Abstract
We report a novel function for BLADE-ON-PETIOLE1 (BOP1) and BOP2 in regulating Arabidopsis thaliana lateral organ cell fate and polarity, through the analysis of loss-of-function mutants and transgenic plants that ectopically express BOP1 or BOP2. 35S:BOP1 and 35S:BOP2 plants exhibit a very short and compact stature, hyponastic leaves, and downward-orienting siliques. We show that the LATERAL ORGAN BOUNDARIES (LOB) domain genes ASYMMETRIC LEAVES2 (AS2) and LOB are upregulated in 35S:BOP and downregulated in bop mutant plants. Ectopic expression of BOP1 or BOP2 also results in repression of class I knox gene expression. We further demonstrate a role for BOP1 and BOP2 in establishing the adaxial-abaxial polarity axis in the leaf petiole, where they regulate PHB and FIL expression and overlap in function with AS1 and AS2. Interestingly, during this study, we found that KANADI1 (KAN1) and KAN2 act to promote adaxial organ identity in addition to their well-known role in promoting abaxial organ identity. Our data indicate that BOP1 and BOP2 act in cells adjacent to the lateral organ boundary to repress genes that confer meristem cell fate and induce genes that promote lateral organ fate and polarity, thereby restricting the developmental potential of the organ-forming cells and facilitating their differentiation.
INTRODUCTION
Continuous lateral organ formation is critical for higher plants to produce their characteristic architectures, but the regulatory pathways that specify organ cell fate are still poorly understood. In angiosperms, leaves are derived from small populations of founder cells set aside on the flanks of the pluripotent shoot apical meristem (SAM). These leaf primordia are separated from the main shoot through the establishment of a boundary between the organ-forming cells and the meristem cells that provides a permissive environment for differentiation. Once leaf primordia initiate, leaves grow rapidly through active cell division and expansion from the initial primordia cells into mature organs with three developmental axes of polarity: the proximodistal, the dorsoventral (adaxial-abaxial), and mediolateral axes (Waites and Hudson, 1995).
A number of mutants displaying leaf formation defects have been isolated in Arabidopsis thaliana. Plants carrying a mutation in either ASYMMETRIC LEAVES1 (AS1) or AS2 form asymmetric, rumpled, lobed leaves with ectopic leaflet-like organs on the petioles (Tsukaya and Uchimiya, 1997; Serrano-Cartagena et al., 1999; Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Sun et al., 2002). AS1 encodes a MYB domain–containing putative transcription factor (Byrne et al., 2000) and is closely related to PHANTASTICA (PHAN) in Antirrhinum majus and ROUGH SHEATH2 in Zea mays (Waites et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999). AS2 encodes a widely expressed protein with a Leu zipper motif (Iwakawa et al., 2002; Xu et al., 2002). Genetic studies have revealed that AS1 and AS2 function in overlapping developmental pathways (Serrano-Cartagena et al., 1999; Ori et al., 2000; Semiarti et al., 2001). Moreover, the AS1 and AS2 proteins interact in yeast two-hybrid assays (Xu et al., 2003). Both genes act in the leaves to repress the expression of three class I knox genes, BREVIPEDICELLUS (BP), KNAT2, and KNAT6, which promote the activity and maintenance of the SAM (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001). Leaf initiation occurs via a reciprocal negative molecular interaction between class I knox genes and AS1 and AS2: the class I knox gene SHOOT MERISTEMLESS negatively regulates AS1 and AS2 expression in the SAM, while AS1 and AS2 in turn repress BP, KNAT2, and KNAT6 activity in the initiating primordia (Byrne et al., 2000, 2002).
AS2 is a member of the plant-specific LATERAL ORGAN BOUNDARIES (LOB) domain (LBD) family of proteins (Iwakawa et al., 2002; Shuai et al., 2002; Xu et al., 2002). LOB, the founding LBD family member, is expressed at the boundary between the SAM and developing organ primordia (Shuai et al., 2002) and is positively regulated by AS1 and AS2 (Byrne et al., 2002; Shuai et al., 2002). lob plants show no developmental defects, most likely because of functional redundancy between LOB and other LBD genes. However, plants that ectopically express LOB are dwarfed and produce small leaves that curl upward slightly at the margins (Shuai et al., 2002). Ectopic expression of the LBD gene LBD36/ASL1 (Iwakawa et al., 2002; Shuai et al., 2002) also causes the formation of upward-curling (hyponastic) leaves and downward-pointing siliques (Nakazawa et al., 2003; Chalfun-Junior et al., 2005). Like lob plants, loss-of-function lbd36/asl1 plants do not display detectable mutant phenotypes, but AS2 and LBD36/ASL1 have overlapping functions during flower formation (Chalfun-Junior et al., 2005).
Following leaf initiation, a number of Arabidopsis genes act to specify leaf polarity along the adaxial-abaxial axis. Five members of the class III homeodomain/Leu zipper (HD-ZIP) family of transcription factors specify adaxial organ identity (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003; Prigge et al., 2005). Their mRNAs are targeted by two microRNAs, miR165 and miR166, leading to their downregulation in abaxial cells (Tang et al., 2003; Bao et al., 2004; Kim et al., 2005; Williams et al., 2005). Conversely, KANADI (KAN) gene family members, along with YABBY (YAB) gene family members, confer abaxial identity and growth (Eshed et al., 1999; Sawa et al., 1999; Siegfried et al., 1999; Kerstetter et al., 2001; Emery et al., 2003; Eshed and Bowman, 2004). 35S:AS2 transgenic plants form nearly radial, adaxialized leaves (Lin et al., 2003; Xu et al., 2003) and as2 alleles in the Landsberg erecta (Ler) background condition the appearance of abaxial cellular morphology in the adaxial region of leaves (Xu et al., 2003). Ectopic AS2 expression leads to upregulation of PHABULOSA (PHB), a class III HD-ZIP gene, and to repression of KAN1, KAN2, and the YAB genes FILAMENTOUS FLOWER (FIL) and YAB3 (Lin et al., 2003).
Arabidopsis as1 leaves show only weakly abaxialized characteristics in contrast with orthologous Antirrhinum phan leaves that are severely abaxialized, suggesting the presence of redundant factors in the AS1-AS2 pathway. Recently, several genes were reported as genetic enhancers of the as1 or as2 polarity phenotype. Combinations between as1 or as2 and mutations in genes that function in trans-acting short-interfering RNA production, for example, RNA-DEPENDENT RNA POLYMERASE6, SUPPRESSOR OF GENE SILENCING3, ARGONAUTE7, and DICER-LIKE4, result in severely enhanced adaxial/abaxial polarity defects (Li et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Xu et al., 2006). Furthermore, mutations in the subunits of the 26S proteasome, such as AE3 (RPN8a), strongly enhance the polarity defects of as1 or as2 leaves (Huang et al., 2006). These data suggest that organ polarity specification is controlled at the transcriptional, posttranscriptional, and posttranslational levels.
Like AS2 and AS1, the Arabidopsis BLADE-ON-PETIOLE1 (BOP1) gene plays an important role in suppressing meristematic activity in developing lateral organ primordia through the repression of class I knox gene expression (Ha et al., 2003). BOP1 acts synergistically with AS1 and AS2 to regulate leaf morphogenesis (Ha et al., 2003). BOP1 and BOP2 encode closely related BTB/POZ domain and ankyrin repeat–containing proteins that have largely overlapping expression patterns (Ha et al., 2004; Hepworth et al., 2005; Norberg et al., 2005). In this study, we report the phenotypic and molecular effects of BOP1 and BOP2 null alleles and overexpression lines in wild-type and various leaf mutant backgrounds. We demonstrate that BOP1 and BOP2 positively regulate the expression of AS2 and LOB and that BOP overexpression leads to upregulation of LBD36/ASL1. Conversely, high levels of BOP1 and BOP2 expression cause downregulation of the class I knox genes BP, KNAT2, and KNAT6. We show that bop1 bop2 leaf petioles exhibit abaxialized vasculature, a defect that is enhanced in the as1 or as2 backgrounds, and that changes in BOP1 and BOP2 activity alter the expression of the organ polarity genes PHAVOLUTA (PHV) and FIL. Finally, we find that KAN1 and KAN2 function in the establishment of adaxial and abaxial organ identity.
RESULTS
BOP1 and BOP2 Have Overlapping Functions during Development
BOP1 and BOP2 are closely related genes with very similar transcription patterns, which are largely restricted to the base of developing lateral organs (Ha et al., 2004; Hepworth et al., 2005; Norberg et al., 2005). To reveal the full spectrum of BOP function in Arabidopsis plants, we isolated bop1 and bop2 null alleles and generated double mutants. The bop1-4 null allele was reported previously (Ha et al., 2003, 2004). Since bop2 null alleles had not been previously identified, we characterized a new BOP2 insertion allele, bop2-11. This allele contains a T-DNA insertion at nucleotide +1414 in the second exon within the sequence encoding the ankyrin repeats. We could not detect full-length BOP2 transcripts in bop2-11 plants (see Supplemental Figure 1 online).
BOP1 and BOP2 have overlapping functions in leaf organogenesis. Compared with wild-type Columbia (Col-0) plants (see Supplemental Figure 2A online), bop1-4 plants developed a single ectopic outgrowths from the leaf petiole region with low penetrance (see Supplemental Figure 2B online). By contrast, bop2-11 plants showed normal rosette leaf morphogenesis (see Supplemental Figure 2C and Supplemental Table 1 online). Interestingly, bop1-4 bop2-11 double mutant plants developed extensive ectopic blade outgrowths along the rosette leaf petiole, like those observed in homozygous bop1-1 plants (see Supplemental Figure 2D online; Ha et al., 2003), revealing a synergistic genetic interaction between BOP1 and BOP2. Cauline leaves of bop1 but not bop2 plants frequently developed ectopic outgrowths from the basal region (see Supplemental Figures 2F and 2G online), while bop1-4 bop2-11 plants displayed an enhanced cauline leaf outgrowth phenotype (see Supplemental Figure 2H online) compared with bop1-4 plants. These data indicate that BOP1 and BOP2 function in a redundant pathway to suppress excess growth in proximal positions during leaf development.
Both bop1 and bop2 plants displayed defects in inflorescence development. bop2-11 plants frequently formed fused and/or fasciated inflorescences and generated multiple flowers from the same node (see Supplemental Figure 2J and Supplemental Table 1 online). These defects were rarely detected either in wild-type or in bop1 plants (see Supplemental Figure 2I and Supplemental Table 1 online). bop1-4 bop2-11 inflorescences displayed slightly enhanced phenotypes compared with those of either single mutant (see Supplemental Figure 2K and Supplemental Table 1 online; Hepworth et al., 2005; Norberg et al., 2005). As reported previously (Hepworth et al., 2005; Norberg et al., 2005), we found that BOP1 and BOP2 act redundantly during reproductive development to control bract suppression, floral patterning, and floral organ number (data not shown). In addition, we observed a novel defect in gynoecium formation: whereas the gynoecium of wild-type flowers consists of two fused carpels (see Supplemental Figure 2L online), ∼50% of bop1-4 bop2-11 gynoecia contained only a single, fertile carpel (see Supplemental Figure 2M online). These data indicate additional functional redundancy between BOP1 and BOP2 in regulating aspects of inflorescence and flower development.
