeblick, W., and Neuhaus, H.E. (2004). Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiol. 136 3524–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivoal, J., Smith, C.R., Moraes, T.F., Turpin, D.H., and Plaxton, W.C. (2002). A method for activity staining after native polyacrylamide gel electrophoresis using a coupled enzyme assay and fluorescence detection: Application to the analysis of several glycolytic enzymes. Anal. Biochem. 300 94–99. [DOI] [PubMed] [Google Scholar]
- Ruuska, S.A., Andrews, T.J., Badger, M.R., Price, G.D., and von Caemmerer, S. (2000). The role of chloroplast electron transport and metabolites in modulating Rubisco activity in tobacco. Insights from transgenic plants with reduced amounts of cytochrome b/f complex or glyceraldehyde 3-phosphate dehydrogenase. Plant Physiol. 122 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska, S.A., Girke, T., Benning, C., and Ohlrogge, J.B. (2002). Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14 1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska, S.A., Schwender, J., and Ohlrogge, J.B. (2004). The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol. 136 2700–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, H. (2005). Mutagenesis of the active site lysine 221 of the pyruvate kinase from Bacillus stearothermophilus. J. Biochem. (Tokyo) 137 141–145. [DOI] [PubMed] [Google Scholar]
- Sangwan, R.S., Gauthier, D.A., Turpin, D.H., Pomeroy, M.K., and Plaxton, W.C. (1992). Pyruvate kinase isoenzymes from zygotic and microspore-derived embryos of Brassica napus - Developmental profiles and subunit composition. Planta 187 198–202. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schramm, A., Siebers, B., Tjaden, B., Brinkmann, H., and Hensel, R. (2000). Pyruvate kinase of the hyperthermophilic crenarchaeote Thermoproteus tenax: Physiological role and phylogenetic aspects. J. Bacteriol. 182 2001–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender, J., Goffman, F., Ohlrogge, J.B., and Shachar-Hill, Y. (2004. a). Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432 779–782. [DOI] [PubMed] [Google Scholar]
- Schwender, J., and Ohlrogge, J.B. (2002). Probing in vivo metabolism by stable isotope labeling of storage lipids and proteins in developing Brassica napus embryos. Plant Physiol. 130 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender, J., Ohlrogge, J.B., and Shachar-Hill, Y. (2003). A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J. Biol. Chem. 278 29442–29453. [DOI] [PubMed] [Google Scholar]
- Schwender, J., Ohlrogge, J., and Shachar-Hill, Y. (2004. b). Understanding flux in plant metabolic networks. Curr. Opin. Plant Biol. 7 309–317. [DOI] [PubMed] [Google Scholar]
- Shen, W.J., and Forde, B.G. (1989). Efficient transformation of Agrobacterium spp by high-voltage electroporation. Nucleic Acids Res. 17 8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.R., Knowles, V.L., and Plaxton, W.C. (2000). Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell cultures - Implications for the integration of glycolysis with nitrogen assimilation. Eur. J. Biochem. 267 4477–4485. [DOI] [PubMed] [Google Scholar]
- Tang, G.Q., Hardin, S.C., Dewey, R., and Huber, S.C. (2003). A novel C-terminal proteolytic processing of cytosolic pyruvate kinase, its phosphorylation and degradation by the proteasome in developing soybean seeds. Plant J. 34 77–93. [DOI] [PubMed] [Google Scholar]
- Thelen, J.J., and Ohlrogge, J.B. (2002). Both antisense and sense expression of biotin carboxyl carrier protein isoform 2 inactivates the plastid acetyl-coenzyme A carboxylase in Arabidopsis thaliana. Plant J. 32 419–431. [DOI] [PubMed] [Google Scholar]
- Tranel, P.J., Froehlich, J., Goyal, A., and Keegstra, K. (1995). A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 14 2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, W.L., Knowles, V.L., and Plaxton, W.C. (2005). Cytosolic pyruvate kinase: Subunit composition, activity, and amount in developing castor and soybean seeds, and biochemical characterization of the purified castor seed enzyme. Planta 222 1051–1062. [DOI] [PubMed] [Google Scholar]
- Verwoerd, T.C., Dekker, B.M.M., and Hoekema, A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigeolas, H., van Dongen, J.T., Waldeck, P., Huhn, D., and Geigenberger, P. (2003). Lipid storage metabolism is limited by the prevailing low oxygen concentrations oilseed rape. Plant Physiol. 133 2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakao, S., and Benning, C. (2005). Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis. Plant J. 41 243–256. [DOI] [PubMed] [Google Scholar]
- Wan, J., Blakeley, S.D., Dennis, D.T., and Ko, K. (1995). Import characteristics of a leucoplast pyruvate kinase are influenced by a 19-amino acid domain within the protein. J. Biol. Chem. 270 16731–16739. [DOI] [PubMed] [Google Scholar]
- Weber, A.P. (2004). Solute transporters as connecting elements between cytosol and plastid stroma. Curr. Opin. Plant Biol. 7 247–253. [DOI] [PubMed] [Google Scholar]
- White, J.A., Todd, J., Newman, T., Focks, N., Girke, T., de Ilarduya, O.M., Jaworski, J.G., Ohlrogge, J.B., and Benning, C. (2000). A new set of Arabidopsis expressed sequence tags from developing seeds. The metabolic pathway from carbohydrates to seed oil. Plant Physiol. 124 1582–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C.C., Härtel, H., Wada, H., Hagio, M., Yu, B., Eakin, C., and Benning, C. (2002). The pgp1 mutant locus of Arabidopsis encodes a phosphatidylglycerolphosphate synthase with impaired activity. Plant Physiol. 129 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.