Abstract
Bread wheat (Triticum aestivum) is a hexaploid species with A, B, and D ancestral genomes. Most bread wheat genes are present in the genome as triplicated homoeologous genes (homoeologs) derived from the ancestral species. Here, we report that both genetic and epigenetic alterations have occurred in the homoeologs of a wheat class E MADS box gene. Two class E genes are identified in wheat, wheat SEPALLATA (WSEP) and wheat LEAFY HULL STERILE1 (WLHS1), which are homologs of Os MADS45 and Os MADS1 in rice (Oryza sativa), respectively. The three wheat homoeologs of WSEP showed similar genomic structures and expression profiles. By contrast, the three homoeologs of WLHS1 showed genetic and epigenetic alterations. The A genome WLHS1 homoeolog (WLHS1-A) had a structural alteration that contained a large novel sequence in place of the K domain sequence. A yeast two-hybrid analysis and a transgenic experiment indicated that the WLHS1-A protein had no apparent function. The B and D genome homoeologs, WLHS1-B and WLHS1-D, respectively, had an intact MADS box gene structure, but WLHS1-B was predominantly silenced by cytosine methylation. Consequently, of the three WLHS1 homoeologs, only WLHS1-D functions in hexaploid wheat. This is a situation where three homoeologs are differentially regulated by genetic and epigenetic mechanisms.
INTRODUCTION
Flower development has been the subject of intensive study over the last decade, particularly in two dicot species, Arabidopsis thaliana and Antirrhinum majus (Jack, 2004). These studies have provided a general understanding of the development of floral organs in higher plants and led to the production of the ABCDE model. This model postulates that floral organ identity is defined by five classes of homeotic genes, named A, B, C, D, and E (Zahn et al., 2006). According to the ABCDE model, class A and E genes specify sepals in the first floral whorl, class A, B, and E genes specify petals in the second whorl, class B, C, and E genes specify stamens in the third whorl, class C and E genes specify carpels in the fourth whorl, and class D and E genes specify the ovule in the pistil. Cloning of ABCDE organ identity genes in Arabidopsis showed that they encode MADS box transcription factors, except for the class A gene APETALA2 (AP2). The class A MADS box gene is AP1, the class B genes are AP3 and PISTILLATA (PI), the class C gene is AGAMOUS (AG), and the class D gene is SEEDSTICK. In Arabidopsis, the class E genes consist of four members, SEPALLATA1 (SEP1), SEP2, SEP3, and SEP4, which show partially redundant functions in identity determination of petals, stamens, and carpels (Pelaz et al., 2000; Honma and Goto, 2001; Ditta et al., 2004). The diversification of the MADS box genes during evolution has contributed to the wide variation of flower shapes in land plants (Irish and Litt, 2005).
Analysis of the ABCDE genes in monocot species, such as rice (Oryza sativa), suggests that the ABCDE model could essentially be extended to monocots, except for the role of the class A genes (Kater et al., 2006; Yamaguchi and Hirano, 2006). Transgenic rice expressing antisense RNA of the class B gene Os MADS4 shows alteration of stamens into a carpel-like organ (Kang et al., 1998). The maize (Zea mays) class B gene–deficient mutant silky1 exhibits male sterility due to homeotic transformation of stamens into carpels (Ambrose et al., 2000). Furthermore, we showed in wheat (Triticum aestivum) that downregulation of the class B genes wheat PISTILLATA (WPI) and wheat APETALA3 (WAP3) induces pistillody, the homeotic transformation of stamens into carpel-like organs (Hama et al., 2004). These findings together with recent progress in understanding of the maize class B gene (Whipple et al., 2004, 2007) suggest that class B genes have a fundamentally conserved function in dicot and monocot species. Although a functional analysis has not been performed, expression analysis and protein–protein interaction analysis suggest that the rice class D gene Os MADS13 is involved in specifying ovule identity (Lopez-Dee et al., 1999; Favaro et al., 2002, 2003). In contrast with the class B and D genes, it has been reported that the duplicated class C genes in rice, Os