Ectopic Expression of BOP1 and BOP2
To further investigate the function of BOP1 and BOP2 in plant development, we generated transgenic plants that ectopically expressed either BOP1 or BOP2 under the control of the constitutive 35S promoter. Transformation of the 35S:BOP1 construct into bop1-4 bop2-11 plants rescued the mutant phenotypes, indicating that this construct is functional in vivo (data not shown).
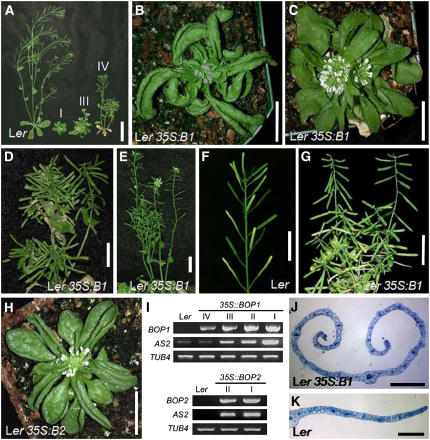
One-third (90/251) of transgenic plants containing 35S:BOP1 in the Ler background displayed significant developmental defects that could be categorized into four classes. Class I plants (22/251) were the most severely affected. They displayed a small, compact overall shape, rounded rosette leaves with short petioles, strongly hyponastic rosette leaves, severely reduced stem elongation, and densely packed clusters of flowers with little internode growth (Figures 1A and 1B, Table 1). These plants produced small numbers of aborted seeds. Class II plants (20/251) displayed all of the phenotypes exhibited by class I plants but had a weak hyponastic leaf character (Figure 1C, Table 1). Class III plants (25/251) developed normal-sized rosettes and leaves but had a strong clustering of flowers with reduced growth of the inflorescence shoot (Figure 1D, Table 1). Class IV plants (23/251) had nearly normal leaf and shoot growth but either displayed a weak clustering of flowers and slightly reduced shoot growth or were indistinguishable from wild-type plants except that their siliques developed in abnormal orientations (Figure 1E, Table 1). More than 80% of class III and >50% of class IV plants also exhibited weak hyponastic leaf morphology. Wild-type plants formed upward-orienting siliques (Figure 1F). By contrast, although class I and class II plants formed shoot apices that were too compact to determine silique orientation, class III and class IV shoots formed siliques that grew sharply downward and/or outward at abnormal angles (Figure 1G, Table 1). The severity of the abnormal silique orientation phenotype was stronger in class III plants than class IV plants. The overall severity of the 35S:BOP1 phenotypes correlated with the level of BOP1 transcription, as BOP1 expression was highest in class I plants and lowest in class IV plants (Figure 1I).
Figure 1.
Phenotypes of BOP1 and BOP2 Overexpressor Plants.
(A) Phenotypic variation among 35S:BOP1 (35S:B1) plants. From left to right: Ler wild-type (Ler), 35S:BOP1 class I (I), class III (III), and class IV (IV) transgenic plants.
(B) to (E) Class I (B), class II (C), class III (D), and class IV (E) 35S:BOP1 transgenic plants.
(F) and (G) Inflorescence development in Ler wild-type (F) and 35S:BOP1 (G) plants.
(H) 35S:BOP2 transgenic plant (35S:B2).
(I) BOP1, BOP2, and AS2 expression in 35S:BOP1 and 35S:BOP2 plants. RNA was isolated from the leaves of 35-d-old 35S:BOP1 T1 plants displaying class I, class II, class III, or class IV phenotypes or from 35-d-old 35S:BOP2 T1 plants displaying class I or class II phenotypes. TUBULIN4 (TUB4) was used as a control.
(J) and (K) Cross section through a 35S:BOP1 (J) and a Ler (K) leaf.
Bars = 40 mm in (A), 10 mm in (B) to (H), and 1 mm in (J) and (K).
Table 1.
Phenotypes of BOP1 and BOP2 Overexpressor Lines (%)
| Downward Silique
|
|||||||
|---|---|---|---|---|---|---|---|
| Transgenic Line (T1) | Totala | Class I | Class II | Class IIIb | Class IVb | Strongc | Weakd |
| Ler 35S:BOP1 | 251 | 8.8 | 8.0 | 10.0 (84) | 9.2 (52) | 3.2 | 11.6 |
| Ler 35S:BOP2 | 117 | 10.3 | 11.1 | 12.8 (87) | 13.7 (31) | 4.3 | 12.8 |
| Lan 35S:BOP1 | 180 | 0 | 2.8 | 4.4 (0) | 8.3 (0) | 0.0 | 0.6 |
| as2-101 35S:BOP1 | 122 | 0 | 8.2 | 11.5 (0) | 13.1 (0) | 0.8 | 16.4 |
| as1-101 35S:BOP1 | 66 | 0 | 12.1 | 10.6 (0) | 13.6 (0) | 0.0 | 13.6 |
| lob 35S:BOP1 | 190 | 15.8 | 4.7 | 6.8 (92) | 5.8 (36) | 6.8 | 9.5 |
| lob 35S:BOP2 | 183 | 13.1 | 4.4 | 12.6 (96) | 13.1 (38) | 8.2 | 13.7 |
Number of T1 plants examined.
Numbers in parenthesis denote the percentage of plants with weakly upward curling morphology among class III and class IV plants, respectively.
Strongly downward-orienting siliques.
Weakly downward-orienting siliques, including those developing at abnormal angles.
We also examined the effects of ectopic BOP2 expression. Transformation of the 35S:BOP2 construct into bop1-4 bop2-11 plants rescued the mutant leaf development phenotype (data not shown). Ectopic expression of BOP2 in the Ler background caused a range of developmental phenotypes similar to those observed in 35S:BOP1 transgenic plants (Figure 1H, Table 1). As observed for 35S:BOP1, the phenotypic severity of 35S:BOP2 transgenic plants correlated with the level of BOP2 transcription (Figure 1I).
To further examine the morphological defects in 35S:BOP1 plants, we examined the internal cellular structure of the rosette leaves from class I plants. Strong hyponastic leaf morphology was clearly observed in transverse sections through 35S:BOP1 leaves (Figures 1J and 1K). However, we were unable to detect a specific difference in the arrangement of vasculature and mesophyll cells along the adaxial-abaxial axis, or in epidermal cell morphology on either the adaxial or the abaxial surface, of 35S:BOP1 rosette leaves compared with those of wild-type plants.
BOP1 and BOP2 Negatively Regulate Class I knox Gene Expression
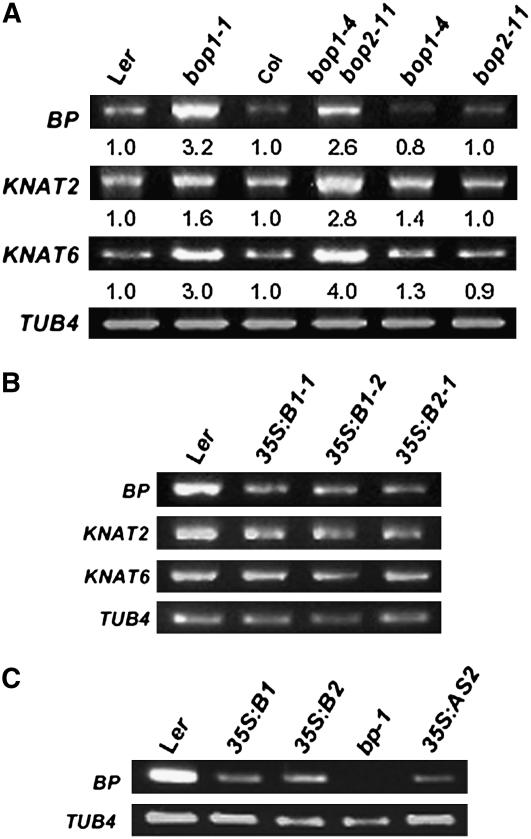
We have previously shown that several class I knox genes were misexpressed in semidominant bop1-1 mutant leaves (Ha et al., 2003). To further explore the role of BOP1 and BOP2 in class I knox gene regulation, we examined BP, KNAT2, and KNAT6 expression in bop1 and bop2 loss-of-function and overexpressing plants. The expression levels of BP, KNAT2, and KNAT6 were relatively unaffected in bop1-4 and bop2-11 seedlings compared with wild-type Col seedlings but were significantly increased in bop1-4 bop2-11 seedlings (Figure 2A). KNAT2 and KNAT6 transcript levels were relatively unaltered in BOP1 and BOP2 overexpressor plants compared with wild-type Ler plants. However, BP mRNA levels were reduced in the shoots of 25-d-old 35S:BOP1 and 35S:BOP2 plants (Figure 2B). 35S:BOP1 and 35S:BOP2 plants displayed a downward-growing silique phenotype similar to that of bp mutant plants (Douglas et al., 2002; Venglat et al., 2002). BP expression was strongly reduced in the stems of BOP transgenic plants compared with those of wild-type plants (Figure 2C), suggesting that the downward-orienting silique phenotype of 35S:BOP1 and 35S:BOP2 plants is caused by a severe decrease in BP activity.
Figure 2.
Expression of Class I knox Genes.
(A) RT-PCR analysis of class I knox gene expression using RNA isolated from the shoot region of 12-d-old seedlings. The relative amount of transcript accumulation in the various genotypes is indicated below each image.
(B) RT-PCR analysis of class I knox gene expression using RNA isolated from the shoot region of 25-d-old plants. 35S:B1-1, 35S:B1-2, and 35S:B2-1 represent independent transgenic lines.
(C) RT-PCR analysis of BP expression in the stem region. TUB4 was used as a control.
The erecta Mutation Conditions Severe BOP1 Overexpressor Phenotypes
The phenotypic defects of our 35S:BOP1 plants were considerably more severe than those previously reported (Norberg et al., 2005). A likely explanation for this is the difference in the genetic backgrounds used in the two analyses, since the prior study used the Col ecotype, whereas we used the Ler ecotype. The erecta (er) mutation is known to influence leaf polarity because the asymmetric leaf differentiation in the adaxial/abaxial axis of as2-101 plants is dependent on the presence of er (Xu et al., 2003). To determine the potential contribution of er to the appearance of 35S:BOP1 phenotypes, we transformed the 35S:BOP1 construct into Landsberg (Lan) plants that are wild-type at the ER locus. We could not detect class I plants among Lan 35S:BOP1 transformants (Table 1), and the percentage of class II, class III, and class IV plants was decreased in Lan 35S:BOP1 plants compared with Ler 35S:BOP1 plants. Furthermore, upward-curling leaf development was not detected in any class of Lan 35S:BOP1 transformants (Table 1). Thus, these data indicate that the er mutation is required for the appearance of the severe phenotypic defects in Ler 35S:BOP1 plants.
