Abstract
Camalexin (3-thiazol-2-yl-indole) is an indole alkaloid phytoalexin produced by Arabidopsis thaliana that is thought to be important for resistance to necrotrophic fungal pathogens, such as Alternaria brassicicola and Botrytis cinerea. It is produced from Trp, which is converted to indole acetaldoxime (IAOx) by the action of cytochrome P450 monooxygenases CYP79B2 and CYP79B3. The remaining biosynthetic steps are unknown except for the last step, which is conversion of dihydrocamalexic acid to camalexin by CYP71B15 (PAD3). This article reports characterization of CYP71A13. Plants carrying cyp71A13 mutations produce greatly reduced amounts of camalexin after infection by Pseudomonas syringae or A. brassicicola and are susceptible to A. brassicicola, as are pad3 and cyp79B2 cyp79B3 mutants. Expression levels of CYP71A13 and PAD3 are coregulated. CYP71A13 expressed in Escherichia coli converted IAOx to indole-3-acetonitrile (IAN). Expression of CYP79B2 and CYP71A13 in Nicotiana benthamiana resulted in conversion of Trp to IAN. Exogenously supplied IAN restored camalexin production in cyp71A13 mutant plants. Together, these results lead to the conclusion that CYP71A13 catalyzes the conversion of IAOx to IAN in camalexin synthesis and provide further support for the role of camalexin in resistance to A. brassicicola.
INTRODUCTION
Phytoalexins are small antimicrobial compounds produced by plants in response to pathogen attack. A wide variety of different compounds serve as phytoalexins in various plant families. They are thought to contribute to resistance, but in most systems, the evidence for this idea consists solely of antimicrobial activity in vitro. Arabidopsis thaliana is thought to produce only a single phytoalexin, camalexin. Very little camalexin is present in healthy Arabidopsis plants, but large amounts are produced in response to attack by pathogens, such as Pseudomonas syringae bacteria and the necrotrophic fungi Alternaria brassicicola and Botrytis cinerea. Camalexin synthesis can also be elicited by silver nitrate treatment. Camalexin-deficient mutants have proven useful for elucidating the pathway of camalexin biosynthesis and for testing the role of camalexin in disease resistance in vivo.
Several phytoalexin-deficient (pad) mutants that produce reduced amounts of camalexin have been described, but these varied with respect to their effects on susceptibility to pathogens, making it difficult to draw conclusions about the role of camalexin in disease resistance (Glazebrook and Ausubel, 1994; Glazebrook et al., 1997). Three of the PAD genes have been identified by map-based cloning. PAD4 encodes a regulator that affects expression of many pathogen-inducible genes, making pad4 mutants unsuitable for studies of the role of camalexin in disease resistance (Jirage et al., 1999; Glazebrook et al., 2003). PAD2 encodes γ-glutamyl cysteine synthase, an enzyme required for synthesis of glutathione (Parisy et al., 2007). The role of glutathione in camalexin synthesis or other aspects of plant defense is not clear, but it seems likely that the reduced glutathione levels in pad2 mutants could affect multiple aspects of plant metabolism. PAD3 encodes the cytochrome P450 monooxygenase CYP71B15. This observation, together with the fact that PAD3 is strongly induced by P. syringae, led to the idea that PAD3 encodes an enzyme required for camalexin biosynthesis (Zhou et al., 1999). This idea was confirmed by the demonstration that PAD3 catalyzes the oxidative decarboxylation of dihydrocamalexic acid to camalexin (Schuhegger et al., 2006).
The pad3-1 allele is null, and there is almost no camalexin in pad3-1 plants, making them useful for testing the contribution of camalexin to resistance against various pathogens (Zhou et al., 1999). Camalexin does not contribute substantially to resistance to the biotrophic oomycte Hyaloperonsopsora parasitica (Glazebrook et al., 1997), the biotrophic ascomycete Erysiphe orontii (Reuber et al., 1998), or the hemibiotrophic bacterium P. syringae (Glazebrook and Ausubel, 1994). By contrast, the enhanced susceptibility of pad3 mutants to the necrotrophic ascomycetes A. brassicicola (Thomma et al., 1999), B. cinerea (Ferrari et al., 2003), and Leptosphaeria maculans (Bohman et al., 2004) suggests that camalexin limits the growth of these pathogens.
Induction of many plant defense responses depends on signaling mediated by oxylipins, such as jasmonic acid, ethylene, or salicylic acid (SA). Control of camalexin synthesis appears to be largely independent of these signals. Mutations in coi1 (which block oxylipin signaling) or in sid2 or npr1 (which block SA synthesis and SA responses, respectively) have no effect on pathogen-induced camalexin production (Glazebrook et al., 1996; Nawrath and Metraux, 1999; van Wees et al., 2003). Ethylene signaling may play a role, as ein2 mutants accumulate reduced amounts of camalexin after P. syringae infection, but ein2 does not compromise A. brassicicola–induced camalexin synthesis (Heck et al., 2003; van Wees et al., 2003). Camalexin production in response to infection by virulent P. syringae strains requires PAD4, but PAD4 is not required for camalexin production induced by A. brassicicola or avirulent P. syringae strains (Zhou et al., 1998; van Wees et al., 2003).
Recent work has defined some of the enzymatic steps in camalexin synthesis. The cytochrome P450 enzymes CYP79B2 and CYP79B3, which catalyze the conversion of Trp to indole-3-acetaldoxime (IAOx) (Hull et al., 2000; Mikkelsen et al., 2000), are required for camalexin biosynthesis (Glawischnig et al., 2004). This was demonstrated genetically by the complete lack of camalexin in a cyp79B2 cyp79B3 background (Glawischnig et al., 2004). IAOx is thus a key metabolic branch point, as it is also a precursor for indole glucosinolate and auxin biosynthesis (Bak et al., 2001; Hansen et al., 2001; Zhao et al., 2002). In the biosynthesis of indole glucosinolates, IAOx is oxidized to the corresponding aci-nitro or nitrile oxide compound by CYP83B1 (Bak et al., 2001; Hansen et al., 2001). Mutants with defects in CYP83B1 accumulate elevated levels of free indole-3-acetic acid (IAA) and have high-auxin phenotypes (Delarue et al., 1998; Barlier et al., 2000; Bak et al., 2001). The route from IAOx to auxin is not known, but it is speculated that indole-3-acetonitrile (IAN) and/or indole-3-acetaldehyde may be involved (Pollmann et al., 2006). No IAOx-metabolizing enzymes in the camalexin biosynthetic pathway have been described. The thiazole ring of camalexin was proposed to be derived from Cys based on incorporation of labeled Cys, but not Met, into camalexin (Zook and Hammerschmidt, 1997). It was proposed that Cys condenses with indole-3-carboxaldehyde to form a product that subsequently cyclizes to form the thiazole ring (Browne et al., 1991). The last step of camalexin biosynthesis is conversion of dihydrocamalexic acid to camalexin, which is catalyzed by CYP71B15 (PAD3) (Schuhegger et al., 2006).
In this article, we demonstrate the involvement of the CYP71A13 enzyme in camalexin biosynthesis and pathogen resistance. We describe genetic and biochemical characterization of the CYP71A13 enzyme and show that mutations in the CYP71A13 gene result in camalexin deficiency and enhanced susceptibility to A. brassicicola. When expressed in two heterologous systems, CYP71A13 catalyzed the dehydration of IAOx to IAN, which is the first committed step in the biosynthesis of camalexin. Application of exogenous IAN restored camalexin synthesis in cyp71A13 knockout mutants.
RESULTS
CYP71A13 Is Required for Resistance to A. brassicicola
In a previous expression profiling experiment, many Arabidopsis genes were found to be induced in response to infection by A. brassicicola (van Wees et al., 2003). To identify genes that contribute to resistance to this pathogen, we isolated mutants homozygous for T-DNA insertions in these genes and tested them for susceptibility to A. brassicicola. One of the genes studied was At2g30770, which encodes cytochrome P450 monooxygenase 71A13.