MADS3 and Os MADS58, show only partial conservation of function with the Arabidopsis class C gene, AG (Yamaguchi et al., 2006). Mutant and transgenic analysis indicated that Os MADS58 regulates floral meristem determinacy and normal carpel morphogenesis and that Os MADS3 predominantly regulates stamen identity and prevents lodicule development. Interestingly, carpel identity is determined by a YABBY gene named DROOPING LEAF in rice (Nagasawa et al., 2003; Yamaguchi et al., 2004). It is not known if monocots have class A genes. Arabidopsis has two class A genes, AP1 and AP2. The AP1 MADS box gene functions in specification of floral meristem identity and in determination of sepal and petal identities. There are two other AP1-like genes, FRUITFULL (FUL) and CAULIFLOWER (CAL), which have redundancy of function in specification of floral meristem identity with AP1 (Ferrandiz et al., 2000). Sequence analysis of monocot AP1-like genes suggests that monocots have only FUL-like proteins, in contrast with dicot species, which have AP1, FUL, and CAL proteins (Litt and Irish, 2003). In wheat, it has been reported that the AP1-like gene WAP1 (sometimes called VRN1) (Murai et al., 1998) has no class A function but acts in phase transition from vegetative to reproductive growth (for diploid wheat, Yan et al., 2003; for hexaploid wheat, Danyluk et al., 2003; Murai et al., 2003; Trevaskis et al., 2003). Based on functional analysis using transgenic plants, the rice AP1-like genes Os MADS14 and Os MADS18 also play a role in the flowering pathway rather than specification of floral organs (Jeon et al., 2000; Fornara et al., 2004).
Recent studies indicated that the class E genes of rice belong to two clades, the SEP clade and the Os MADS1 clade (Malcomber and Kellogg, 2004; Agrawal et al., 2005). Os MADS24 and Os MADS45 show high sequence similarity to Arabidopsis SEP genes and exhibit similar expression and interaction properties as SEP proteins, indicating that they are rice orthologs of SEP genes (Favaro et al., 2002; Pelucchi et al., 2002; Malcomber and Kellogg, 2004; Prasad et al., 2005). However, mutation of Os MADS1 in rice causes the leafy hull sterile1 (lhs1) mutant phenotype that has leaf-like lemma and palea (Jeon et al., 2000). Furthermore, loss-of-function of Os MADS1 induces the homeotic transformation of lemma and palea into leaf-like structures (Prasad et al., 2005), indicating that Os MADS1 functions in lemma and palea differentiation. These facts suggest that the class E genes have diverged into two groups during rice evolution and that the mechanism of floral organ specification in rice could be complicated by the duplicated class E genes.
Wheat is a hexaploid species with the genome constitution AABBDD that originated from three diploid ancestral species: the A genome came from Triticum urartu, the B genome from Aegilops speltoides or another species classified in the Sitopsis section, and the D genome from Aegilops tauschii (Feldman, 2001; Feldman and Levy, 2005). Allopolyploidization leads to the generation of duplicated homoeologous genes (homoeologs), as opposed to paralogous genes (paralogs). Consequently, the hexaploid wheat genome contains triplicated homoeologs derived from the ancestral diploid species. In previous studies, we identified three homoeologs of the wheat AG-like MADS box gene wheat AG (WAG), situated on chromosomes 1A, 1B, and 1D (Meguro et al., 2003), and three homoeologs of the wheat AP1-like MADS box gene WAP1 on chromosomes 5A, 5B, and 5D (Murai et al., 2003). There are three possible evolutionary fates for homoeologous genes in polyploids: functional diversification, gene silencing, and retention of original or similar function (Wendel, 2000). Functional diversification of homoeologs is one of the important factors in the evolutionary success of polyploid species. Furthermore, the evolutionary success of allopolyploids is due to the retention of function of all homoeologs in many loci and to the gene silencing in other loci. The former facilitates positive intergenomic interactions that are maintained in a self-pollinating plants like wheat as permanent heterosis, and the latter prevents the intergenomic interactions with deleterious effect.