BOP1 and BOP2 Positively Regulate LBD Gene Expression
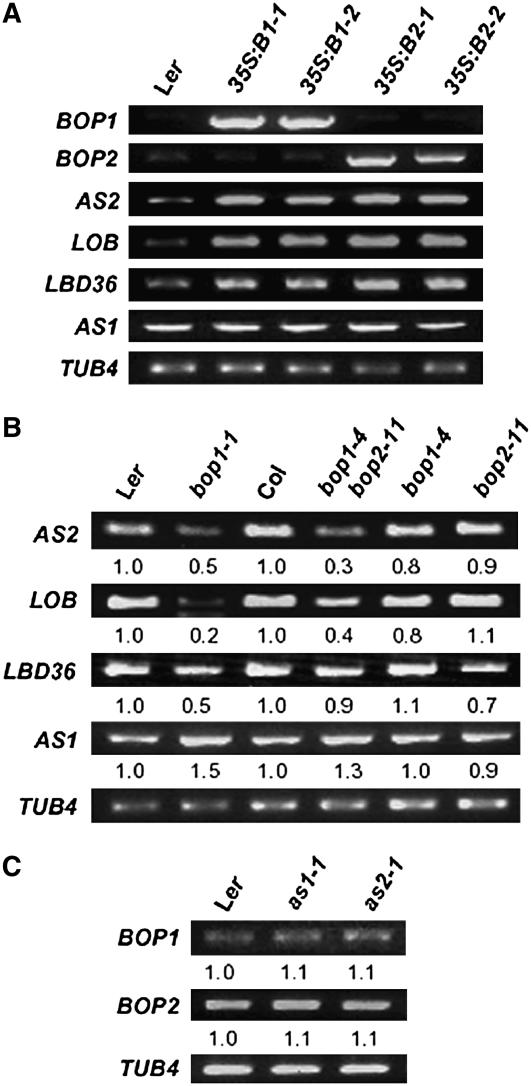
Ectopic expression of either AS2 or LOB has been shown to cause phenotypic defects, such as hyponastic leaf development (Shuai et al., 2002; Xu et al., 2003), similar to those observed in 35S:BOP1 and 35S:BOP2 plants. Thus, one interpretation of our results is that the phenotypic defects observed in the BOP1 and BOP2 overexpression lines are caused by increased AS2 and/or LOB expression. To test this hypothesis, we examined the transcript levels of both genes in the BOP overexpression lines. Expression levels of both AS2 and LOB were highly upregulated in 35S:BOP1 and 35S:BOP2 plants (Figure 3A). Moreover, the elevated AS2 expression level in 35S:BOP1 and 35S:BOP2 plants was correlated with the phenotypic severity of the transgenic plants, with plants displaying the strongest phenotypes also exhibiting the highest levels of AS2 transcripts (Figure 1I). Conversely, LOB expression was decreased in bop1-1 and bop1-4 bop2-11 plants, especially strongly in the bop1-1 dominant-negative mutant background (Figure 3B). AS2 transcript levels were also reduced in bop1-1 and bop1-4 bop2-11 plants. Taken together, these results indicate that BOP1 and BOP2 are necessary and sufficient to induce high-level expression of AS2 and LOB.
Figure 3.
Expression of AS1 and LOB Domain Genes.
(A) RT-PCR analysis of BOP, AS1, and LOB domain gene expression in 35S:BOP plants. RNA was isolated from the shoots of 15-d-old plants. 35S:B1-1, 35S:B1-2, 35S:B2-1, and 35S:B2-2 represent independent transgenic lines.
(B) RT-PCR analysis of AS1 and LBD gene expression in bop plants. RNA was isolated from the shoots of 12-d-old plants.
(C) RT-PCR analysis of BOP1 and BOP2 expression in 12-d-old as1 and as2 plants. The relative amount of transcript accumulation in the various genotypes is indicated below each image. TUB4 was used as a control.
LBD36/ASL1 encodes a LOB domain protein with the highest sequence similarity to AS2 (Iwakawa et al., 2002; Shuai et al., 2002), with which it has overlapping functions in the flower (Chalfun-Junior et al., 2005). Misexpression of LBD36/ASL1 resulted in a hyponastic leaf phenotype (Chalfun-Junior et al., 2005) like that observed in BOP1 and BOP2 overexpression lines. We found that LBD36/ASL1 transcript levels were elevated in 35S:BOP1 and 35S:BOP2 plants (Figure 3A) and were reduced in bop1-1 but not bop1-4 bop2-11 plants (Figure 3B). Thus, we conclude that BOP1 and BOP2 can also upregulate LBD36/ASL1 when they are ectopically expressed.
AS1 has overlapping functions with AS2 in regulating leaf morphogenesis and interacts with the AS2 protein. In contrast with AS2 and the other two LOB domain genes, AS1 expression levels were unchanged in both 35S:BOP1 and 35S:BOP2 plants. However, AS1 transcript levels were slightly elevated in bop1-1 and bop1-4 bop2-11 plants compared with the wild type. AS1 is expressed in the initiating leaf primordia region (Byrne et al., 2000), and bop mutant plants develop many ectopic leaf primordia along the petiole. We therefore interpret these results to indicate that the increased level of AS1 transcription in bop mutant plants might be derived not from negative regulation by BOP1 and BOP2 but from its induction in the ectopic leaf primordia formed on bop mutant leaves. We detected no change in BOP1 or BOP2 expression levels in as1 and as2 mutant plants compared with wild-type plants (Figure 3C); thus, neither AS1 nor AS2 is required for the activation of the BOP genes.
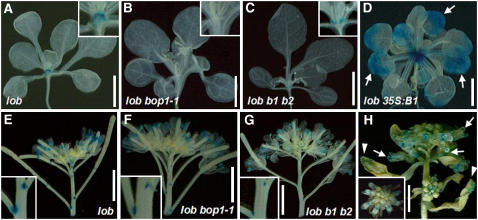
Neither AS2 transcripts nor LOB transcripts have been detected in seedling tissues using in situ hybridization, but a LOB enhancer trap line (lob:ET22) with an (ET):β-glucuronidase (GUS) reporter gene insertion in the 3′ end of the LOB gene exists that allows examination of LOB transcriptional activity via GUS assay (Shuai et al., 2002). These lob mutant plants do not show any detectable phenotypes, likely because of redundancy with other LBD family genes (Shuai et al., 2002). To observe the spatial effect of BOP1 and BOP2 activity on LOB expression, we crossed plants carrying the lob:ET22 GUS reporter construct to bop1-1 and bop1-4 bop2-11 plants and also transformed the 35S:BOP1 construct into lob:ET22 plants.
In lob plants, the lob:ET22 enhancer trap drove GUS reporter gene expression at the shoot apex, in the hypocotyl, at the base of all lateral organs, and in the roots (Figure 4A; Shuai et al., 2002). In bop1-1 plants containing lob:ET22, we could not detect GUS gene activity in the shoot apex (Figure 4B). In bop1-4 bop2-11 plants containing lob:ET22, GUS expression was observed in the shoot apex but more weakly than in lob plants (cf. Figures 4A and 4C). Conversely, ectopic expression of BOP1 resulted in increased and expanded lob:ET22 GUS expression in the shoot apex (Figure 4D). We also detected ectopic GUS activity broadly across the blade region of 35S:BOP1 leaves (Figure 4D, arrows), a pattern that was not observed in lob leaves. In lob flowers, LOB GUS activity was detected in the anthers and at the base of siliques and pedicels, respectively (Figure 4E; Shuai et al., 2002). In bop1-1 and bop1-4 bop2-11 flowers, GUS expression from lob:ET22 was also observed in these organs but more weakly than in lob flowers (Figures 4F and 4G). In the flowers of 35S:BOP1 plants, GUS activity was detected in the wild-type expression domain and also ectopically in the stigmatic tissues at the tip of the gynoecia (Figure 4H, arrows) and at the distal end of cauline leaves (Figure 4H, arrowheads). Thus, ectopic BOP1 activity is sufficient to induce ectopic LOB transcription outside the context of its normal expression domain in both leaves and flowers.
Figure 4.
Expression Analysis of GUS Activity in a lob Enhancer Trap Line.
(A) to (C) lob (A), bop1-1 (B), and bop1-4 bop2-11 (C) seedlings carrying a lob:ET22 enhancer trap construct, showing GUS expression at the shoot apex of each plant. Insets show a higher magnification of the shoot apex in each plant.
(D) 35S:BOP1 seedling carrying a lob:ET22 enhancer trap construct, showing extensive GUS expression around the SAM. Arrows indicate ectopic GUS expression in the leaf blades.
(E) to (G) lob (E), bop1-1 (F), and bop1-4 bop2-11 (G) inflorescences carrying a lob:ET22 enhancer trap construct, showing GUS expression in the anthers, the abscission zone, and the base of the pedicels. Insets show a higher magnification of the base of the pedicels in each plant.
(H) 35S:BOP1 inflorescence carrying a lob:ET22 enhancer trap construct, showing GUS expression in the anthers, the abscission zone, and the base of the pedicels. GUS expression is also detected ectopically in the stigmatic tissue (arrows) and cauline leaves (arrowheads). Inset shows a top view.
Bars = 5 mm.
35S:BOP1 Phenotypes Require AS2 and AS1 but Not LOB
Previous genetic and molecular analyses indicated that BOP1 and AS2 act in overlapping developmental pathways (Ha et al., 2003). 35S:BOP1 and 35S:BOP2 transgenic plants display a similar range of phenotypes as 35S:AS2 plants, and AS2 is strongly upregulated in 35S:BOP1 and 35S:BOP2 plants. These observations suggested that AS2 might be required to condition the phenotypes of 35S:BOP1 and 35S:BOP2 transgenic plants.
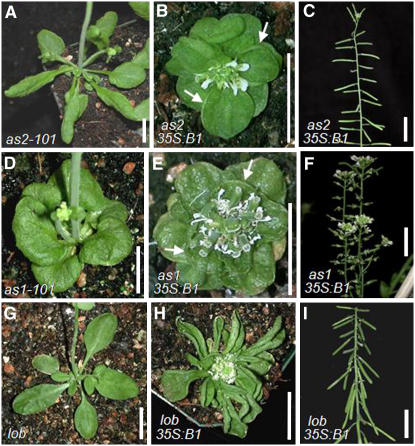
To test this hypothesis, we transformed the 35S:BOP1 construct into as2-101 plants. The as2-101 allele was derived from ethyl methanesulfonate–treated Ler plants, and plants carrying this allele displayed weak phenotypes (Figure 5A; Xu et al., 2002). as2-101 35S:BOP1 plants displayed the same short stature and reduced internode elongation as Ler 35S:BOP1 plants, and their siliques were pointed outward or slightly downward (Figures 5B and 5C, Table 1). However, the strongly downward-orienting silique phenotype observed in Ler 35S:BOP1 plants was very rarely detected in as2-101 35S:BOP1 plants (Figure 5C, Table 1). We also could not find plants with strongly upward-curling leaf morphology among the as2-101 35S:BOP1 plants (Table 1). In addition, weakly upward-curling leaves were not detected among class III and class IV as2-101 35S:BOP1 plants, although they were readily observed in class III and class IV Ler 35S:BOP1 plants. Conversely, when the 35S:AS2 construct was transformed into bop1-1 plants, none of the 175 T1 transgenic plants showed rescue of the bop1-1 leaf phenotypes (data not shown).