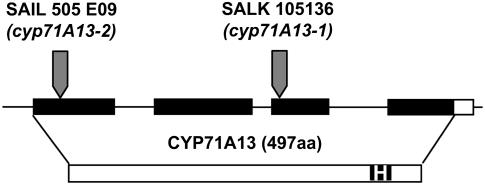
We analyzed two T-DNA insertion alleles of cyp71A13, SALK 105136 (cyp71A13-1) and SAIL 505 E09 (cyp71A13-2) (Sessions et al., 2002; Alonso et al., 2003). Plants homozygous for each insertion were identified using a PCR test. The mutant plants did not show any obvious morphological phenotypes. The positions of the insertions were verified by DNA sequencing of PCR products spanning the junctions of the left borders of the T-DNAs and the genome. Figure 1 shows that the insertions in cyp71A13-1 and cyp71A13-2 lie in the third and first exons, respectively. Both insertions are upstream from the Prosite heme-iron ligand signature (amino acids 432 to 441). This heme binding domain is essential for the functions of cytochrome P450 monooxygenases (Schuler and Werck-Reichhart, 2003). Consequently, both insertion alleles are likely to be null mutations.
Figure 1.
Positions of T-DNA Insertions in At2g30770.
The gene model from The Arabidopsis Information Resource (www.arabidopsis.org) is shown. Exons are represented by rectangles and introns and untranscribed regions by lines. Black fill indicates translated sequences. The protein sequence is represented by the bottom rectangle. The filled region labeled “H” represents the heme-iron ligand signature. Positions of the T-DNA insertions are indicated by arrows. aa, amino acids.
Both cyp71A13 mutants were tested for susceptibility to A. brassicicola. Droplets (10 μL) containing105 spores/mL were placed on the third, fourth, and fifth true leaves of each plant. Disease was assessed 3 d later. As shown in Figure 2, resistant wild-type plants developed necrotic spots at the sites of the inoculation droplets, while both cyp71A13 mutants developed spreading necrotic lesions similar to those on pad3-1 plants. At least 20 infected leaves of each genotype were assigned to one of four disease severity classes, as shown in Figure 3. Clearly, both cyp71A13 mutants are much more susceptible to A. brassicicola than wild-type plants.
Figure 2.
Susceptibility of Various Arabidopsis Mutants to A. brassicicola.
Leaves 3 d after inoculation. Plants were 24 d old when they were inoculated with A. brassicicola. The fifth true leaves from two plants of each genotype are shown.
Figure 3.
Disease Index Scoring of A. brassicicola–Infected Plants.
Plants (21 d old) were inoculated as above. After 3 d, 20 to 24 leaves per genotype were assigned a disease index score. 1, necrosis confined to the area of the inoculation droplet; 2, some chlorosis around the necrotic spot; 3, spreading chlorosis and/or some necrosis beyond the inoculation droplet; 4, spreading necrosis.
CYP71A13 Is Required for Camalexin Production
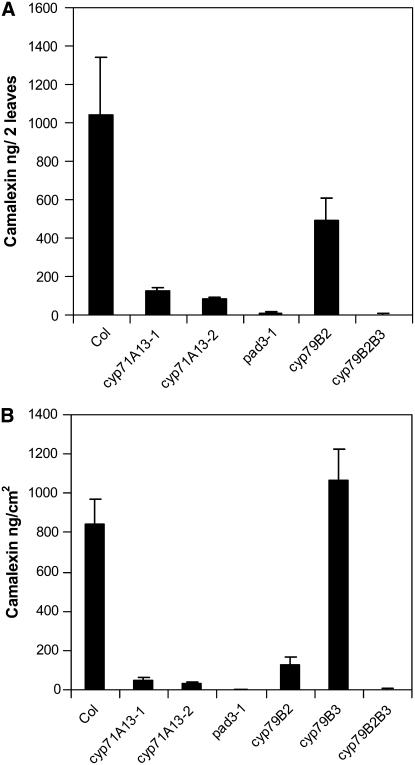
The fact that camalexin-deficient pad3-1 plants are susceptible to A. brassicicola led us to suspect that the A. brassicicola–susceptible cyp71A13 mutants might also be camalexin deficient. We first used A. brassicicola infection to induce camalexin accumulation. After 3 d, entire infected leaves were excised, and camalexin levels were determined. Figure 4A shows that camalexin levels in infected cyp71A13 mutants were <20% of wild-type levels, even though the ratio of infected to uninfected leaf area was much larger in the mutants than in the wild type. Camalexin was not detected in pad3-1 plants. We next used P. syringae infection to induce camalexin accumulation and assayed camalexin 2 d later. Again, camalexin levels in both cyp71A13 mutants were much lower than in wild-type plants, and no camalexin was detected in pad3-1 plants (Figure 4B). Camalexin was not detected in uninfected plants of any genotype (data not shown).
Figure 4.
Camalexin Levels in Various Arabidopsis Mutants after Infection by Pathogens.
(A) Infection by A. brassicicola. Plants were inoculated with A. brassicicola spores as described in Methods. After 3 d, entire leaves were excised and camalexin was determined. Each sample consisted of two leaves, and each bar represents the mean and sd of eight replicate samples. Col, Columbia.
(B) Infection by P. syringae. Plants were inoculated with bacteria at a starting density of 4 × 104 colony-forming units/cm2 (OD600 = 0.004). After 48 h, samples consisting of 1 cm2 of leaf were excised using a cork borer, and camalexin was determined. Bars represent the means and sd of eight replicates. No camalexin was observed in uninfected leaves of any genotype (data not shown).
Plants carrying T-DNA insertion mutations may carry unknown second-site mutations that are the real causes of observed phenotypes attributed to T-DNA insertions in genes of interest. It is unlikely that the A. brassicicola–susceptible and camalexin-deficient phenotypes that we observed in the cyp71A13 mutants are due to second-site mutations because we observed these phenotypes in plants containing two independent mutant alleles. We conclude that CYP71A13 is required for wild-type levels of camalexin synthesis and A. brassicicola resistance in Arabidopsis. It is likely that the camalexin deficiency causes the A. brassicicola susceptibility because pad3-1 plants, which cannot perform the last step of camalexin synthesis, are also susceptible to A. brassicicola.
Camalexin synthesis is not completely abolished in cyp71A13 mutants, so there is likely another enzyme that can perform the same function. A reasonable candidate is CYP71A12 (At2g30750) because it is the Arabidopsis CYP450 most closely related to CYP71A13 (89% identical amino acids). We obtained a line homozygous for a T-DNA insertion in this gene (GABI-Kat 127 H03; Rosso et al., 2003). When camalexin was determined 2 d after infection with P. syringae, no reduction in camalexin levels relative to wild-type plants was observed (see Supplemental Figure 1 online). It remains possible that CYP71A12 and CYP71A13 have similar activities, but that the contribution of CYP71A12 to camalexin synthesis is small relative to that of CYP71A13, and so no diminution of camalexin levels can be observed in cyp71A12 mutants when CYP71A13 is present. Construction of a double mutant is impractical because the two genes are closely linked. We cannot exclude the possibility that another enzyme is responsible for the residual camalexin in cyp71A13 mutants.
CYP79B2 and CYP79B3 Are Required for A. brassicicola Resistance and Pathogen-Induced Camalexin Production
CYP79B2 and CYP79B3 convert Trp to IAOx (Hull et al., 2000; Mikkelsen et al., 2000), which is known to be a precursor to camalexin because a cyp79B2 cyp79B3 double mutant does not produce camalexin after silver nitrate treatment (Glawischnig et al., 2004). We studied the roles of these enzymes in camalexin production and disease resistance during interactions with P. syringae and A. brassicicola. Following P. syringae infection, camalexin levels in cyp79B2 were approximately half of wild-type levels, cyp79B3 was indistinguishable from the wild type, and camalexin was undetectable in cyp79B2 cyp79B3 double mutants (Figure 4B). Similar results for cyp79B2 and cyp79B2 cyp79B3 were observed following A. brassicicola infection (Figure 4A). These results agree with those obtained by Glawischnig et al. (2004) following elicitation with silver nitrate and Kliebenstein et al. (2005) following infection with B. cinerea.
The cyp79B2 cyp79B3 double mutant was as susceptible to A. brassicicola as pad3-1 and cyp71A13 mutants (Figures 2 and 3). This observation strengthens the conclusion of Thomma et al. (1999) that the A. brassicicola susceptibility of pad3-1 indicates that camalexin plays a critical role in A. brassicicola resistance. Taken together with these previous results, all of four mutants with severe reductions in camalexin levels are susceptible to A. brassicicola. Susceptibility to A. brassicicola was not observed in cyp79B2 or cyp79B3 single mutants (Figures 2 and 3; data not shown). Apparently, the modest reduction in camalexin in cyp79B2 plants does not compromise resistance to A. brassicicola.