Here, we describe the identification of three homoeologs in the two class E–type genes wheat SEPALLATA (WSEP) and wheat LEAFY HULL STERILE1 (WLHS1) of wheat. Analyses of gene structure, expression patterns, and protein functions showed that no alterations were present in the WSEP homoeologs. By contrast, the three WLHS1 homoeologs showed genetic and epigenetic alterations. The A genome WLHS1 homoeolog (WLHS1-A) contained a large novel sequence in place of the K domain sequence. A yeast two-hybrid analysis and a transgenic experiment indicated that the WLHS1-A protein had no function. WLHS1-B and WLHS1-D, located in the B and D genomes, respectively, have a complete MADS box gene structure, but WLHS1-B was predominantly silenced by cytosine methylation. Consequently, of the three homoeologs, only WLHS1-D functions in hexaploid wheat.
RESULTS
Identification of Class E MADS Box Genes in Wheat
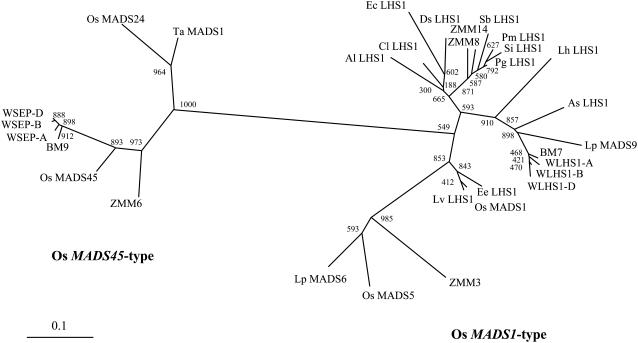
Wheat MADS box genes were isolated from a wheat EST database (Ogihara et al., 2003). By screening all the EST contigs through a BLASTN search, we identified 57 putative MADS box sequences. Among 57 MADS box genes, wheat homologs of the rice Os MADS45and Os MADS1 genes, named WSEP and WLHS1, respectively, were identified by their sequence similarity. The sequences of three homoeologs of WSEP and two homoeologs of WLHS1 were detected in the EST database. The third homoeolog of WLHS1 was cloned by RT-PCR using cDNA from young wheat spikes. The chromosomal locations of these clones were determined by homoeolog-specific PCR in combination with Chinese Spring (CS) nulli-tetrasomics lines and CS ditelosomic lines (Sears, 1966). The CS nulli-tetrasomics lines are defined as a series of lines missing a pair of chromosomes that are replaced by an extra pair of homoeologous chromosomes, and the CS ditelosomic lines are defined as a series of lines lacking pairs of the half arms of each chromosome. This mapping exercise showed that the three WSEP clones were located on chromosomes 7A, 7B, and 7D, and we named the genes WSEP-A, WSEP-B, and WSEP-D, respectively. The three WLHS1 homoeologs were found to be located on chromosomes 4A, 4B, and 4D, and we named the genes WLHS1-A, WLHS1-B, and WLHS1-D, respectively. Wheat chromosome 4 is syntenic to rice chromosome 3, on which the Os MADS1 gene is located, suggesting that the WLHS1 genes are putative orthologs of Os MADS1. Using the deduced amino acid sequences (see Supplemental Figure 1 online), a phylogenetic tree of class E genes of monocot species together with WSEP and WLHS1 was constructed (Figure 1). The tree indicated that the class E gene family in monocots was separated into two groups, Os MADS45-type and Os MADS1-type. The WSEP homoeologs belong to the Os MADS45-type cluster, and the WLHS1 homoeologs belong to the Os MADS1-type cluster. In the Os MADS45-type cluster, there are two rice genes, Os MADS24 and Os MADS45 (Greco et al., 1997). WSEP genes formed a subcluster with Os MADS45 that was distinct from Os MADS24. In the subcluster, barley (Hordeum vulgare) BM9 is the sister to WSEP. The expression of BM9 is localized to the primordia of lodicule, stamen, and carpel, suggesting that it is involved in floral organ identity (Schmitz et al., 2000). Another wheat SEP-like gene, Ta MADS1, belongs to the Os MADS24 subcluster. Ta MADS1 is expressed in wheat floret primordia, suggesting that it functions in floret development (Zhao et al., 2006). In the Os MADS1-type cluster, WLHS1 genes are closest to the barley BM7 gene. BM7 expression is restricted to the primordia of lemma, palea, lodicule, and ovary, suggesting that it functions in the formation of these organs (Schmitz et al., 2000). The maize MADS box genes ZMM8 and ZMM14, which belong to the Os MADS1 type, are hypothesized to act as selector genes that are involved in distinguishing the upper from the lower floret in the maize spikelet (Cacharron et al., 1999). Os MADS5 is another rice gene in the Os MADS1-type cluster. The loss-of-function mutation of Os MADS5 showed almost no effect on flower development, suggesting that it does not have class E function (Agrawal et al., 2005). However, the wheat homolog of Os MADS5 remains to be characterized.