Figure 5.
Effects of BOP1 Overexpression in Leaf Mutants.
(A), (D), and (G) Forty-day-old as2-101 (A), as1-101 (D), and lob plants (G).
(B), (E), and (H) Forty-day-old as2-101 35S:BOP1 (B), as1-101 35S:BOP1 (E), and lob 35S:BOP1 plants (H). Arrows in (B) and (E) indicate weak upward-curling leaf morphology.
(C), (F), and (I) Inflorescences of as2-101 35S:BOP1 (C), as1-101 35S:BOP1 (F), and lob 35S:BOP1 plants (I).
Bars = 10 mm.
Molecular, genetic, and biochemical data indicate that AS1 and AS2 act together in leaf development (Serrano-Cartagena et al., 1999; Ori et al., 2000; Semiarti et al., 2001; Byrne et al., 2002; Xu et al., 2003). To determine whether AS1 is required for the 35S:BOP1 phenotypes, we transformed the 35S:BOP1 construct into the weak as1-101 allele in the Ler background (Figure 5D; Sun et al., 2002; Xu et al., 2003). as1-101 35S:BOP1 plants exhibited a similar range of phenotypes as Ler 35S:BOP1 plants (Figures 5E and 5F, Table 1). However, in as1-101 35S:BOP1 plants, strongly upward-curling leaves were not detected and siliques developed at abnormal angles rather than in a strongly downward orientation (Figure 5F, Table 1). Thus, both AS1 and AS2 are required to condition the 35S:BOP1 leaf morphology and silique orientation phenotypes.
AS1 and AS2 positively regulate the expression of LOB (Byrne et al., 2002; Shuai et al., 2002), and we have shown here that LOB is also positively regulated by BOP1 and BOP2. To determine whether LOB is required for the acquisition of the BOP1 and BOP2 overexpression phenotypes, the 35S:BOP1 and 35S:BOP2 constructs were introduced into lob:ET22 mutant plants (Figure 5G). lob 35S:BOP1 and lob 35S:BOP2 plants exhibited identical phenotypes to Ler 35S:BOP1 and Ler 35S:BOP2 plants, both with respect to the range of phenotypes and to the ratio of each phenotypic class (Figures 5H and 5I, Table 1). The number of plants with the class I hyponastic leaf shape was even slightly increased in lob 35S:BOP1 and lob 35S:BOP2 plants compared with Ler 35S:BOP1 and Ler 35S:BOP2 plants. Taken together, these results indicate that LOB activity is not necessary to condition the 35S:BOP1 and 35S:BOP2 phenotypes.
Genetic Interaction between bop1 bop2 and Organ Polarity Mutants
The bop1 bop2 leaf phenotypes are reminiscent of those of as1 or as2 plants, which form lobed and rumpled leaves (Figures 6A, 6B, 6G, and 6H; Tsukaya and Uchimiya, 1997; Serrano-Cartagena et al., 1999; Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Sun et al., 2002). In addition, we have determined that AS2 is positively regulated by BOP1 and BOP2 and that AS1 and AS2 function is required for the appearance of many 35S:BOP1 phenotypes. These data suggested that BOP1 and BOP2 might play roles in leaf morphogenesis events mediated by AS1 and AS2.
Figure 6.
Genetic Interaction between bop1 bop2 Mutants and Organ Polarity Mutants.
(A), (B), (G), and (H) Forty-day-old as2-1 ([A] and [B]) and as1-1 plants ([G] and [H]).
(C), (D), (I), and (J) Forty-five-day-old bop1 bop2 as2-1 ([C] and [D]) and bop1 bop2 as1-1 plants ([I] and [J]).
(E), (F), (K), and (L) Forty-eight-day-old kan1 kan2 as2-15 ([E] and [F]) and kan1 kan2 as1-1 plants ([K]) and [L]).
(B), (D), (F), (H), (J), and (L) each show a detached single rosette leaf.
(M) to (O) Forty-five-day-old kan1 kan2 plants (M) and bop1 bop2 kan1 kan2 plants ([N] and [O]). (O) shows a detached rosette leaf.
(P) to (R) Detached rosette leaf from bop1 bop2 introgressed once into Ler (x1L) plants (P), kan1 kan2 (Q), and bop1 bop2 kan1 kan2 plants (R). Left images show the adaxial view, while right images show the abaxial view. In (Q) and (R), insets show ectopic blade outgrowths on the abaxial side of the rosette leaf.
(S) Detached radialized leaves of bop1 bop2 kan1 kan2 plants.
Arrows in (D), (J), (N), and (O) indicate ectopically developed radialized leaves. r, rosette leaf; el, ectopic leaf; b1 b2, bop1 bop2; k1 k2, kan1 kan2. Bars = 10 mm.
We investigated this possibility by examining the phenotypic effects of combining the bop1-4 bop2-11 alleles with severe as1-1 or as2-1 alleles. The rosette leaf petioles of bop1 bop2 as1 and bop1 bop2 as2 plants showed extensive ectopic outgrowth formation in the marginal region (Figures 6C, 6D, 6I, and 6J), and some outgrowths exhibited a strongly radialized character (Figures 6D and 6J, arrows). To identify the origin of these ectopic organ outgrowths, we performed anatomical analysis of young seedlings. The leaf petioles of 11-d-old Col, as1-1, and as2-1 seedlings were fully differentiated and consisted of highly vacuolated cells lacking a detectable nucleus (see Supplemental Figures 3A to 3C online). By contrast, bop1 bop2, bop1 bop2 as1, and bop1 bop2 as2 leaf petioles displayed clusters of densely cytoplasmic, undifferentiated cells on the adaxial side, coincident with the development of ectopic outgrowths (see Supplemental Figures 3D to 3F online, arrows). These data show that the double and triple mutant leaf petioles acquired ectopic meristematic activity in the adaxial regions.
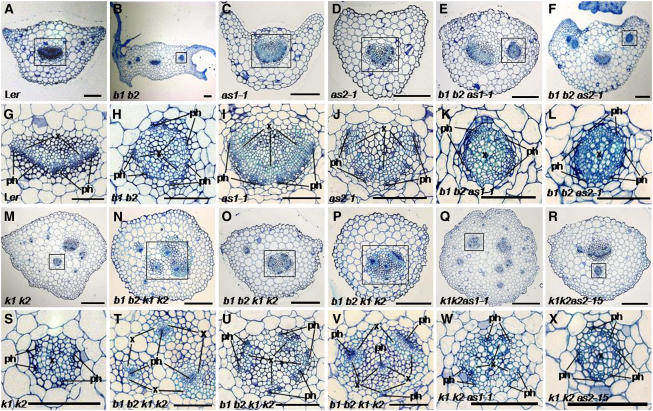
To further characterize the bop1 bop2, bop1 bop2 as1, and bop1 bop2 as2 phenotypes, we analyzed the internal morphology of the leaf petioles. The petioles of wild-type leaves contain polarized vascular bundles consisting of xylem on the adaxial side and phloem on the abaxial side (Figures 7A and 7G). Interestingly, some bop1 bop2 leaf petioles (22.2%) had xylem surrounded by phloem (Figures 7B and 7H, Table 2), which is characteristic of the abaxialized lateral organs produced by Antirrhinum phan plants and by Arabidopsis phb phv rev and as2 enhancer3 (ae3) plants (Waites and Hudson, 1995; Emery et al., 2003; Huang et al., 2006). More than 55% of bop1 bop2 petioles formed half-moon-shaped vasculature, with xylem developing on the inside and phloem on the outside. We interpret this as partially abaxialized vasculature (Table 2). These Col bop1 bop2 phenotypes were observed at nearly the same ratio in bop1 bop2 plants introgressed into Ler (Table 2), indicating that er does not significantly contribute to the phenotypes. In as1 and as2 leaf petioles, partially abaxialized vein morphology was observed at a high frequency (Figures 7C, 7D, 7I, and 7J, Table 2). However, completely abaxialized vasculature consisting of xylem surrounded by phloem was not detected in as1 and as2 petioles (Table 2). The vascular patterning defects in the bop1 bop2 as1 or bop1 bop2 as2 leaf petioles were greatly enhanced compared with those of the parental plants (Figures 7E, 7F, 7K, and 7L, Table 2). These data indicate that BOP1 and BOP2 play redundant roles in adaxial-abaxial polarity specification in the leaf petiole and that their activity largely overlaps with that of AS1 and AS2.
Figure 7.
Histological Analysis of Leaf Petioles in Genetic Combinations between bop1 bop2 and Organ Polarity Mutants.
Transverse sections were performed on rosette leaf petioles from wild-type ([A] and [G]), bop1 bop2 ([B] and [H]), as1-1 ([C] and [I]), as2-1 ([D] and [J]), bop1 bop2 as1-1 ([E] and [K]), bop1 bop2 as2-1 ([F] and [L]), kan1 kan2 ([M] and [S]), kan1 kan2 as1-1 ([Q] and [W]), and kan1 kan2 as2-1 plants ([R] and [X]). bop1 bop2 kan1 kan2 radial leaf petiole ([N] to [P]) and ([T] to [V]). (G) to (L) and (S) to (X) are the magnified views of the regions boxed in (A) to (F) and (M) to (R), respectively. ph, phloem; x, xylem; b1 b2, bop1 bop2; k1 k2, kan1 kan2. Bars = 200 μm in (A) to (F) and (M) to (R) and 100 μm in (G) to (L) and (S) to (X).
Table 2.