Camalexin Deficiency Does Not Compromise Resistance to P. syringae
While camalexin-deficient pad4 and pad2 plants display enhanced susceptibility to P. syringae, pad3 plants do not. These observations led to the conclusion that camalexin itself does not play a major role in limiting P. syringae growth and that enhanced susceptibility of pad2 and pad4 plants likely results from pleiotropic effects on other defense responses. To test this idea further, the susceptibility of cyp71A13 and cyp79B mutants to P. syringae was examined (see Supplemental Figure 2 online). While pad4 plants displayed characteristic extreme susceptibility to this pathogen, no increase in pathogen growth relative to wild-type plants was observed in either cyp71A13 mutant, cyp79B2, cyp79B3, or the cyp79B2 cyp79B3 double mutant. This is consistent with the idea that camalexin does not play a major role in resistance to P. syringae. The cyp79B2 cyp79B3 double mutant also fails to produce indole glucosinolates, so these compounds must not play major roles in limiting P. syringae growth.
PAD3 and CYP71A13 Are Coregulated
PAD3 is rapidly induced in response to P. syringae infection (Zhou et al., 1999). To determine whether this is true of other genes involved in camalexin synthesis, the expression levels of PAD3, CYP71A13, CYP71A12, CYP79B2, and CYP79B3 were examined in expression profiling data of wild-type plants infected with Psm ES4326 obtained using the ATH1 GeneChip array from Affymetrix. Figure 5A shows that PAD3, CYP71A13, and CYP79B2 were all strongly induced by 9 h after infection, and their expression levels increased further over the course of the experiment. CYP79B3 was not induced by infection. Expression of CYP71A12 was induced by infection, although its induced level appeared to be substantially lower than PAD3, CYP71A13, and CYP79B2 (note that gene-to-gene comparisons are unreliable due to variations in the hybridization efficiency of different oligonucleotides on the array).
Figure 5.
Expression Patterns of Selected Genes during P. syringae Infection as Measured by Microarray Experiments.
(A) Expression changes in response to infection by Psm ES4326. Expression values were determined using robust multichip average and divided by 1000 for convenience. Each bar represents the mean and sd of three independent replicates.
(B) Comparison of PAD3 and CYP71A13 expression patterns. Data points represent log2-transformed values from the Genevestigator database. The underlying data are provided as Supplemental Table 1 online.
The Genevestigator database of Arabidopsis expression profiling data was used to test for coregulation among pairs of these five genes (Zimmermann et al., 2004). Arrays in which expression of both PAD3 and CYP71A13 was significantly above background (P < 0.05) were selected from the 1432 ATH1 arrays in the database. Then, for each of the resulting 459 arrays, the log2 expression values for each gene were plotted as shown in Figure 5B. The correlation between the expression levels of these two genes was quite strong (r2 = 0.72 by linear regression). This coregulation was much weaker in the case of CYP79B2. The r2 values for CYP79B2 versus PAD3 and CYP79B2 versus CYP71A13 were 0.20 and 0.27, respectively. The P values for all comparisons were smaller than the lower limit for calculation in R, P < 2.2 × 10−16, indicating that all the r2 values were significantly different from 0. While high expression levels of PAD3 and CYP71A13 were associated with high expression levels of CYP79B2, there were many arrays in which expression of CYP79B2 was high but PAD3 and CYP71A13 were not. This likely reflects the role of CYP79B2 in biosynthesis of other compounds in addition to camalexin.
To further investigate coregulation of PAD3 and CYP71A13, we used quantitative RT-PCR (qRT-PCR) to monitor expression of these genes over the course of infection in plants with defense signaling mutations and in wild-type plants. We included dde2, in which jasmonic acid signaling is blocked due to a mutation in allene oxide synthase; ein2, in which ethylene signaling is blocked; pad4, in which SA signaling and expression of many other pathogen-responsive genes is reduced; and sid2, in which SA synthesis is greatly reduced. Levels of PAD3, CYP71A13, and CYP79B2 mRNAs were determined immediately after infection with Psm ES4326 and 6, 9, and 24 h later. The data are provided as Supplemental Table 2 online and summarized in Table 1. The expression patterns of PAD3 and CYP71A13 were very similar. Both genes were strongly induced in wild-type plants (see Supplemental Table 2 online) and expressed at lower levels in pad2, pad4, and sid2 mutants at 6 and 9 h after infection, in ein2 mutants at 9 h after infection, and at levels higher than wild-type in sid2 plants 24 h after infection. By contrast, none of the regulatory mutations affected CYP79B2 expression except pad4, where the effect was only evident at 9 h after infection. These results extend the conclusion that PAD3 and CYP71A13 are tightly coregulated, while CYP79B2 is not.
Table 1.
Ratios of mRNA Levels in P. syringae–Infected Mutant Plants Relative to Infected Wild-Type Plants Determined by qRT-PCR
| Gene
|
||||
|---|---|---|---|---|
| Genotype | HAIa | PAD3 | CYP71A13 | CYP79B2 |
| dde2 | 0 | 0.650 | 0.280 | 0.33 |
| 6 | 1.910 | 1.860 | 0.38 | |
| 9 | 1.350 | 1.120 | 1.52 | |
| 24 | 1.520 | 1.150 | 1.32 | |
| ein2 | 0 | 0.460 | 0.210 | 0.92 |
| 6 | 1.360 | 1.350 | 0.67 | |
| 9 | 0.330 | 0.400 | 0.96 | |
| 24 | 0.930 | 0.740 | 1.04 | |
| pad2 | 0 | 0.370 | 0.150 | 1.02 |
| 6 | 0.430 | 0.230 | 0.87 | |
| 9 | 0.360 | 0.180 | 0.63 | |
| 24 | 1.730 | 0.880 | 1.64 | |
| pad4 | 0 | 0.190 | 0.170 | 1.02 |
| 6 | 0.190 | 0.280 | 0.66 | |
| 9 | 0.077 | 0.057 | 0.45 | |
| 24 | 1.000 | 0.720 | 0.49 | |
| sid2 | 0 | 0.110 | 0.240 | 0.55 |
| 6 | 0.390 | 0.430 | 0.73 | |
| 9 | 0.110 | 0.028 | 0.60 | |
| 24 | 2.260 | 2.270 | 1.95 | |
Data from qRT-PCR analysis of three biologically independent experiments, each consisting of three replicate samples. Ratios <0.5 or >2 for which the difference between wild-type and mutant is statistically significant by Duncan's multiple range test (P < 0.01) are shown in bold.
HAI, hours after infection.
Recombinant CYP71A13 Metabolizes IAOx to IAN
The camalexin-deficient phenotype of cyp71A13 mutants and the correlation of CYP71A13 expression with PAD3 led us to investigate whether the enzyme was involved in the biosynthesis of camalexin. We hypothesized that CYP71A13 would metabolize IAOx, as it belongs to the CYP71 family, whose members include the oxime-metabolizing P450s CYP71E1, involved in dhurrin biosynthesis, as well as CYP83A1 and CYP83B1, involved in glucosinolate biosynthesis (Bak et al., 1998b, 2001; Hansen et al., 2001; Naur et al., 2003). Historically, the CYP83s were assigned to their own family based on a short EST sequence. They have since been assigned to the CYP71 family based on similarity of the full-length sequences.
CYP71A13 was heterologously expressed in Escherichia coli to test it for the ability to metabolize IAOx. The coding region of CYP71A13 was PCR amplified from cDNA generated from rosette leaves and cloned into the expression vector pSP19g10L, which is optimized for expression of cytochromes P450 in E. coli (Barnes et al., 1991). The construct consisted of the CDS of CYP71A13 except that the second codon, for Gly, was changed to an Ala codon, which is preferred for expression in E. coli (Barnes et al., 1991). CYP71A13 was functionally expressed in E. coli as evidenced by a carbon monoxide difference spectrum with the characteristic peak of 450 nm, as shown in Figure 6 (Omura and Sato, 1964). Based on the peak at 450 nm, the expression level of CYP71A13 was estimated to be 500 nmol cytochrome P450 (liter of culture)−1. When spheroplasts from E. coli expressing CYP71A13 were reconstituted in lipid micelles with recombinant NADPH-cytochrome P450-reductase from Arabidopsis (ATR1) and fed with IAOx, a compound accumulated in the reaction mixture, as shown by the liquid chromatography–mass spectrometry (LC-MS) spectra in Figure 7. LC-MS analysis showed that the compound was IAN based on comparison to an authentic standard (Figure 7). This result shows that CYP71A13 catalyzes the dehydration of IAOx to IAN.