Figure 1.
Phylogenetic Tree of Deduced Amino Acid Sequences of Class E MADS Box Genes of Wheat and Other Monocot Species.
WSEP-A, WSEP-B, and WSEP-D are homoeologs located on chromosomes 7A, 7B, and 7D. WLHS1-A, WLHS1-B, and WLHS1-D are homoeologs located on chromosomes 4A, 4B, and 4D. The phylogenetic tree was constructed by the neighbor-joining method using deduced amino acid sequences. The numbers at the nodes show bootstrap values after 1000 replicates.
The Spatial and Temporal Expression Patterns of WSEP and WLHS1 in Wheat Inflorescences
The wheat inflorescence (spike, ear, or head) develops at the tip of a stem and is composed of spikelets (Murai et al., 2002). The spikelet is composed of florets and encompassed by two small bract leaves called glumes. In each floret, the reproductive organs are enveloped by two leaf-like structures, a lemma and a palea. An individual wheat flower contains one pistil, three stamens, and two lodicules. Data from maize and rice suggest that the lodicule in monocots is a modified petal (Kang et al., 1998; Ambrose et al., 2000; Kyozuka et al., 2000).
In situ hybridization analyses were performed to determine the localizations of WSEP and WLHS1 transcripts during flower development in wheat (Figure 2). Antisense probes were synthesized from gene-specific regions at the 3′ regions of WSEP-D and WLHS1-D. These 3′ regions have high sequence similarities among the homoeologs. Thus, the hybridization signals should be a mixture of transcripts of all three homoeologs. Expression of WSEP was not detectable during the spikelet differentiation stage (Figure 2A); signals were initially detected in whorls 2, 3, and 4, at the stage just before initiation of lodicule/stamen/carpel formation (Figure 2B). The expression pattern of WSEP was quite similar to that of barley BM9 (Schmitz et al., 2000). Expression signals were detectable in all subsequent stages of floral organ maturation (Figure 2C) and were detected not only in the inner three whorls but also in the palea of the floret before the booting stage (Figure 2D). These observations suggest that WSEP expression is related to floral organ differentiation, as it is similar to that for typical class E genes, such as SEP3 in Arabidopsis (Pelaz et al., 2000).
Figure 2.
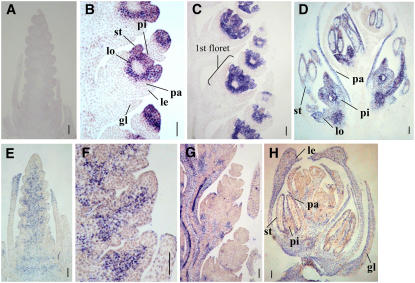
In Situ Hybridization Analysis of Class E MADS Box Genes in Wheat.
(A) In situ localization of WSEP transcripts in a young spike at the spikelet differentiation stage. Note that transcripts were not detected.
(B) to (D) In situ localization of WSEP transcripts in young spikelets: early floral organ differentiation stage (B), late floral organ differentiation stage (C), and floral organ developing stage (D).
(E) In situ localization of WLHS1 transcripts in a young spike at the spikelet differentiation stage.