Ratio of Vasculature Phenotypes in Rosette Leaf and Stem Organs
| Ratio of Organ Vasculature Phenotypes (%)
|
||||||
|---|---|---|---|---|---|---|
| Lines | Totala | Normalb | Abaxialized | Partially Abaxialized | Adaxialized | Mixed: Ab- and Adaxialized |
| Rosette Leaf Petiole | ||||||
| Ler | 30 | 100 | 0 | 0 | 0 | 0 |
| Col | 29 | 100 | 0 | 0 | 0 | 0 |
| bop1-4 | 31 | 100 | 0 | 0 | 0 | 0 |
| bop2-11 | 31 | 100 | 0 | 0 | 0 | 0 |
| bop1 bop2 (Col)c | 27 | 22.2 | 22.2 | 55.6 | 0 | 0 |
| bop1 bop2 (Ler)d | 21 | 19.0 | 23.8 | 57.1 | 0 | 0 |
| as1-1 | 27 | 18.8 | 0 | 81.3 | 0 | 0 |
| as2-1e | 31 | 25.8 | 0 | 74.2 | 0 | 0 |
| as2-15 | 18 | 33.3 | 0 | 66.7 | 0 | 0 |
| bop1 bop2 as1-1 | 25 | 8.0 | 40.0 | 52.0 | 0 | 0 |
| bop1 bop2 as2-1 | 30 | 0 | 93.3 | 6.7 | 0 | 0 |
| bop1 bop2 as2-15 | 35 | 11.4 | 20.0 | 68.6 | 0 | 0 |
| kan1 kan2 | 34 | 26.5 | 8.8 | 0 | 55.9 | 8.8 |
| bop1 bop2 kan1 kan2f | 32 | 6.3 | 87.5 | 0 | 0 | 6.3 |
| bop1 bop2 kan1 kan2g | 25 | 4.0 | 32.0 | 12.0 | 36.0 | 16.0 |
| as1-1 kan1 kan2 | 21 | 19.0 | 61.9 | 9.5 | 0 | 9.5 |
| as2-15 kan1 kan2 | 21 | 19.0 | 19.0 | 61.9 | 0 | 0 |
| Stem | ||||||
| Ler | 18 | 100 | 0 | 0 | 0 | 0 |
| Col | 22 | 100 | 0 | 0 | 0 | 0 |
| bop1-4 | 21 | 100 | 0 | 0 | 0 | 0 |
| bop2-11 | 22 | 100 | 0 | 0 | 0 | 0 |
| bop1 bop2 (Col)c | 18 | 100 | 0 | 0 | 0 | 0 |
| bop1 bop2 (Ler)d | 17 | 100 | 0 | 0 | 0 | 0 |
| as1-1 | 17 | 100 | 0 | 0 | 0 | 0 |
| as2-1e | 23 | 100 | 0 | 0 | 0 | 0 |
| as2-15 | 21 | 100 | 0 | 0 | 0 | 0 |
| bop1 bop2 as1-1 | 23 | 100 | 0 | 0 | 0 | 0 |
| bop1 bop2 as2-1 | 22 | 100 | 0 | 0 | 0 | 0 |
| bop1 bop2 as2-15 | 18 | 100 | 0 | 0 | 0 | 0 |
| kan1 kan2 | 28 | 0 | 0 | 0 | 100 | 0 |
| bop1 bop2 kan1 kan2 | 30 | 13.3 | 6.7 | 0 | 66.7 | 13.3 |
| as1-1 kan1 kan2 | 20 | 40.0 | 0 | 0 | 60.0 | 0 |
| as2-15 kan1 kan2 | 20 | 40.9 | 0 | 0 | 59.1 | 0 |
Number of organs examined.
Normal in stem means phloem in the peripheral outside region and xylem in the central inside region of the vasculature.
bop1 bop2 in the Col background.
bop1 bop2 backcrossed into Ler one time.
as2-1 backcrossed into Col one time.
Rosette leaf petioles were examined.
Radialized leaf petioles were examined.
To better understand BOP1 and BOP2 function with respect to lateral organ polarity establishment, we examined the genetic interaction between bop1 bop2 and kan1 kan2 alleles. KAN1 and KAN2 function redundantly to promote abaxial organ identity, and mutations in these genes cause adaxialized lateral organ development (Eshed et al., 2001, 2004). kan1 kan2 plants had narrow leaves and developed ectopic outgrowths on their abaxial lamina (Figures 6M and 6Q; Eshed et al., 2001). bop1 bop2 kan1 kan2 plants developed narrow leaves with ectopic blade outgrowth along the petioles, like bop1 bop2 leaves, as well as ectopic outgrowths on their abaxial lamina, like kan1 kan2 leaves (Figures 6N and 6R). However, all of the bop1 bop2 kan1 kan2 leaves also showed extended, radialized petiole development that was not observed in either parental genotype (Figures 6N, 6O, and 6S, arrows). These data reveal a synergistic interaction between the bop1 bop2 and kan1 kan2 alleles in controlling lateral organ polarity in the basal regions of the leaf.
We better characterized the organ polarity phenotypes by analyzing the internal vascular patterning of the double and quadruple mutant leaves. The vasculature in more than half (55.9%) of kan1 kan2 leaf petioles exhibited a pattern of phloem surrounded by xylem, representing an adaxialized phenotype (Table 2). This is consistent with the reported role of KAN1 and KAN2 in specifying abaxial organ identity (Eshed et al., 2001, 2004; Kerstetter et al., 2001). However, very surprisingly, 17.6% of kan1 kan2 leaf petioles had abaxialized vasculature or a mixture of adaxialized and abaxialized vasculature (Figures 7M and 7S, Table 2). This observation suggests that KAN1 and KAN2 also contribute to the establishment of adaxial cell identity. We next examined the vascular patterning of bop1 bop2 kan1 kan2 rosette leaf petioles and found that about nine-tenths of them exhibited abaxialized vasculature (Table 2). The exclusively adaxialized vasculature observed in kan1 kan2 petioles was not detected in bop1 bop2 kan1 kan2 petioles. The radialized portion of bop1 bop2 kan1 kan2 leaves showed various kinds of organ polarity defects. The vascular bundles of nearly half of the leaves (44.0%) consisted of phloem surrounded by xylem (Figure 7T, Table 2), while one-third (36.0%) displayed xylem surrounded by phloem (Figures 7O and 7U, Table 2), and a small percentage (16.0%) had a mixture of both types of vasculature (Figures 7P and 7V, Table 2).
To know whether the vascular polarity defects occurred in other tissues, we examined the stem vasculature. The wild-type stem consists of multiple vascular bundles (see Supplemental Figure 4A online), each of which has xylem oriented toward the center in the adaxial position and phloem toward the periphery in the abaxial position (see Supplemental Figure 4E online). The stem vasculature in bop1 bop2 plants showed normal vein morphology (see Supplemental Figures 4B and 4F online), but all kan1 kan2 stem vascular bundles displayed phloem surrounded by xylem, indicating adaxialized polarity (see Supplemental Figures 4C and 4G online; Table 2; Izhaki and Bowman, 2007). bop1 bop2 kan1 kan2 stems had a decreased ratio of adaxialized vascular phenotypes compared with kan1 kan2 stems (see Supplemental Figures 4D and 4H online; Table 2). Similar to what was observed in petioles, 6.7% of bop1 bop2 kan1 kan2 stems exhibited an abaxialized phenotype of xylem surrounded by phloem, while others (13.3%) had both types of vasculature in a single stem (see Supplemental Figures 4I, 4J, 4M, and 4N online; Table 2).
Next, we analyzed genetic interactions between the as1-1 or as2-15 alleles and the kan1 kan2 alleles. Approximately three-fourths of as1 kan1 kan2 and as2 kan1 kan2 leaf petiole vascular bundles were partially or fully abaxialized (71.4 and 80.9%, respectively; Figures 7Q, 7R, 7W, and 7X, Table 2). In addition, 9.5% of the as1 kan1 kan2 leaf petioles displayed both adaxialized and abaxialized vascular bundles (Table 2). The stem vascular morphology of as1 and as2 plants is normal (data not shown), whereas 40% of as1 kan1 kan2 and as2 kan1 kan2 stem veins exhibited xylem surrounded by phloem, which was not observed in the vasculature of kan1 kan2 stems (see Supplemental Figures 4K, 4L, 4O, and 4P online; Table 2). Thus, the as1 and as2 mutations decreased the frequency of adaxialized vascular bundle formation in kan1 kan2 leaves and stems. In contrast with the clear defects in internal tissue polarity observed in the double, triple, and quadruple mutants, we could not detect any significant polarity defects in epidermal cell morphogenesis using scanning electron microscopy (data not shown).
Regulation of Adaxial and Abaxial Polarity Genes by BOP1 and BOP2
Our morphological and anatomical analyses implicate BOP1 and BOP2 in the specification of leaf polarity, suggesting that BOP1 and BOP2 might affect organ polarity gene expression. To investigate this idea, we used RT-PCR to examine the expression levels of several key organ polarity genes. PHB is one of the class III HD-ZIP genes that specify adaxial cell fate, and its steady state transcript levels were increased in 8-d-old 35S:BOP1 seedlings compared with wild-type seedlings (see Supplemental Figure 5 online). At this stage we could not detect a significant change in the expression levels of FIL and KAN1, which are restricted to abaxial leaf cells and promote their identity and/or growth. However, we found that PHB transcription was slightly upregulated, and FIL and KAN1 transcription was slightly reduced in the developing young leaves of 22-d-old 35S:BOP1 leaves compared with wild-type leaves (see Supplemental Figure 5 online). Neither gene showed detectable changes in its mRNA transcript levels in bop1-4, bop2-11, or bop1-4 bop2-11 plants (data not shown). These results show that PHB, FIL, and KAN1 transcript levels are misregulated in 35S:BOP1 leaves, although not sufficiently to cause a detectable polarity phenotype in these organs.
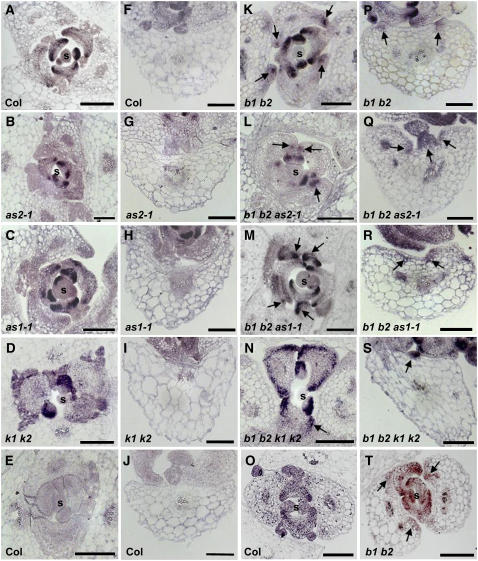
Although the transcript levels of the organ polarity genes appeared unaltered in bop mutant plants, it remained possible that the spatial expression patterns of these genes were perturbed. Thus, we examined the expression patterns of the polarity genes using RNA in situ hybridization. In wild-type plants, FIL expression is restricted to the abaxial side of the developing leaf (Figure 8A; Sawa et al., 1999; Siegfried et al., 1999). In as2, as1, and kan1 kan2 leaves, FIL transcripts are detected in the correct abaxial domain (Figures 8B to 8D). Similarly, in bop1 bop2, bop1 bop2 as2, bop1 bop2 as1, and bop1 bop2 kan1 kan2 plants, FIL expression was detected on the abaxial side of initiating and young leaf primordia (Figures 8K to 8N). However, more mature bop1 bop2, bop1 bop2 as2, bop1 bop2 as1, and bop1 bop2 kan1 kan2 leaves exhibited ectopic FIL transcription in the adaxial domain (Figures 8K to 8N, arrows). The differentiated leaves of bop1 bop2 plants exhibited FIL expression on the adaxial side of the primordial ectopic organ outgrowths (Figure 8P, arrows), while FIL transcripts were not detected in differentiated leaves of Col plants (Figure 8F). FIL transcripts were likewise detected on the adaxial side of bop1 bop2 as2, bop1 bop2 as1, and bop1 bop2 kan1 kan2 differentiated leaves but with the expression in the more central region of the leaf (Figures 8Q to 8S, arrows). By contrast, no FIL expression was detected in as2, as1, or kan1 kan2 differentiated leaves (Figures 8G to 8I). Like FIL transcripts, KAN1 transcripts are restricted to the abaxial domain of wild-type leaves (Figure 8O). However, in bop1 bop2 leaves, KAN1 expression is detected in the adaxial region in the area in which the ectopic organ outgrowths develop (Figure 8T). Taken together, these data demonstrate that BOP1 and BOP2 play a role in regulating the spatial expression domains of the abaxial polarity regulatory genes FIL and KAN1.