Figure 6.
Carbon Monoxide Difference Spectrum of CYP71A13.
CYP71A13 was functionally expressed in E. coli as observed by the CO-difference spectrum of purified membranes. The dotted and solid lines represent the spectrum before and 15 min after CO exposure. The spectra were recorded at room temperature.
Figure 7.
The Catalytic Properties of CYP71A13 toward IAOx as Analyzed by LC-MS.
LC-MS chromatograms from E. coli spheroplasts reconstituted with recombinant NADPH:cytochrome P450 reductase from Arabidopsis (ATR1) and incubated with IAOx. The reaction mixtures were extracted with ethyl acetate, and the organic phase was analyzed by LC-MS. IAOx is indicated by arrows. The two geometric isomers appear as separate peaks but with an elevated baseline in between due to partial on-column equilibration.
IAN did not accumulate in reaction mixtures with spheroplasts from E. coli carrying the empty vector (Figure 7). Although product formation was strictly dependent on CYP71A13, it was not always dependent on exogenous NADPH or reductase.
Spectral Characterization of Recombinant CYP71A13
We analyzed the binding of different compounds to the active site of CYP71A13 spectroscopically (Schenkman et al., 1967). Detection of direct binding of oximes to P450 enzymes has been reported to be difficult (Boucher et al., 1994; Kahn et al., 1999; Bak et al., 2001). As an alternative method that has been applied in similar situations, we tested if the binding of substrate could be measured as the ability to displace tryptamine from the active site (Jefcoate, 1978; Bak et al., 2001). Figure 8 shows that addition of tryptamine to recombinant CYP71A13 resulted in the formation of a typical type II spectrum, typically seen for amines. The spectrum was gradually reversed by addition of increasing concentrations of IAOx, resulting in a reverse type II spectrum (Figure 8). The minimum concentration of IAOx required to displace tryptamine was 2.5 μM, showing that low concentrations of IAOx can displace tryptamine from the active site. Displacement of tryptamine was also observed after addition of phenylacetaldoxime (Figure 8). However, under the conditions used, the amplitude of the reverse type II spectrum was not as strong as with IAOx even at higher substrate concentrations, suggesting that phenylacetaldoxime is not as good of a substrate as IAOx since amplitude of the spectrum has been correlated with rate of turnover (Kahn et al., 1999). Neither the Tyr-derived oxime, p-hydroxyphenylacetaldoxime, nor IAN displaced the inhibitor (data not shown).
Figure 8.
Analysis of CYP71A13 by Optical Difference Spectroscopy.
A saturated type II spectrum was obtained with 100 μM tryptamine in the sample cuvette (thick solid line). The addition of 100 μM tryptamine to the reference cuvette gave a baseline (dotted line). The spectra were recorded at room temperature and did not increase over time.
(A) Increasing concentrations of IAOx in the sample cuvette (2.5, 10, and 100 μM) gradually displaced tryptamine, giving the reverse type II spectrum.
(B) Increasing concentrations of phenylacetaldoxime in the sample cuvette (20, 40, and 100 μM) gradually displaced tryptamine, giving the reverse type II spectrum.
Transient Expression of CYP71A13 in Tobacco
To determine whether CYP71A13 converts IAOx to IAN in plants, we expressed CYP71A13 together with CYP79B2 transiently in tobacco (Nicotiana benthamiana) plants. We took advantage of the fact that several enzymes can be transiently coexpressed in high amounts in tobacco cells in the presence of the suppressor of silencing, p19 (Voinnet et al., 2003). Tobacco plants were infiltrated with Agrobacterium tumefaciens transformed with expression constructs of CYP79B2, CYP71A13, or both. In all cases, the suppressor of silencing construct, p19, was coinfiltrated. The expression of CYP79B2 and CYP71A13 were driven by the 35S promoter. No CYP79 homologs have been identified in tobacco (Bak et al., 1998a), and neither IAOx nor IAN has been reported. When microsomes were incubated with Trp, IAOx was detected in reaction mixtures with microsomes from plants expressing CYP79B2, whereas both IAOx and IAN were detected in reaction mixtures with microsomes from plants expressing CYP79B2 and CYP71A13, as shown in Figure 9. Neither IAOx nor IAN was detected in reaction mixtures from microsomes expressing p19. These results show that recombinant CYP71A13 expressed in E. coli or in plant cells can catalyze the dehydration of IAOx to IAN in vitro.
Figure 9.
Transient Expression of CYP79B2 and CYP71A13 in N. benthamiana Results in Conversion of Trp to IAN via IAOx.
Microsomes from leaves infiltrated with p19 (thin solid line), CYP79B2 (dotted line), or CYP79B2 and CYP71A13 (thick solid line) in combination with p19 were incubated with Trp. After incubation, the reaction mixtures were extracted with ethyl acetate, and the organic phases were analyzed by LC-MS. m/z, mass-to-charge ratio.
(A) Reconstructed ion chromatogram of m/z 175 (IAOx) from the different microsomal assays.
(B) Reconstructed ion chromatogram of m/z 155 (IAN) from the different microsomal assays.
The Role of IAN in Camalexin Biosynthesis
In vivo feeding of IAN to camalexin biosynthetic mutants was performed to address the question of whether IAN is a precursor in the camalexin pathway. Figure 10 shows that when IAN was applied to silver nitrate–treated cyp71A13 leaves, camalexin levels were approximately sixfold higher than when water was applied. Similarly, IAN treatment restored camalexin synthesis in cyp79B2 cyp79B3 plants that are unable to produce camalexin due to failure to synthesize IAOx (Glawischnig et al., 2004). By contrast, the camalexin deficiency of pad3 was not corrected by IAN application, consistent with PAD3 (CYP71B15) catalyzing the final step in camalexin biosynthesis (Schuhegger et al., 2006). These results are in accordance with the idea that IAN is an intermediate in camalexin synthesis, and the reduced camalexin synthesis in cyp71A13 mutants results from inadequate IAN production.
Figure 10.
In Vivo Feeding of IAN to AgNO3-Treated Arabidopsis Mutants.
After spraying with silver nitrate, rosette leaves of Col-0, cyp71A13-1, cyp79B2 cyp79B3, and pad3 plants were incubated with 250 μM IAN (black bars) or water (gray bars). Bars represent means and sd of six replicates. Significance of differences between water- and IAN-treated plants was tested using an unpaired, two-tailed t test. The difference for Col-0 was not significant (P = 0.16), while the differences for cyp71A13-1 and cyp79B2 cyp79B3 were highly significant (P = 0.00028 and 0.0000049, respectively).
DISCUSSION
We have shown that CYP71A13 is required for pathogen-induced camalexin synthesis in Arabidopsis and that like camalexin-deficient pad3 mutants, cyp71A13 mutants are susceptible to A. brassicicola. We found that CYP71A13 catalyzes the first committed step in camalexin biosynthesis by dehydrating IAOx to IAN as evidenced by biochemical characterization of recombinant CYP71A13. In addition, we have shown that the cyp71A13 knockout phenotype can be restored by in vivo feeding with IAN, strongly suggesting that IAN is an intermediate in camalexin biosynthesis.
The Role of Camalexin in Disease Resistance
After identification of PAD3 as CYP71B15, the disease phenotypes of pad3 plants were taken as an indication of whether or not camalexin is important for resistance to various pathogens. However, there remained a small possibility that these conclusions were incorrect due to other possible activities of PAD3, antimicrobial activity of the PAD3 substrate, or other unanticipated effects of the mutation. Our observation that cyp71A13 and pad3 mutants are unaffected in resistance to P. syringae and are dramatically more susceptible to A. brassicicola substantially strengthens the conclusion that camalexin does not play a major role in resistance to P. syringae but is very important for resistance to A. brassicicola (Glazebrook and Ausubel, 1994; Thomma et al., 1999).