(F) to (H) In situ localization of WSEP transcripts in young spikelets: early floral organ differentiation stage (F), late floral organ differentiation stage (G), and floral organ developing stage (H).
pi, pistil; st, stamen; lo, lodicule; pa, palea; le, lemma; gl, glume. Bars = 100 μm.
In contrast with WSEP, expression of WLHS1 was first detected in the inflorescence axis at the stage when the inflorescence meristem is initiated (Figure 2E). Expression signals were then detected in the spikelet axis at the stage of floral organ differentiation (Figure 2F). During the floral organ development stage, WLHS1 signals were detected at the most proximal position of the spikelet (Figure 2G); at later stages, the signals were observed in the glume, lemma, and palea until the floral organs were fully mature (Figure 2H). The expression pattern of WLHS1 was similar to that of rice Os MADS1 and that of barley BM7, whose transcripts are confined to the lemma, palea, lodicule, and carpel (Chung et al., 1994; Schmitz et al., 2000; Prasad et al., 2001).
Genomic Structures of the Homoeologs of WSEP and WLHS1
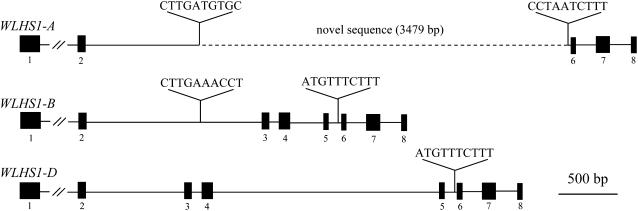
Comparison of the cDNA and genomic DNA sequences of the three WSEP homoeologs indicated that they had no insertions or deletions (indels) and that they had the typical MIKC-type MADS box gene structure. In contrast with WSEP, molecular size differences were found among the cDNAs of three WLHS1 homoeologs (data not shown). WLHS1-B and WLHS1-D cDNAs contained MIKC domains, but WLHS1-A cDNA lacked the K domain. Sequence analysis of WLHS1-A revealed that exons 3, 4, and 5 had been replaced by a novel sequence (Figure 3). A BLAST search against the DNA database of Japan (DDBJ) database found that this region showed no homology to any other region of the wheat genome.
Figure 3.
Genomic Structures of Three WLHS1 Homoeologs.
The WLHS1-A has an altered section between the second and fifth introns, which is possibly the result of a historical event. This region was substituted by a sequence of unknown origin that has no homology to any other DNA sequence determined. Numbered black boxes indicate exons, and lines indicate introns. The location of the unknown sequence is indicated by a broken line.
We sought to identify the origin of the novel sequence in WLHS1-A by studying the genetic diversity of WLHS1-A among Triticum species. A total of 90 lines of diploid, tetraploid, and hexaploid species and synthetic hexaploids were screened (see Supplemental Table 1 online). WLHS1-A containing the novel sequence (WLHS1-Anovel) was found in several lines of Triticum dicoccum (genome constitution AABB) and T. aestivum (AABBDD) and in all lines of Triticum macha (AABBDD) (Table 1). T. dicoccum is a primitive (hulled-grain type) domesticated tetraploid species, and T. macha is a hexaploid species that is endemic in Transcaucasia and closely related to bread wheat (T. aestivum). Interestingly, we did not find WLHS1-Anovel in any lines of Triticum dicoccoides and T. urartu. T dicoccoides is the wild progenitor of domesticated tetraploid wheat, and T. urartu is the A genome donor of the tetraploid wheat. Also, WLHS1-Anovel was not found in eight lines of synthetic hexaploids produced by crossing tetraploid wheat (AABB) and Ae. tauschii, the D genome donor of hexaploid wheat.
Table 1.