Figure 8.
RNA in Situ Hybridization Analysis of FIL and KAN Expression.
(A) to (N) and (P) to (S) RNA in situ hybridization analysis of FIL expression in 9-d-old Col ([A], [E], [F], and [J]), as2-1 ([B] and [G]), as1-1 ([C] and [H]), kan1 kan2 ([D] and [I]), bop1 bop2 ([K] and [P]), bop1 bop2 as2-1 ([L] and [Q]), bop1 bop2 as1-1 ([M] and [R]), and bop1 bop2 kan1 kan2 ([N] and [S]). (E) and (J) show the sense control.
(O) and (T) RNA in situ hybridization analysis of KAN expression in 9-d-old Col (O) and bop1 bop2 plants (T). Arrows indicate FIL or KAN ectopic expression.
s, shoot meristem region; b1 b2, bop1 bop2; k1 k2, kan1 kan2. Bars = 100 μm.
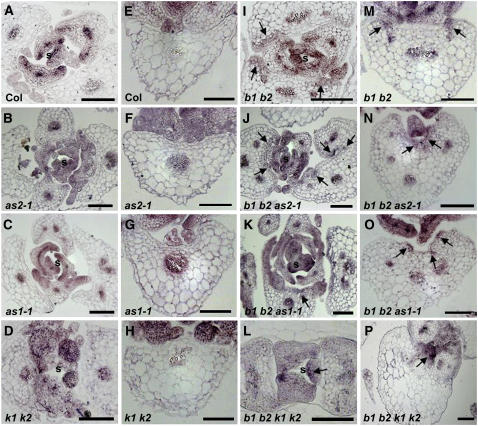
In wild-type seedlings, PHV expression becomes restricted to the vasculature and the adaxial region of initiating rosette leaf primordia (Figure 9A; Eshed et al., 2001; McConnell et al., 2001; Emery et al., 2003). The domain of PHV expression is unaltered in as2 and as1 seedlings (Figures 9B and 9C). In kan1 kan2 seedlings, PHV expression is detected throughout the leaf primordia, but a stronger signal is observed on the adaxial side than on the abaxial side (Figure 9D), suggesting that these organs still maintain some residual asymmetry. Developing young leaves of bop1 bop2, bop1 bop2 as2, bop1 bop2 as1, and bop1 bop2 kan1 kan2 seedlings exhibited PHV expression in the normal adaxial domain (Figures 9I to 9L). However, in bop1 bop2 kan1 kan2 leaves, PHV expression is restricted to a smaller area of the adaxial domain than in kan1 kan2 leaves (Figures 9D and 9L, arrow). This result indicates that, whereas the young leaves of kan1 kan2 plants have adaxialized organ identity, the young leaves of quadruple mutant plants acquire abaxialized organ identity, which is consistent with histological data. PHV expression is not detected in the differentiated leaves of Col, as2, as1, and kan1 kan2 plants (Figures 9E to 9H) but can be observed on the adaxial side of the differentiated leaves of bop1 bop2, bop1 bop2 as2, bop1 bop2 as1, and bop1 bop2 kan1 kan2 plants (Figures 9I to 9P, arrows). Furthermore, PHV was expressed more centrally in bop1 bop2 as2, bop1 bop2 as1, and bop1 bop2 kan1 kan2 leaves than in bop1 bop2 leaves (Figures 9M to 9P, arrows). In summary, in situ hybridization data from bop1 bop2, bop1 bop2 as2, bop1 bop2 as1, and bop1 bop2 kan1 kan2 rosette leaves indicate that BOP1 and BOP2 act to specify adaxial organ identity.
Figure 9.
RNA in Situ Hybridization Analysis of PHV Expression.
RNA in situ hybridization analysis of PHV expression in 9-d-old Col ([A] and [E]), as2-1 ([B] and [F]), as1-1 ([C] and [G]), kan1 kan2 ([D] and [H]), bop1 bop2 ([I] and [M]), bop1 bop2 as2-1 ([J] and [N]), bop1 bop2 as1-1 ([K] and [O]), and bop1 bop2 kan1 kan2 ([L] and [P]). Arrows indicate ectopic expression of PHV. s, shoot meristem region; b1 b2, bop1 bop2; k1 k2, kan1 kan2. Bars = 100 μm.
DISCUSSION
Functional Relationship between BOP1 and BOP2
Our data show that BOP1 and BOP2 have unique and shared functions in Arabidopsis development. bop1 and bop2 single mutants display phenotypes in distinct tissues, with bop1 plants exhibiting defects in rosette leaf development and bop2 plants showing phenotypes exclusively in inflorescence and flower development (see Supplemental Figure 2 and Supplemental Table 1 online). This suggests that although both BOP1 and BOP2 have high sequence homology and almost identical expression patterns (Ha et al., 2004; Hepworth et al., 2005; Norberg et al., 2005), the requirement for the two genes differs in different tissues, with BOP1 being more important for leaf development and BOP2 more important for flower development.
The bop1-1 allele was formally characterized as having a dominant-negative genetic character (Ha et al., 2004). A straightforward explanation for the behavior of this allele is that the mutant bop1-1 protein interferes with the activity of the related BOP2 protein, which can otherwise largely compensate for the lack of BOP1 activity during lateral organ formation. However, bop1-1 plants have much weaker inflorescence and flower phenotypes than bop1 bop2 plants (Ha et al., 2003; Hepworth et al., 2005; Norberg et al., 2005), suggesting that bop1-1 protein might not completely abolish BOP2 function in these tissues.
Role of BOP1 and BOP2 in Lateral Organ Formation
The formation of a lateral organ from the flanks of the SAM requires the repression of the class I knox genes by AS1 and AS2 (Byrne et al., 2000, 2002) and the establishment of a boundary between the meristem and the primordium that demarcates their separate fates. AS1 and AS2, along with BP, have been shown to positively regulate LOB expression along this boundary (Byrne et al., 2002). Based on genetic interactions, AS1 and AS2 are proposed to act in a common hierarchy that promotes lateral organ cell fate by repressing BP and KNAT2 and inducing LOB (Byrne et al., 2002). Here, we have demonstrated a key role for BOP1 and BOP2 in this cell fate regulatory hierarchy.
Expression of the class I knox genes BP, KNAT2, and KNAT6 is increased in bop1-1 (Ha et al., 2003) and bop1-4 bop2-11 plants (Figure 2A), implicating BOP1 and BOP2 in the negative regulation of knox gene activity. 35S:BOP1 and 35S:BOP2 plants show a corresponding decrease in BP transcript levels (Figures 2B and 2C), indicating that BP responds quantitatively to BOP1 and BOP2. Expression of both BOP1 and BOP2 is detected at the base of flowers, including the pedicel (Ha et al., 2003; Hepworth et al., 2005; Norberg et al., 2005), and BOP1:GUS is expressed in nodes and stems (C.M. Ha and J.C. Fletcher, unpublished data). These expression domains overlap with those of BP (Douglas et al., 2002; Venglat et al., 2002), consistent with BOP1 potentially repressing BP in nodes and stems as well as leaf primordia. bp plants form shortened internodes and downward-orienting siliques, indicating a role for BP in stem and pedicel differentiation (Douglas et al., 2002; Venglat et al., 2002). The downward-orienting silique phenotype of 35S:BOP1 and 35S:BOP2 plants may therefore result from strong suppression of BP transcription by highly expressed BOP1 or BOP2.
We identified three LOB domain genes as targets of positive regulation by BOP1 and BOP2: AS2, LOB, and LBD36/ASL1. AS2 expression levels are reduced in bop mutant plants and elevated in 35S:BOP1 and 35S:BOP2 plants (Figures 3A and 3B), indicating that BOP1 and BOP2 are required for AS2 induction. However, AS2 activation does not depend entirely on the action of BOP1 and BOP2, since a small amount of AS2 transcript is present in bop1-4 bop2-11 plants (Figure 3B). However, we could not rule out the possibility that the BTB/POZ domain regions of BOP1 and BOP2 might be transcribed in bop1-4 bop2-11 plants, which could activate AS2. To test this possibility, we examined AS2 expression in bop1-7 bop2-12 plants that contain T-DNA insertions in the BTB/POZ domains of BOP1 and BOP2. These plants lack either partial- or full-length transcripts of BOP1 and BOP2 and show the same morphological defects as bop1-4 bop2-11 plants. We could still detect AS2 transcripts at a low level in bop1-7 bop2-12 plants (C.M. Ha and J.C. Fletcher, unpublished data), indicating that a BOP-independent pathway also contributes to AS2 activation. BOP1 and BOP2 expression is unchanged in as2 mutants, revealing that AS2 is downstream of BOP1 and BOP2 at the level of transcriptional regulation.
Does AS2 mediate all the functions of BOP1 and BOP2 in leaf development? Like BOP1 and BOP2, AS2 negatively regulates class I knox gene expression in leaves (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Sun et al., 2002). Ectopic expression of AS2 causes a similar, though not identical, effect to that of ectopic BOP1 and BOP2 (Lin et al., 2003; Xu et al., 2003), and some 35S:BOP1 phenotypes are attenuated in an as2-101 background. It is therefore possible that reduced AS2 function causes the bop1-1 and bop1 bop2 leaf phenotypes. However, bop1-1 as2-1 and bop1 bop2 as2-1 plants have synergistic phenotypes (Ha et al., 2003; this study), rather than the epistatic phenotypes that would be expected if loss of AS2 expression were the sole cause of the bop leaf phenotypes. Further, the 35S:AS2 construct does not rescue the bop1-1 leaf phenotypes. Thus, although AS2 acts downstream of BOP1 and BOP2, the regulation of leaf development by BOP1 and BOP2 is not exclusively mediated through AS2.