Regulation of Camalexin Synthesis
We found that mRNA levels of both PAD3 and CYP71A13 are greatly increased in response to P. syringae infection. Expression of these two genes is tightly correlated over a large number of microarray experiments and similarly affected by mutations in canonical defense regulatory genes. This suggests that it might be possible to identify the missing enzymes in camalexin synthesis by searching for genes coregulated with PAD3 and CYP71A13. However, we have not found other genes as tightly coregulated with PAD3 and CYP71A13 as they are with each other, and some testing of candidate genes has not yet yielded additional camalexin-deficient mutants. Perhaps the missing enzymes participate in biosynthetic processes in addition to camalexin synthesis and so do not show tight coregulation, as is the case with CYP79B2. Alternatively, they may be absent from the ATH1 array, which does not include probes for every presently annotated Arabidopsis gene.
We observed coregulated expression of PAD3 and CYP71A13 in a set of defense signaling mutants. Expression of PAD3 and CYP71A13 was transiently reduced in ein2, pad2, pad4, and sid2 mutants. Among these, ein2, pad2, and pad4 display reduced camalexin levels. PAD2 encodes γ-glutamyl cysteine synthase, which is required for synthesis of glutathione (Parisy et al., 2007). Glutathione levels in pad2 plants are reduced, while cysteine levels are fivefold higher than in the wild type. Based on in vivo feeding studies, Zook and Hammerschmidt (1997) proposed that cysteine is the sulfur donor for camalexin, but perhaps the sulfur donor is actually glutathione. Glutathione is also involved in regulation of NPR1 activity through redox changes (Mou et al., 2003). Thus, the camalexin deficiency of pad2 plants may result from causes other than or in addition to the reduced expression of PAD3 and CYP71A13. PAD4 encodes a regulator that affects expression of many defense-related genes (Jirage et al., 1999; Glazebrook et al., 2003). The phenotypes of sid2 plants are difficult to explain. While the impact of sid2 on expression of PAD3 and CYP71A13 at 9 h after infection is comparable to that of pad4, there is no reduction of camalexin in sid2 plants (Nawrath and Metraux, 1999). Perhaps a more detailed time course would show that the depression of PAD3 and CYP71A13 is more transient in sid2 than in pad4 plants. None of the mutations studied compromised expression of PAD3 or CYP71A13 at 24 h after infection, so even though there is a requirement for SA signaling at early times, some other signaling process must be sufficient for wild-type expression levels at later time points. It seems likely that regulation of camalexin synthesis involves the action of multiple regulatory factors and that a complete understanding will require considerably more work.
CYP71A13 Converts IAOx to IAN
CYP71A13 expressed in E. coli and in leaves of N. benthamiana metabolized IAOx to IAN. In E. coli spheroplasts, the dehydration of IAOx was not dependent on exogenous P450 reductase, which suggests that the reaction could be supported by the endogenous E. coli flavodoxin/NADPH-flavodoxin reductase system, which can support other heterologously expressed P450 enzymes (Jenkins and Waterman, 1994; Halkier et al., 1995). Furthermore, the IAN formation was not always dependent on exogenous NADPH, suggesting that residual amounts of NADPH may occasionally be present in the spheroplast preparation. For P450-catalyzed dehydration of oximes, it has been suggested that once NADPH reduces cytochrome P450 to the ferrous state, the reaction proceeds without further action by NADPH (DeMaster et al., 1992). Similar conditions might apply to CYP71A13. Dehydration of oxime to nitrile is not a redox reaction and therefore not a typical P450-catalyzed reaction. However, other P450s have been shown to catalyze dehydration reactions, including members of the CYP3A family (Boucher et al., 1994; Mathews et al., 1998) and the multifunctional CYP71E1 (Bak et al., 1998b).
The substrate binding spectra showed that oximes derived from Trp and Phe, but not Tyr, could displace tryptamine from the active site of CYP71A13. This indicates that the range of compounds that fit into the substrate binding pocket is limited. The spectral data are in accordance with reconstitution experiments with labeled oximes, in which IAOx, but not the Tyr-derived p-hydroxyphenylacetaldoxime, was metabolized by CYP71A13. The binding of phenylacetaldoxime to the active site of CYP71A13 indicates that this compound is a substrate for the enzyme, although it remains to be determined whether it can be metabolized.
In the biosynthesis of the Tyr-derived cyanogenic glucoside dhurrrin in Sorghum bicolor, CYP71E1 catalyzes the metabolism of p-hydroxyphenylacetaldoxime to the corresponding hydroxynitrile, p-hydroxyphenylmandelonitrile, via a nitrile intermediate (Bak et al., 1998b; Kahn et al., 1999). Both oxime and nitrile fed to sorghum microsomes or to micelles with recombinant CYP71E1 reconstituted with sorghum reductase were converted to the hydroxynitrile. Reconstitution of recombinant CYP71E1 with E. coli flavodoxin/NADPH-flavodoxin reductase drove the reaction only to the corresponding nitrile, p-hydroxyphenylacetonitrile, suggesting a specific redox requirement (Bak et al., 1998b). In our study, IAN was the only product formed when IAOx was incubated with recombinant CYP71A13 from either E. coli or tobacco. In contrast with CYP71E1 in the cyanogenic pathway, CYP71A13 was not able to metabolize IAN further. The role of IAN as a product of CYP71A13 in plants was supported by the ability of exogenous IAN to rescue the camalexin-deficient phenotype of cyp71A13 knockout mutants.
When tobacco microsomes were incubated with Trp, IAN accumulated in reaction mixtures from plants expressing CYP79B2 and CYP71A13. In addition, trace amounts of IAN were observed in reaction mixtures from plants that expressed CYP79B2, whereas no IAN was observed in the reaction mixture from plants expressing the control, p19. This suggests that tobacco can convert some of the accumulated IAOx to IAN. We do not know whether this occurs enzymatically or nonenzymatically.
IAOx constitutes a metabolic branch point among indole glucosinolates, camalexin, and IAA biosynthesis. How these pathways are organized and regulated around IAOx is currently unclear (Hansen and Halkier, 2005). However, as camalexin is produced only under inducing conditions, IAOx is presumed to be funneled into indole glucosinolates under normal conditions (Bak et al., 2001).
IAN in Camalexin Biosynthesis
The role of IAN as an intermediate in camalexin biosynthesis was supported by the ability of exogenous IAN to rescue the camalexin-deficient phenotype of cyp71A13 knockout mutants. Plants overexpressing CYP79B2 or PAD3 did not have increased accumulation of camalexin (Glawischnig et al., 2004; Schuhegger et al., 2006). The photosynthetic mutant dgd1 (Dörmann et al., 1995), which contains 45-fold elevated levels of IAN (Fiehn et al., 2000), does not accumulate more camalexin than the wild type in response to silver nitrate (R. Schuhegger and E. Glawischnig, unpublished data). Thus, the rate-limiting step in camalexin synthesis is unclear. Whether IAN functions as an intermediate in the biosynthesis of other IAOx-derived phytoalexins from Brassica remains to be investigated. IAN has been recognized as a phytoalexin that is induced upon fungus attack in Brassica juncea (Pedras et al., 2002).
IAN and Auxin
Our data show that IAN produced by CYP71A13 is an intermediate in the biosynthesis of camalexin. In addition, IAN has often been suggested as an intermediate in IAA biosynthesis in crucifers (reviewed in Pollmann et al., 2006) and also in maize (Zea mays), a plant that does not produce camalexin or indole glucosinolates (Park et al., 2003). Auxin biosynthesis via IAN requires the action of nitrilases (NIT1-3) that hydrolyze IAN to IAA (Bartling et al., 1992). Generally, nitrilases have Km values for IAN in the millimolar range (Vorwerk et al., 2001), indicating that IAN is not a very good substrate. Nevertheless, nit1 plants did not show the auxin effect seen in wild-type plants upon treatment with exogenous IAN (Normanly et al., 1997), and NIT2-overexpressing lines from Arabidopsis and tobacco show increased sensitivity to exogenous IAN (Schmidt et al., 1996; Normanly et al., 1997). However, attempts to rescue the high-auxin phenotype of sur2 by crossing sur2 to nit1-1 were unsuccessful (Bak et al., 2001), raising further questions about the role of IAN in auxin biosynthesis.
IAN might be an intermediate in both camalexin and auxin biosynthesis, suggesting that either the IAN-metabolizing enzymes are competing for substrate or IAN is highly channeled within each pathway (i.e., IAN is produced and metabolized in situ). Alternatively, IAN is primarily involved in camalexin biosynthesis and only converted to IAA when excess IAN accumulates. The latter idea may explain why the expression of NIT1 and NIT2 is induced by exogenous IAN (Grsic et al., 1998). Another source of IAN is the turnover of indole glucosinolates (Bones and Rossiter, 2006). A basal level of IAN is present in healthy plants (Ilic et al., 1996).