The Distribution of WLHS1-Anovel among Triticum Species
| Ploidy | Chromosome Number | Species | Genome Constitution | No. of Lines Examined | No. of Lines with WLHS1-Anovel | No. of Lines with WLHS1-Aintact |
|---|---|---|---|---|---|---|
| Diploid | 2n = 14 | T. urartu | AA | 15 | 0 | 15 |
| Tetraploid | 2n = 28 | T. dicoccoides | AABB | 6 | 0 | 6 |
| T. dicoccum | AABB | 14 | 7 | 7 | ||
| T. durum | AABB | 6 | 0 | 6 | ||
| T. turgidum | AABB | 1 | 0 | 1 | ||
| T. carthlicum | AABB | 1 | 0 | 1 | ||
| T. polonicum | AABB | 1 | 0 | 1 | ||
| T. timopheevi | AABB | 1 | 0 | 1 | ||
| Hexaploid | 2n = 42 | T. sphaerococcum | AABBDD | 1 | 0 | 1 |
| T. spelta | AABBDD | 1 | 0 | 1 | ||
| T. macha | AABBDD | 5 | 5 | 0 | ||
| T. compactum | AABBDD | 1 | 0 | 1 | ||
| T. aestivum | AABBDD | 29 | 4 | 25 | ||
| Synthetic hexaploid | 2n = 42 | AABBDD | 8 | 0 | 8 |
WLHS1-Anovel is the novel form of WLHS1-A containing the novel sequence instead of the K domain. WLHS1-Aintact is the intact form of WLHS1-A with the MIKC domain.
Interaction between WSEP or WLHS1 and Other MADS Box Proteins
Protein–protein interactions among MADS box proteins are central to the ABCDE model of flower formation (Kaufmann et al., 2005). Here, we used yeast two- or three-hybrid systems to investigate interactions between the WSEP or WLHS1 proteins and other wheat MADS box proteins. In wheat, there are two PI orthologs, WPI-1 and WPI-2 (Hama et al., 2004), and two AG orthologs, WAG-1 and WAG-2 (Meguro et al., 2003; our unpublished data). We found that all WSEP homoeologs showed similar patterns of protein–protein interaction (Table 2). The WSEP proteins interacted with WAP1 (putative class A), WAP3/WPI-2 (class B), WAG-1 and WAG-2 (class C), and all class E genes except for WLHS1-A. In comparison, WLHS1-B and WLHS1-D interacted with WAP3/WPI-2 (class B) and all class E genes except for WLHS1-A. The lack of interaction between WLHS1-A and other MADS box proteins may be due to the loss of the K domain in WLHS1-A; this domain is associated with protein–protein interactions (Davies et al., 1996).
Table 2.
Two- and Three-Hybrid Interactions of MADS Box Proteins of Wheat in the Yeast GAL4 System
| AD\BD | WAP1 | WAP3/WPI-1 | WAP3/WPI-2 | WAG-1 | WAG-2 | WLHS1-A | WLHS1-B | WLHS1-D | WSEP-A | WSEP-B | WSEP-D |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WSEP-A | + | − | + | + | + | − | + | + | ++ | ++ | ++ |
| WSEP-B | + | − | + | + | + | − | + | + | ++ | ++ | ++ |
| WSEP-D | + | − | + | + | + | − | + | + | ++ | ++ | ++ |
| WLHS1-A | − | − | − | − | − | − | − | − | − | − | − |
| WLHS1-B | − | − | + | − | − | − | + | + | ++ | ++ | ++ |
| WLHS1-D | − | − | + | − | − | − | + | + | ++ | ++ | ++ |
Protein interactions are assessed by the viability of yeast transformants on selective medium. Relative levels of protein interaction denoted by the following: ++, strong; +, moderate; −, not detectable. AD indicates a chimeric protein with the transcriptional activation domain, and BD indicates a chimeric protein with the DNA binding domain.