Another target of positive regulation by the BOP1 and BOP2 genes is LOB, which is thought to play a role in establishing a boundary between the SAM and initiating lateral organs (Shuai et al., 2002). BOP1 and BOP2 act redundantly to promote LOB expression at the meristem-leaf boundary (Figures 3A and 3B), and BOP1 is sufficient to induce LOB transcription in certain developmental contexts (Figure 4). LOB is activated by AS1 and AS2, so the simplest scenario would be that BOP1 and BOP2 activate AS2 expression, and then AS2 activates LOB expression. However, BOP1 overexpression but not AS2 overexpression is sufficient to induce ectopic LOB activation (Figure 3A), suggesting either that BOP1 can induce LOB through an AS2-independent mechanism or that BOP1 functions together with AS2 to activate LOB specifically at the meristem-organ boundary. Interestingly, although 35S:LOB plants are small and exhibit hyponastic leaf morphology (Shuai et al., 2002) similar to that of 35S:BOP1 and 35S:BOP2 plants, LOB does not condition the BOP overexpression phenotypes. Potentially other LBD family genes, such as LBD36/ASL1, may contribute along with AS2 to generating the 35S:BOP phenotypes.
The expression pattern of LBD36/ASL1 is broad and overlaps with that of BOP1 and BOP2 (Chalfun-Junior et al., 2005), and like LOB, its transcription responds positively as BOP1 and BOP2 expression increases (Figure 3A). Dominant, activation-tagged isoginchaku-3D (iso-3D), iso-4D, and downwards siliques-1D plants that overexpress LBD36/ASL1 display bp-like phenotypes and have reduced BP mRNA levels (Nakazawa et al., 2003; Chalfun-Junior et al., 2005), suggesting that the repression of BP transcription in 35S:BOP1 and 35S:BOP2 plants might be mediated, at least in part, via their induction of LBD36/ASL1. It remains to be seen whether additional genes in the 42-member LBD family are also regulated by BOP1 and BOP2.
BOP1 and BOP2 Act Redundantly with AS1 and AS2 to Regulate Lateral Organ Polarity
Our study has shown that bop1 bop2 double mutants have abaxialized leaf petiole vasculature (Figure 7H, Table 2). This lateral organ polarity defect was strongly enhanced by mutations in either as1 or as2 (Figures 7K and 7L, Table 2), demonstrating that BOP1 and BOP2 function redundantly with the AS1-AS2 pathway to regulate adaxial organ identity. A question that then arises is why ectopic expression of BOP1 or BOP2 does not cause widespread adaxialization of structures such as stem vasculature and leaf mesophyll and vascular tissue. A trivial possibility is that the level of ectopic BOP1 or BOP2 transcription in the 35S lines is not sufficient to trigger a detectable polarity shift. Alternatively, BOP1/2 activity may require the presence of localized cofactors that are not themselves misexpressed in the 35S lines, thus leading to a limited set of overexpression phenotypes.
Our data suggest that BOP1 and BOP2 promote adaxial cell fate together with AS1 or AS2 during cellular differentiation in the leaf petiole, after the initial establishment of the adaxial-abaxial polarity axis via interaction between class III HD-ZIP and KAN family genes. In bop1 bop2 as2 and bop1 bop2 as1 plants, radialized organs develop extensively along the petioles of more mature rosette leaves (Figures 6D and 6F). Furthermore, the initiating leaf primordia of bop1 bop2, bop1 bop2 as2, bop1 bop2 as1, and bop1 bop2 kan1 kan2 plants display normal FIL and PHV expression patterns, yet ectopic FIL and reduced PHV expression is detected in more mature rosette leaves in which petiole differentiation is clearly visible (Figures 8L and 8M). These results indicate that BOP1 and BOP2 play an important role in maintaining adaxial cell fate at this later stage. The lack of alteration in the FIL and PHV expression patterns in initiating leaf primordia of bop1 bop2 plants may be explained by redundant functions of other genes, such as AE3 (Huang et al., 2006), resulting in nearly normal blade morphogenesis in bop1 bop2 plants.
A longstanding model holds that the juxtaposition of adaxial and abaxial cellular domains is required for outgrowth of the leaf blade, or lamina (Waites and Hudson, 1995). This outgrowth seems to be mediated, at least in part, though YAB gene activity (Eshed et al., 2004). Mutations in BOP1 and BOP2 activate and expand the transcription of the YAB gene FIL into the adaxial marginal regions of the leaf, demonstrating that BOP1 and BOP2 are required to repress FIL expression in this domain. Ectopically expressed FIL, juxtaposed with ectopically expressed PHV on the upper side of bop1 bop2, bop1 bop2 as2, bop1 bop2 as1, and bop1 bop2 kan1 kan2 petioles (Figures 8 and 9), may be sufficient to generate new boundaries of adaxial and abaxial gene expression. In wild-type plants, the margins of the petiole display a small and undifferentiated cellular character, indicating the maintenance of proliferative activity in these regions. Thus, in bop1 bop2 plants, the combination of a less differentiated cellular state with the juxtaposition of adaxial and abaxial gene expression in the petiole margin would result in the development of a new boundary region sufficient for ectopic organ outgrowth in that region. Taken together, these data suggest that BOP1, BOP2, AS1, and AS2 act redundantly to repress abaxial organ identity gene expression in the rosette leaf petiole and, as a result, promote adaxial organ identity. This in turn maintains the cellular fate of the leaf petiole in a differentiated state.
Our data reveal that BOP1 and BOP2 can also contribute to patterning the stem vasculature because mutations in both BOP1 and BOP2 partially reversed the kan1 kan2 adaxialized stem vasculature phenotypes (see Supplemental Figure 4 online; Table 2). Mutations in AS1 or AS2 likewise decreased the frequency of adaxialized vasculature formation in kan1 kan2 stems. Currently, we have no evidence that either BOP1 or BOP2 is expressed in the provascular tissue of the shoot meristem, suggesting that a downstream component(s) of the regulatory pathway may act non-cell-autonomously to control adaxial-abaxial polarity during stem vascular patterning.
KAN1 and KAN2 Promote Both Abaxial and Adaxial Organ Identity
Loss of KAN activity results in leaf adaxialization, implicating the KAN genes in specification of abaxial cell identity (Eshed et al., 2001, 2004; Kerstetter et al., 2001; Emery et al., 2003). In this study, we have shown that some kan1 kan2 rosette leaf petioles develop abaxialized vasculature (Figure 7S). Furthermore, this novel role for KAN1 and KAN2 is strongly enhanced in the bop1 bop2, as1, and as2 mutant backgrounds (Figures 7T to 7X, Table 2). Thus, KAN1 and KAN2 may function to promote adaxial organ identity redundantly with BOP1, BOP2, AS1, and AS2 in addition to their well-known role in establishing abaxial organ identity.
A dual function in adaxial-abaxial polarity regulation has also been observed for the YAB gene GRAMINIFOLIA (GRAM), the Antirrhinum ortholog of FIL (Golz et al., 2004; Navarro et al., 2004). Like FIL, GRAM is expressed abaxially in developing Antirrhinum organs and is required for abaxial cell identity and growth at the leaf margins. Interestingly, the gram mutation also causes adaxial mesophyll cells away from the leaf margin to partially resemble abaxial spongy mesophyll cells, and gram plants occasionally develop needle-like leaves in which xylem is surrounded by phloem. Thus, GRAM is also needed to establish adaxial organ identity in some regions of the Antirrhinum leaf. In contrast with gram mutants, Arabidopsis fil yab3 mutants only show adaxialized lateral organ development (Siegfried et al., 1999). It will thus be interesting to examine whether FIL and YAB3 have lost GRAM-like functions or if GRAM acquired new functions during evolution.
METHODS
Plant Materials and Genetics
The bop1-4 allele in the Col ecotype was described previously (Ha et al., 2004). The bop2-11 (N55227) allele was obtained from the Nottingham Arabidopsis Stock Centre and is in the Col ecotype. as1-1 Col (CS3374), as2-1 ER (CS3117), bp-1 (CS30), and Lan (CS1304) seeds were obtained from the ABRC. as1-101 and as2-101 seeds in the Ler ecotype were kindly provided by Hai Huang (Chinese Academy of Sciences, China), as2-15 seeds in the Col ecotype by Gerco Angenent (Plant Research International, The Netherlands), and kan1-2 kan2-1 seeds in the Ler ecotype by John Bowman (University of California, Davis). The lob:ET22 enhancer-trap line was a gift from Patty Springer (University of California, Riverside). Arabidopsis thaliana plants were grown as described previously (Ha et al., 2004).
To generate bop1 bop2 as1 or bop1 bop2 as2 plants, bop1 bop2 plants were crossed with as1 or as2 plants. bop1 bop2 kan1 kan2, as1 kan1 kan2, or as2 kan1 kan2 plants were isolated from crosses between bop1 bop2, as1, or as2 and kan1 kan2/+ plants. Plants with novel phenotypes in the F2 generation of the kan1 kan2/+ crosses were selected and genotyped using the primers KAN1-F (5′-TGAGCTCAAGAACCGACTT-3′) and KAN2-F (5′-TTAGCTGGGAATCTTGGCA-3′). When crosses were performed between mutants in different ecotypes, such as bop1-4 bop2-11 in Col to as2-1 in the ER background, control crosses were performed of the relevant mutant into Col to control for background genetic variation.
Construction of Transgenic Plants
Constructs for BOP overexpression were generated by amplifying the BOP1 and BOP2 coding regions from Ler seedling cDNA using the primers BOP1-F (5′-AGATCTAA-ATCAACAAAGGAGCTATGAGC-3′) and BOP1-R (5′-GGTCACCGATGATAGGG-ACTACAAAAAGAC-3′) or BOP2-F (5′-CCATGGAGATGAGCAATCTTGAAGAA-3′) and BOP2-R (5′-GGTCCACCGAGAACTAGAAGTGATGTTG-3′). Each PCR product was sequenced and placed downstream of the 35S promoter in the pCAMBIA1302 binary vector (CAMBIA). The 35S:AS2 construct was kindly provided by Patty Springer (University of California, Riverside). For each construct, the binary vector in the Agrobacterium tumefaciens strain GV3101 was introduced into Ler plants by the floral dip method (Clough and Bent, 1998). Transformants were selected on Murashige and Skoog medium containing 50 μM hygromycin.