Camalexin Biosynthesis Downstream of IAN
So far, the metabolic route from IAN to dihydrocamalexic acid in camalexin biosynthesis is unknown. A proposed candidate intermediate is indole-3-carboxaldehyde (Browne et al., 1991), although this compound does not appear to be a precursor of camalexin based on in vivo feeding studies (Schuhegger et al., 2006). Further studies are needed to identify the enzymes catalyzing the conversion of IAN to dihydrocamalexic acid in the biosynthetic pathway of camalexin, the model compound for studying the bioactive cruciferous indole alkaloids.
METHODS
Plant Growth
Nicotiana benthamiana plants used for transient expression were grown at 24°C (day) and 17°C (night) in a greenhouse. For pathogen experiments, Arabidopsis thaliana was grown in a controlled-environment chamber at 22°C, 70% relative humidity, 12 h light (100 μM m−2 s−1 fluorescent illumination), and 12 h dark. All genotypes were in the Col-0 background. pad3 was pad3-1 (Glazebrook and Ausubel, 1994), cyp79B2, cyp79B3, and the double mutant were from Zhao et al. (2002), dde2 was dde2-2 (von Malek et al., 2002), ein2 was ein2-1 (Alonso et al., 1999), pad2 was pad2-1 (Parisy et al., 2007), pad4 was pad4-1 (Jirage et al., 1999), and sid2 was sid2-2 (Wildermuth et al., 2001).
Characterization of cyp71A13 Mutant Alleles
Plants homozygous for cyp71A13-1 and cyp71A13-2 were identified by PCR as described (Sessions et al., 2002). For cyp71A13-1, gene-specific primers were 5′-CGAGTAGAGTTGCGTTGGGAAGA-3′ (105136LP) and 5′-CCATGTGGCCTAATAGTTGACCG-3′ (105136RP), while the T-DNA left border primer was 5′-GGAACAACACTCAACCCTATCTCG-3′ (LBe). For cyp71A13-2, gene-specific primers were 5′-TTCTCCATAGGGAGCAAACAC-3′ (505E09LP) and 5′-TGACGTCCTGCACTATTGACA-3′ (505E09RP), while the T-DNA left border primer was 5′-TTCATAACCAATCTCGATACAC-3′ (LB3). The positions of the T-DNA insertions were determined by DNA sequencing of the PCR products obtained with primers LBe and 105136RP (for cyp71A13-1), and LB3 and 505E09 (for cyp71A13-2).
Infection with Pathogens
Infection with Pseudomonas syringae strain Psm ES4326 and determination of bacterial growth was as described (Parisy et al., 2007). Infection with Alternaria brassicicola strain ATCC 96866 was achieved by placing 10-μL droplets of water containing 105 spores/mL of water onto the abaxial surface of the third, fourth, and fifth true leaves of 3-week-old plants. Plants were kept tightly covered for the first day, and symptoms were assessed after 3 d. Camalexin in pathogen-infected plants was determined as described (Glazebrook and Ausubel, 1994).
Determination of Expression Levels
For ATH1 microarray analysis, wild-type Col-0 plants were infected with Psm ES4326 by syringe infiltration to a starting bacterial titer of 104 colony-forming units/cm2 of leaves (OD600 = 0.002) or mock-infected and sampled after 9, 24, or 32 h. Three independent experiments were performed. RNA was purified using Trizol reagent (Invitrogen) according to the manufacturer's instructions with modifications for tissues with high starch content. Labeled cRNA was produced using the Affymetrix one-cycle target labeling kit (Affymetrix). Raw data were converted to expression values for each gene using robust multichip average (Irizarry et al., 2003). Complete data sets are available from the Nottingham Arabidopsis Stock Centre (accession number NASCARRAYS-414; http://affymetrix.arabidopsis.info/). The values shown in Figure 5 were obtained by combining the expression values from the three replicate data sets to obtain the means and the standard deviations.
For qRT-PCR analysis, plants were infected as above and sampled immediately or 6, 9, or 24 h later. Total RNA was isolated using TRIzol (Invitrogen) following the manufacturer's protocol. qRT-PCR was performed using the Superscript III Platinum SYBR Green One-Step qRT-PCR kit (Invitrogen) and the Applied Biosystems 7500 Real Time PCR machine. The thermal cycling program was as follows: 50°C for 10 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 1 min, and a one-cycle dissociation stage at 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. The primers were as follows: Actin2 (At3g18780), 5′-AGTGTCTGGATCGGTGGTTC-3′ and 5′-CCCCAGCTTTTTAAGCCTTT-3′; PAD3 (At3g26830), 5′-TGCTCCCAAGACAGACAATG-3′ and 5′-GTTTTGGATCACGACCCATC-3′; CYP71A13 (At2g30770), 5′-TAAAGAGGTGCTTCGGTTGC-3′ and 5′-TATCGCAGTGTCTCGTTGGA-3′; and CYP79B2 (At4g39950), 5′-GTTTCTGGCTAAACCGTTGG-3′ and 5′-TCTGGTAACCGGAATTGACC-3′. Actin2 was used as the internal reference gene. Samples from each of three biological replicates were assayed in triplicate, for a total of nine measurements per data point. Expression values were normalized to those of actin2. Data were then subjected to analysis of variance in a completely randomized design, and the treatment means separated by Duncan's multiple range test. The values in Supplemental Table 2 online are the means and standard deviations from all nine measurements per data point.
Synthesis of IAOx
IAOx was synthesized from indole-3-acetaldehyde as described (Rausch et al., 1985). The structure was verified by nuclear magnetic resonance.
Heterologous Expression of CYP71A13 in Escherichia coli
The coding sequence of CYP71A13 (NM 128630.2; www.ncbi.nlm.nih.gov) was amplified from cDNA with the following primers: forward, 5′-TACGCCATATGGCAAATATTCAAGAAATGGAAATGATATTGAGTA-3′; reverse, 5′-AATAACAAAGCTTACACAACCGAAGATGGAAA-3′. The primers introduce 5′ NdeI and 3′ HindIII sites (underlined), which were used to clone the fragment into NdeI/HindIII-digested pSP19g10L (Barnes et al., 1991). The forward primer changes the second codon from Gly to Ala (in italics). The expression construct was transformed into E. coli strain C43 (DE3). For expression of CYP71A13, one colony was grown overnight in Luria-Bertani medium + ampicillin (100 μg/mL), and 1 mL of the culture was used to inoculate 100 mL of modified terrific broth medium, pH 7.5, with 100 μg/mL of ampicillin, 1 mM isopropylthio-β-galactoside, 1 mM δ-aminolevulinic acid, and 1 mM thiamine. The culture was grown at 28°C at 110 rpm for 40 h. Spheroplasts were made in accordance with Halkier et al. (1995), except that glycerol was omitted from the final buffer. A Triton X-114–induced phase partitioning was done as described previously (Halkier et al., 1995).
Measurement of Carbon Monoxide Difference Spectrum
CYP71A13 (0.24 μM) dissolved in 50 mM KPi, pH 7.9, with a few grains of solid sodium dithionite was distributed in two cuvettes. When a stable baseline was obtained, the sample cuvette was bubbled with CO (30 s), and spectral changes were recorded for 15 min on a Lambda 800 UV-Vis spectrophotometer (Perkin-Elmer) at room temperature. The amount of expressed functional cytochrome P450 was monitored by Fe2+·CO versus Fe2+ difference spectroscopy and quantified using an extinction coefficient of 91 mM−1 cm−1.
Reconstitution of Functional CYP71A13
The activity of CYP71A13 was measured by reconstituting spheroplasts from E. coli expressing CYP71A13 with recombinant NADPH:cytochrome P450 reductase from Arabidopsis (ATR1). The Arabidopsis NADPH:cytochrome P450 reductase was expressed and purified as described previously (Naur et al., 2003). For a typical reconstitution experiment, 5 μL of CYP71A13 was incubated with 5 μL (10 to 20 units) NADPH-cytochrome P450 reductase from Arabidopsis, ATR1, 5 μL 14C-substrate, 25 μg dilauroyl phosphatidyl choline, and 3 mM NADPH in 50 mM KPi, pH 7.9, in a total volume of 50 μL. The reactions were incubated at 30°C for 1 h. For LC-MS analysis, 5× 50-μL reaction mixtures were made as above, except 200 μM cold IAOx was used in place of 14C-labeled IAOx. The reactions were extracted with 200 μM ethyl acetate; the ethyl acetate phases were combined, dried in vacuo, and resuspended in 50 μL 50% ethanol.