Homoeolog-Specific Expression Patterns of WSEP and WLHS1
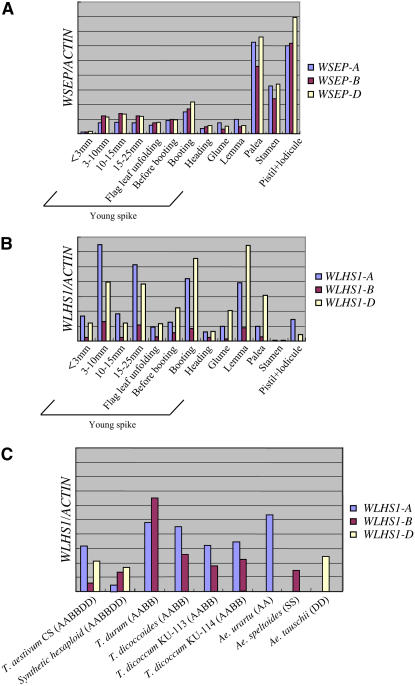
We used gene-specific real-time PCR to examine the expression profiles of the WSEP and WLHS1 homoeologs at different stages of inflorescence development and in different parts of the floral organ (Figures 4A and 4B). The specificity of the primers was confirmed using plasmids containing full-length cDNA sequences of the homoeologs, and the expression level of each gene was normalized against ACTIN gene expression and the amplification efficiency of each primer. The transcription levels of WSEP genes were low in young spikes (from 3 to 10 mm in length to booting), covering all stages of floral organ differentiation through to floral organ maturation (Figure 4A). WSEP transcripts were predominantly expressed in the stamen, the pistil with attached lodicule, and in the palea. There was no significant difference in the amounts of transcript of the three homoeologous genes, WSEP-A, WSEP-B, and WSEP-D, suggesting that all have similar function.
Figure 4.
Expression Analysis of WSEP and WLHS1 Homoeologous Genes by Real-Time PCR.
(A) and (B) Real-time PCR was performed using homoeolog-specific primers for WSEP-A, WSEP-B, and WSEP-D (A) and for WLHS1-A, WLHS1-B, and WLHS1-D (B). ACTIN was used as endogenous control. Total RNAs were isolated from spikes of CS plants at various developmental stages and from various floral organs at the booting stage.
(C) Transcript levels of the WLHS1 genes in young spikes (10 to 25 mm in length) of diploid, tetraploid, and hexaploid species and in immature spikes (booting stage) of a synthetic hexaploid. The genome constitution of each species is indicated in parenthesis. Note that the silencing of WLHS1-B was observed only in hexaploid CS wheat (T. aestivum). ACTIN was used as endogenous control.
By contrast, we observed significant differences in the expression levels of the three WLHS1 homoeologs (Figure 4B). WLHS1-A and -D were highly expressed in spikes and floral organs, except for the stamen, but the transcript level of WLHS1-B was clearly lower than its counterparts. As shown in Figure 3, WLHS1-A contains an insertion of a novel sequence that replaced exons 3, 4, and 5. Thus, the sequence change in the WLHS1-A gene did not cause a significant change in expression levels. To examine gene-specific silencing of WLHS1-B homoeologs in diploid and tetraploid species, we performed expression analysis of WLHS1 genes in Triticum durum, T. dicoccoides, T. dicoccum (all AABB), T. urartu (AA), Ae. speltoides (SS, possibly modified BB), Ae. tauschii (DD), and newly synthesized synthetic hexaploid (F2 line crossed between T. durum and Ae. tauschii) (Figure 4C). Silencing of WLHS1-B was not observed in tetraploid species that have a B genome or in Ae. speltoides that belongs to the same Sitopsis section as the putative B genome donor of tetraploid and hexaploid wheat. T. dicoccum strain KU-113 contains WLHS1-A with the novel sequence (WLHS1-Anovel), and T. dicoccum strain KU-114 does not. Thus, the sequence change in WLHS1-A did not affect the silencing of WLHS1-B in tetraploid wheat. Furthermore, we found that WLHS1-B was expressed in the synthetic hexaploid, suggesting that the silencing of WLSH1-B does not occur soon after the formation of the hexaploid.