Expression Analysis
For RT-PCR studies, cDNA was synthesized from 5 μg of total RNA using an oligo(dT15) primer and SuperScript III reverse transcriptase (Invitrogen). One microliter of the first-strand cDNA reaction was used as a template for PCR amplification. PCR conditions for BOP1 and BOP2 amplification were as follows: denaturation at 94°C for 3 min, followed by 27 to 33 cycles of 94°C for 35 s, 55°C for 45 s, and 72°C for 1 min, using primers BOP1-RT-76R (5′-GTGAATCTGATCCTTCGCAACC-3′), BOP1-24-76F (5′-ATCCAAACTACTTCC-GCTCGTG-3′), BOP2-RT-2R (5′-TATAGACCCGACCCAACATGG G-3′), and BOP2-RT-2F (5′-TCATTCATATGAGGGTAAATCCCG-3′). PCR conditions for BP, KNAT2, and KNAT6 amplification were as follows: denaturation at 94°C for 3 min, followed by 33 cycles of 94°C for 35 s, 53°C for 50 s, and 72°C for 1 min, using primers KNAT1-F (5′-GATGATCCCATATTGTCACTCTTCCC-3′), KNAT1-R (5′-ATGGAAGAATACCAGCAT-GACAAC-3′), KNAT2-F (5′-CCGAAGGCTTCCAATGGCG-3′), KNAT2-R (5′-GCGGCGATCACTGATCGTATC-3′), KNAT6-F (5′-TCATTCCTCGGTAAAGAATGATCCACTAG-3′), and KNAT6-R (5′-ATCTACAATTTCCATTCGGCCGGTG-3′) (Semiarti et al., 2001). PCR conditions for AS1, AS2, and LOB amplification were as follows: denaturation at 94°C for 3 min, followed by 28 to 38 cycles of 94°C for 35 s, 54°C for 45 s, and 72°C for 1 min. Gene-specific primers for LOB and AS1 were as previously published (Shuai et al., 2002; Lin et al., 2003).
For amplification of PHB, FIL, and KAN1 cDNAs, the following gene-specific primers were used: PHB-5 (5′-TGATGGTCCATTCGATGAGC-3′), PHB-3 (5′-TCTAAACTCACGAGGCCGCA-3′), FIL-3 (5′-GCTATGTCCAATGCAACTTT-3′), FIL-4 (5′-TTCTTGGCAGCAGCACTAAA-3′), KAN1-5 (5′-ACAACAACGCTTACCGATCA-3′), and KAN1-R (5′-ATTTCTCGTGCCAATCTGGT-3′) (Lin et al., 2003). PCR conditions for these genes were as follows: denaturation at 94°C for 3 min, followed by 33 cycles of 94°C for 35 s, 56°C for 45 s, and 72°C for 1 min. LBD36/ASL1 cDNA was amplified using the same PCR conditions used for LOB cDNA amplification, with the primers LBD36-F2 (5′-TAATTGAGGCTCTCAAGTCT-3′) and LBD36-R2 (5′-ACCAATGACATTCCTTCTAC-3′). Arabidopsis TUB4 was used as an internal control (Ha et al., 2003). Quantification of RT-PCR was performed using the Scion Image program.
Histology and Microscopy
lob bop1-1 and lob bop1-4 bop2-11 homozygous plants and lob 35S:BOP1 and lob 35S:BOP2 T1 plants were selected and stained for GUS activity as described previously (Jun et al., 2002). Samples for histological analysis were fixed and prepared as described previously (Ha et al., 2003). Plant tissue sections (3 to 4 μm thick) were cut with a rotary microtome (MICROM International) and stained with methylene blue.
In Situ Hybridization
Plant fixation and in situ hybridization were performed as described previously (Jackson, 1992). For the BOP1 and KAN1 probes, the full-length cDNA sequences were amplified and cloned in the pBluescript KS+ plasmid (Stratagene). PHV and FIL probes were generated as previously described (Eshed et al., 2001). Probes for in situ hybridization were transcribed using the digoxigenin labeling mix (Roche).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. BOP1 and BOP2 Expression in Wild-Type and bop Mutant Plants.
Supplemental Figure 2. Phenotypes of bop Mutant Plants.
Supplemental Figure 3. Histological Analysis of Vegetative Shoot Apices.
Supplemental Figure 4. Histological Analysis of Stem Vasculature in Genetic Combinations between bop1 bop2 and Organ Polarity Mutants.
Supplemental Figure 5. Expression of Polarity Genes in BOP1 Overexpressor Plants.
Supplemental Table 1. Phenotypes of bop Mutant Plants.
Supplementary Material
Acknowledgments
We thank Patty Springer for the lob:ET22 seeds and 35S:AS2 construct, Hai Huang for the as1-101 and as2-101 seeds, Gerco Angenent for the as2-15 seeds, and John Bowman for the kan1-2 kan2-1 seeds. We also thank the ABRC for supplying mutant seeds and the Nottingham Arabidopsis Stock Centre for providing insertion lines. We thank Cristel Carles and Ludmila Tyler for critical reading of the manuscript. This work was supported by a USDA Current Research Information System grant to J.C.F. H.G.N. was supported by the National Core Research Center for Systems Biodynamics and the Crop Functional Genomics Center, Korea.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jennifer C. Fletcher (fletcher@nature.berkeley.edu).
Online version contains Web-only data.
References
- Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouche, N., Gasciolli, V., and Vaucheret, H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16 927–932. [DOI] [PubMed] [Google Scholar]
- Bao, N., Lye, K.W., and Barton, M.K. (2004). MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 7 653–662. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957–1965. [DOI] [PubMed] [Google Scholar]
- Chalfun-Junior, A., Franken, J., Mes, J.J., Marsch-Martinez, N., Pereira, A., and Angenent, G. (2005). ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal-distal patterning in Arabidopsis petals. Plant Mol. Biol. 57 559–575. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Douglas, S.J., Chuck, G., Dengler, R.E., Pelecanda, L., and Riggs, C.D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 1768–1774. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11 1251–1260. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., and Bowman, J.L. (2004). MicroRNAs guide asymmetric DNA modifications guiding asymmetric organs. Dev. Cell 7 629–630. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Izhaki, A., Baum, S.F., Floyd, S.K., and Bowman, J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131 2997–3006. [DOI] [PubMed] [Google Scholar]
- Fahlgren, N., Montgomery, T.A., Howell, M.D., Allen, E., Dvorak, S.K., Alexander, A.L., and Carrington, J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16 939–944. [DOI] [PubMed] [Google Scholar]
- Garcia, D., Collier, S.A., Byrne, M.E., and Martienssen, R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16 933–938. [DOI] [PubMed] [Google Scholar]
- Golz, J.F., Roccaro, M., Kuzoff, R., and Hudson, A. (2004). GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development 131 3661–3670. [DOI] [PubMed] [Google Scholar]
- Ha, C.M., Jun, J.H., Nam, H.G., and Fletcher, J.C. (2004). BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 45 1361–1370. [DOI] [PubMed] [Google Scholar]
- Ha, C.M., Kim, G.-T., Kim, B.C., Jun, J.H., Soh, M.S., Ueno, Y., Machida, Y., Tsukaya, H., and Nam, H.G. (2003). The BLADE-ON-PETIOLE1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130 161–172. [DOI] [PubMed] [Google Scholar]
- Hepworth, S.R., Zhang, Y., McKim, S., Li, X., and Haughn, G. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Development 17 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Pi, L., Liang, W., Xu, B., Wang, H., Cai, R., and Huang, H. (2006). The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell 18 2479–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43 467–478. [DOI] [PubMed] [Google Scholar]
- Izhaki, A., and Bowman, J.L. (2007). KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D. (1992). In situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach, D.J. Bowles, S.J. Gurr, and R. McPherson, eds (Oxford, UK: Oxford University Press), pp. 163–174.
- Jun, J.H., Ha, C.M., and Nam, H.G. (2002). Involvement of the VEP1 gene in vascular strand development in Arabidopsis thaliana. Plant Cell Physiol. 43 323–330. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411 706–709. [DOI] [PubMed] [Google Scholar]
- Kim, J., Jung, J.-H., Reyes, J.L., Kim, Y.-S., Kim, S.-Y., Chung, K.-S., Kim, J.A., Lee, M., Lee, Y., Kim, V.N., Chua, N.H., and Park, C.M. (2005). MicroRNA cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 42 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Xu, L., Wang, H., Yuan, Z., Cao, X., Yang, Z., Zhang, D., and Huang, H. (2005). The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and microRNA165/166 in Arabidopsis leaf development. Plant Cell 17 2157–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W., Shuai, B., and Springer, P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and adaxial-abaxial patterning. Plant Cell 15 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J.L., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713. [DOI] [PubMed] [Google Scholar]
- Nakazawa, M., Ichikawa, T., Ishikawa, A., Kobayashi, H., Tsuhara, Y., Kawashima, M., Suzuki, K., Muto, S., and Matsui, M. (2003). Activation tagging: A novel tool to dissect the functions of a gene family. Plant J. 34 741–750. [DOI] [PubMed] [Google Scholar]
- Navarro, C., Efremova, N., Golz, J.F., Rubiera, R., Kuckenberg, M., Castillo, R., Tietz, O., Saedler, H., and Schwarz-Sommer, Z. (2004). Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 131 3649–3659. [DOI] [PubMed] [Google Scholar]
- Norberg, M., Holmlund, M., and Nilsson, O. (2005). The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132 2203–2213. [DOI] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Prigge, M.J., Otsuga, D., Alonso, J.M., Ecker, J.R., Drews, G.N., and Clark, S.E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E.H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Serrano-Cartagena, J., Robles, P., Ponce, M.R., and Micol, J.L. (1999). Genetic analysis of leaf form mutants from the Arabidopsis Information Service collection. Mol. Gen. Genet. 261 725–739. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Pena, C.G., and Springer, P.S. (2002). The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Zhou, Q., Zhang, W., Fu, Y., and Huang, H. (2002). ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 214 694–702. [DOI] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans, M.C.P., Hudson, A., Becraft, P.W., and Nelson, T. (1999). ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Nature 284 151–153. [DOI] [PubMed] [Google Scholar]
- Tsiantis, M., Schneeburger, R., Golz, J.F., Freeling, M., and Langdale, J.A. (1999). The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Nature 284 154–156. [DOI] [PubMed] [Google Scholar]
- Tsukaya, H., and Uchimiya, H. (1997). Genetic analyses of the formation of the serrated margin of leaf blades in Arabidopsis: Combination of a mutational analysis of leaf morphogenesis with the characterization of a specific marker gene expressed in hydathodes and stipules. Mol. Gen. Genet. 256 231–238. [DOI] [PubMed] [Google Scholar]
- Venglat, S.P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., Martienssen, R., Selvaraj, G., and Datla, R. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121 2143–2154. [Google Scholar]
- Waites, R., Selvadurai, H.R.N., Oliver, I.R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93 779–789. [DOI] [PubMed] [Google Scholar]
- Williams, L., Grigg, S.P., Xie, M., Christensen, S., and Fletcher, J.C. (2005). Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132 3657–3668. [DOI] [PubMed] [Google Scholar]
- Xu, L., Xu, Y., Dong, A., Sun, Y., Pi, L., Xu, Y., and Huang, H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying adaxial identity. Development 130 4097–4107. [DOI] [PubMed] [Google Scholar]
- Xu, L., Yang, L., Pi, L., Liu, Q., Ling, Q., Wang, H., Poethig, R.S., and Huang, H. (2006). Genetic interaction between the AS1-AS2 and RDR6-SGS3-AGO7 pathways for leaf morphogenesis. Plant Cell Physiol. 47 853–863. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Sun, Y., Liang, W., and Huang, H. (2002). The Arabidopsis AS2 gene encoding a predicted leucine-zipper protein is required for the leaf polarity formation. Acta Bot. Sin. 44 1194–1202. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.