Substrate Binding Spectra
CYP71A13 (0.1 μM) dissolved in 50 mM KPi, pH 7.9, was distributed in two cuvettes. The reverse type II spectrum was obtained by adding 100 μM tryptamine to the sample cuvette. The spectral change was recorded, and 100 μM tryptamine was added to a reference cuvette, giving a baseline. IAOx was titrated (2.5 to 100 μM) to the sample cuvette. An equal amount of solvent (50% ethanol) was added to the reference cuvette. Curves for CO spectrum and binding spectra were plotted and smoothed using SigmaPlot 2001 software (Systat Software).
LC-MS Analysis
LC-MS was performed using a HP1100 liquid chromatograph (Agilent Technologies) coupled to a Bruker Esquire 3000+ ion trap mass spectrometer (Bruker Daltonics). An XTerra MS C18 column (Waters; 3.5 μM, 2.1 × 100 mm) was used at a flow rate of 0.2 mL min−1. The mobile phases were as follows: A, 0.1% (v/v) HCOOH; B, methanol. The gradient program was as follows: 0 to 2 min, isocratic 35% B; 2 to 20 min, linear gradient 35 to 100% B; 20 to 25 min, isocratic 100% B. The column temperature was kept at 20°C. The spectrometer was run in atmospheric pressure chemical ionization mode, and positive ions were observed.
Transient Expression of CYP71A13 in N. benthamiana
The coding sequence of CYP71A13 was amplified from cDNA (Mikkelsen et al., 2003) with the following primers: forward, 5′-GGCTTAAUATGAGCAATATTCAAGAAAATGGA-3′; reverse, 5′-CCTTTAAUTTACACAACCGAAGATGGAAA-3′. The coding sequence of CYP79B2 was amplified with the following primers: forward, 5′-GGCTTAAUATGAACACTTTTACCTCAAACTCT-3′; reverse, 5′-GGTTTAAUTCACTTCACCGTCGGGTAGAGAT-3′. The sequences were cloned with the USER technology (Nour-Eldin et al., 2006) into USER-compatible pCAMBIA1300 (CYP71A13) or pCAMBIA2300 (CYP79B2) vectors, where expression was driven by the 35S promoter. The constructs were transformed into Agrobacterium strain C58 and grown overnight in YEP with the appropriate antibiotics (p19, kanamycin + tetracycline; CYP71A13 and CYP79B2, kanamycin + rifampicin), pelleted by centrifugation, and resuspended in 10 mM MES, 10 mM MgCl2, and 100 μM acetosyringone to OD 0.5 to 0.8 and left shaking at room temperature for 3 h. Three-week-old tobacco leaves were infiltrated with a given expression construct in combination with the suppressor of silencing, p19 (Voinnet et al., 2003) in a ratio of 3:1. When several expression constructs were used in combination with p19, the ratio was equal among all constructs used.
Microsomal Preparation from Tobacco Leaves
Microsomes were made 4 d after infiltration according to Porchia et al. (2002). Briefly, tobacco leaves were homogenized with a polytron in homogenization buffer (50 mM KPi, pH 7.2, 400 mM sucrose, 4 mM DTT, and Complete proteinase inhibitor cocktail [Roche]) and filtered through muslin. Tissue was pelleted by centrifugation at 5000g for 15 min, and the supernatant was centrifuged at 50,000g for 45 min. The microsomes were resuspended in homogenization buffer and homogenized, and glycerol was added to a final concentration of 15%. To measure CYP71A13 activity, ∼35-μL microsomes (equal amount of protein) were incubated in a total volume of 50 μL with 200 μM substrate and 3 mM NADPH at 30 C for 1 h. The reaction mixture was extracted with 200 μL ethyl acetate. The organic phases were dried under vacuum and resuspended in 50% ethanol. For structural analysis, five reaction mixtures were combined and resuspended in 50 μL 50% ethanol.
In Vivo Feeding of IAN to Arabidopsis Mutants
The Col-0 wild type, cyp71A13, and cyp79B2 cyp79B3 knockout, as well as pad3 mutants, were sprayed with 5 mM silver nitrate to test complementation of the pathway with IAN. After 8 h, rosette leaves were excised at the petiole and incubated in 200 μL 250 μM IAN or water for an additional 16 h. Camalexin was extracted and quantified as described (Schuhegger et al., 2006).
Accession Numbers
Sequence data for genes from this article can be found in the GenBank/EMBL data libraries under the following Arabidopsis Genome Initiative identifiers: CYP71A13 (At2g30770), CYP71A12 (At2g30750), CYP79B2 (At4g39950), CYP79B3 (At2g22330), and PAD3 (At3g26830).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. A Mutation in CYP71A12 (At2g30750) Does Not Cause Camalexin Deficiency.
Supplemental Figure 2. Mutations in Camalexin Biosynthetic Genes Do Not Cause Enhanced Susceptibility to P. syringae.
Supplemental Figure 3. Camalexin Biosynthetic Pathway.
Supplemental Table 1. Genevestigator Data for Expression Levels of PAD3 (At3g26830), CYP71A13 (At2g30770), CYP79B2 (At4g39950), CYP79B3 (At2g22330), and CYP71A12 (At2g30750).
Supplemental Table 2. Quantitative RT-PCR Mean Expression Values for PAD3, CYP71A13, and CYP79B2 after Infection with P. syringae.
Supplementary Material
Acknowledgments
We thank Joanne Chory for cyp79B2, cyp79B3, and cyp79B2 cyp79B3 seed, Fred Ausubel for sid2-2, Beat Keller for dde2-2, and the ABRC for SALK T-DNA lines. ATH1 microarray data were provided by Raka Mitra. This project was initiated by J.G. and S.G. at the Torrey Mesa Research Institute with funding from Syngenta. Support to J.G. and E.G. was from Grant DE-FG02-05ER15670 from the Department of Energy Biosciences and from Grant GL346/1 from the Deutsche Forschungsgemeinschaft, respectively. M.N. thanks the Royal Veterinary and Agricultural University for a PhD stipend.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jane Glazebrook (jglazebr@umn.edu).
Online version contains Web-only data.
References
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 2148–2152. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bak, S., Kahn, R., Nielsen, H., Møller, B., and Halkier, B. (1998. b). Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol. Biol. 36 393–405. [DOI] [PubMed] [Google Scholar]
- Bak, S., Nielsen, H., and Halkier, B. (1998. a). The presence of CYP79 homologues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol. Biol. 38 725–734. [DOI] [PubMed] [Google Scholar]
- Bak, S., Tax, F.E., Feldmann, K.A., Galbraith, D.W., and Feyereisen, R. (2001). CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., Sandberg, G., and Bellini, C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 97 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, H.J., Arlotto, M.P., and Waterman, M.R. (1991). Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc. Natl. Acad. Sci. USA 88 5597–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartling, D., Seedorf, M., Mithofer, A., and Weiler, E.W. (1992). Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone, indole-3-acetic acid. Eur. J. Biochem. 205 417–424. [DOI] [PubMed] [Google Scholar]
- Bohman, S., Staal, J., Thomma, B.P., Wang, M., and Dixelius, C. (2004). Characterisation of an Arabidopsis-Leptosphaeria maculans pathosystem: Resistance partially requires camalexin biosynthesis and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J. 37 9–20. [DOI] [PubMed] [Google Scholar]
- Bones, A.M., and Rossiter, J.T. (2006). The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry 67 1053–1067. [DOI] [PubMed] [Google Scholar]
- Boucher, J.L., Delaforge, M., and Mansuy, D. (1994). Dehydration of alkyl- and arylaldoximes as a new cytochrome P450-catalyzed reaction: Mechanism and stereochemical characteristics. Biochemistry 33 7811–7818. [DOI] [PubMed] [Google Scholar]
- Browne, L.M., Conn, K.L., Ayer, W.A., and Tewari, J.P. (1991). The camalexins: New phytoalexins produced in the leaves of Camelina sativa (cruciferae). Tetrahedron 47 3909–3914. [Google Scholar]
- Delarue, M., Prinsen, E., Onckelen, H.V., Caboche, M., and Bellini, C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14 603–611. [DOI] [PubMed] [Google Scholar]
- DeMaster, E.G., Shirota, F.N., and Nagasawa, H.T. (1992). A Beckmann-type dehydration of n-butyraldoxime catalyzed by cytochrome P-450. J. Org. Chem. 57 5074–5075. [Google Scholar]
- Dörmann, P., Hoffmann-Benning, S., Balbo, I., and Benning, C. (1995). Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7 1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari, S., Plotnikova, J.M., De Lorenzo, G., and Ausubel, F.M. (2003). Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 35 193–205. [DOI] [PubMed] [Google Scholar]
- Fiehn, O., Kopka, J., Dormann, P., Altmann, T., Trethewey, R.N., and Willmitzer, L. (2000). Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18 1157–1161. [DOI] [PubMed] [Google Scholar]
- Glawischnig, E., Hansen, B.G., Olsen, C.E., and Halkier, B.A. (2004). Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., and Ausubel, F.M. (1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 91 8955–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Chen, W., Estes, B., Chang, H.S., Nawrath, C., Metraux, J.P., Zhu, T., and Katagiri, F. (2003). Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34 217–228. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Zook, M., Mert, F., Kagan, I., Rogers, E.E., Crute, I.R., Holub, E.B., Hammerschmidt, R., and Ausubel, F.M. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grsic, S., Sauerteig, S., Neuhaus, K., Albrecht, M., Rossiter, J.T., and Ludwig-Müller, J. (1998). Physiological analysis of transgenic Arabidopsis thaliana plants expressing one nitrilase isoform in the sense or antisense direction. J. Plant Physiol. 153 446–456. [Google Scholar]
- Halkier, B.A., Nielsen, H.L., Koch, B., and Møller, B.L. (1995). Purification and characterization of recombinant cytochrome P450TYR expressed at high levels in Escherichia coli. Arch. Biochem. Biophys. 322 369–377. [DOI] [PubMed] [Google Scholar]
- Hansen, B., and Halkier, B. (2005). New insight into the biosynthesis and regulation of indole compounds in Arabidopsis thaliana. Planta 221 603–606. [DOI] [PubMed] [Google Scholar]
- Hansen, C.H., Du, L., Naur, P., Olsen, C.E., Axelsen, K.B., Hick, A.J., Pickett, J.A., and Halkier, B.A. (2001). CYP83B1 Is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. J. Biol. Chem. 276 24790–24796. [DOI] [PubMed] [Google Scholar]
- Heck, S., Grau, T., Buchala, A., Metraux, J.P., and Nawrath, C. (2003). Genetic evidence that expression of NahG modifies defence pathways independent of salicylic acid biosynthesis in the Arabidopsis-Pseudomonas syringae pv. tomato interaction. Plant J. 36 342–352. [DOI] [PubMed] [Google Scholar]
- Hull, A.K., Vij, R., and Celenza, J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic, N., Normanly, J., and Cohen, J.D. (1996). Quantification of free plus conjugated indoleacetic acid in Arabidopsis requires correction for the nonenzymatic conversion of indolic nitriles. Plant Physiol. 111 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Jefcoate, C.R. (1978). Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol. 52 258–279. [DOI] [PubMed] [Google Scholar]
- Jenkins, C.M., and Waterman, M.R. (1994). Flavodoxin and NADPH-flavodoxin reductase from Escherichia coli support bovine cytochrome P450c17 hydroxylase activities. J. Biol. Chem. 269 27401–27408. [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, R.A., Fahrendorf, T., Halkier, B.A., and Møller, B.L. (1999). Substrate specificity of the cytochrome P450 enzymes CYP79A1 and CYP71E1 involved in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch. Biochem. Biophys. 363 9–18. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Rowe, H.C., and Denby, K.J. (2005). Secondary metabolites influence Arabidopsis/Botrytis interactions: Variation in host production and pathogen sensitivity. Plant J. 44 25–36. [DOI] [PubMed] [Google Scholar]
- Mathews, J., Black, S., and Burka, L. (1998). Disposition of butanal oxime in rat following oral, intravenous and dermal administration. Xenobiotica 28 767–777. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Hansen, C.H., Wittstock, U., and Halkier, B.A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275 33712–33717. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Petersen, B.L., Glawischnig, E., Jensen, A.B., Andreasson, E., and Halkier, B.A. (2003). Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 131 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944. [DOI] [PubMed] [Google Scholar]
- Naur, P., Petersen, B.L., Mikkelsen, M.D., Bak, S., Rasmussen, H., Olsen, C.E., and Halkier, B.A. (2003). CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 133 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., and Metraux, J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly, J., Grisafi, P., Fink, G.R., and Bartel, B. (1997). Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin, H.H., Hansen, B.G., Norholm, M.H., Jensen, J.K., and Halkier, B.A. (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34 e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura, T., and Sato, R. (1964). The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J. Biol. Chem. 239 2379–2385. [PubMed] [Google Scholar]
- Parisy, V., Poinssot, B., Owsianowski, L., Buchala, A., Glazebrook, J., and Mauch, F. (2007). Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 49 159–172. [DOI] [PubMed] [Google Scholar]
- Park, W.J., Kriechbaumer, V., Müller, A., Piotrowski, M., Meeley, R.B., Gierl, A., and Glawischnig, E. (2003). The nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid. Plant Physiol. 133 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedras, M.S., Nycholat, C.M., Montaut, S., Xu, Y., and Khan, A.Q. (2002). Chemical defenses of crucifers: Elicitation and metabolism of phytoalexins and indole-3-acetonitrile in brown mustard and turnip. Phytochemistry 59 611–625. [DOI] [PubMed] [Google Scholar]
- Pollmann, S., Müller, A., and Weiler, E.W. (2006). Many roads lead to “auxin”: Of nitrilases, synthases, and amidases. Plant Biol. (Stuttg.) 8 326–333. [DOI] [PubMed] [Google Scholar]
- Porchia, A.C., Sorensen, S.O., and Scheller, H.V. (2002). Arabinoxylan biosynthesis in wheat. Characterization of arabinosyltransferase activity in Golgi membranes. Plant Physiol. 130 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch, T., Helmlinger, J., and Hilgenberg, W. (1985). High-performance liquid-chromatographic separation and some properties of (E)-3-indoleacetaldoxime and (Z)-3-indoleacetaldoxime. J. Chromatogr. 318 95–102. [Google Scholar]
- Reuber, T.L., Plotnikova, J.M., Dewdney, J., Rogers, E.E., Wood, W., and Ausubel, F.M. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16 473–485. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Schenkman, J.B., Remmer, H., and Estabrook, R.W. (1967). Spectral studies of drug interaction with hepatic microsomal cytochrome. Mol. Pharmacol. 3 113–123. [PubMed] [Google Scholar]
- Schmidt, R., Müller, A., Hain, R., Bartling, D., and Weiler, E. (1996). Transgenic tobacco plants expressing the Arabidopsis thaliana nitrilase II enzyme. Plant J. 9 683–691. [DOI] [PubMed] [Google Scholar]
- Schuhegger, R., Nafisi, M., Mansourova, M., Petersen, B.L., Olsen, C.E., Svatos, A., Halkier, B.A., and Glawischnig, E. (2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, M.A., and Werck-Reichhart, D. (2003). Functional genomics of P450s. Annu. Rev. Plant Biol. 54 629–667. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P., Nelissen, I., Eggermont, K., and Broekaert, W.F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19 163–171. [DOI] [PubMed] [Google Scholar]
- van Wees, S.C., Chang, H.S., Zhu, T., and Glazebrook, J. (2003). Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 132 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- von Malek, B., van der Graaff, E., Schneitz, K., and Keller, B. (2002). The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216 187–192. [DOI] [PubMed] [Google Scholar]
- Vorwerk, S., Biernacki, S., Hillebrand, H., Janzik, I., Müller, A., Weiler, E.W., and Piotrowski, M. (2001). Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta 212 508–516. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Hull, A.K., Gupta, N.R., Goss, K.A., Alonso, J., Ecker, J.R., Normanly, J., Chory, J., and Celenza, J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., and Glazebrook, J. (1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., Tsui, F., Klessig, D.F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook, M., and Hammerschmidt, R. (1997). Origin of the thiazole ring of camalexin, a phytoalexin from Arabidopsis thaliana. Plant Physiol. 113 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.