Methylation Analysis of WLHS1 Homoeologs
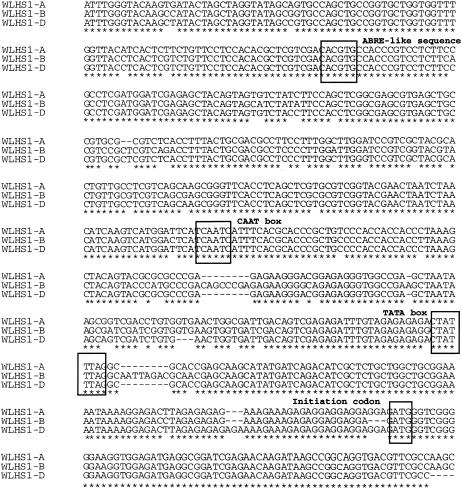
To investigate the mechanisms of gene-specific silencing of the WLHS1-B homoeolog, we first isolated the 5′ regions of the three WLHS1 homoeologs. The 5′ regions showed high sequence similarities with each other, and no specific point mutations nor specific indels of known cis-elements were detected (Figure 5). Sequence analyses indicated that the WLHS1 genes had a >50% GC content and contained the minimal criteria for CpG islands in the 5′ region, including exon 1. The 700-bp region upstream of the ATG initiation codon of WLHS1-A, WLHS1-B, and WLHS1-D were cloned, and transient promoter activities were tested. We found that the promoter region of each of the three homoeologous genes had transcriptional activity in immature wheat inflorescences (Figure 6A).
Figure 5.
Alignment of the 5′ Upstream Region of Three WLHS1 Homoeologs.
Initiation codon and putative cis-elements (ABRE-like sequence, CAAT box, and TATA box) are shown in the boxes.
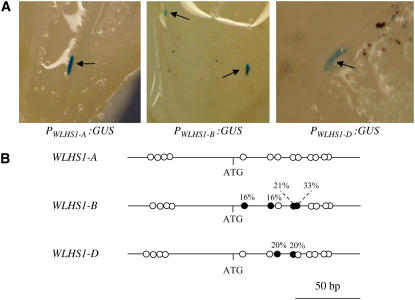
Figure 6.
Transient Promoter Activity Assay of WLHS1 Promoters and Distributions of Methylated Cytosine in the 5′ Region of WLHS1.
(A) Transient expression of GUS in immature inflorescences of wheat. The upstream region of WLHS1-A, WLHS1-B, or WLHS1-D was ligated to the reporter gene GUS and introduced into immature inflorescences by particle bombardment. Arrows indicate the GUS spot stained by X-gluc solution.
(B) The methylation status of CpG/CpNpG sites in the 5′ regions of WLHS1 homoeologous genes. Comparison of the distribution of methylated cytosines in the 5′ regions shows that the WLHS1-B promoter is hypermethylated compared with WLHS1-A or WLHS1-D. Genomic DNA was treated by sodium bisulfite and used for PCR with primers that amplify the first exon and promoter regions of the three WLHS1 homoeologous genes. The sequences were determined in >10 clones of each gene, and the ratios of methylated cytosines are indicated by percentages. Closed circles, methylated; open circles, unmethylated.
The methylation status of WLHS1 was examined using bisulfite PCR analysis of CpG/CpNpG sites of exon 1 and the 5′ upstream region (Figure 6B). Genomic DNA was isolated from leaves of bread wheat plants that were in the process of floral organ differentiation and treated with sodium bisulfite. The bisulfite-treated DNA was subjected to PCR, and the clones were randomly sequenced. We then determined the ratio of methylated to unmethylated sites, and the data were transformed into percentages. The 5′ region of WLHS1-B was highly methylated (Figure 6B), which may be the reason for the specific silencing of this gene.
Ectopic Expression Analysis of WSEP and WLHS1 Using an Arabidopsis Transgenic System
To gain further insight into the functional divergence of SEP-like genes in wheat, constructs containing WSEP-A, WSEP-B, WSEP-D, WLHS1-A, WLHS1-B, or WLHS1-D cDNA, driven by the cauliflower mosaic virus 35S promoter (P35S), were transformed into Arabidopsis. In comparison with control P35S:β-glucuronidase (GUS) plants, P35S:WSEP-A plants showed earlier flowering with four to five small curled leaves (Figure 7A). No alterations in floral organs were present in the terminal flower of the transformants. Arabidopsis plants transformed with either P35S:WSEP-B or P35S:WSEP-D constructs exhibited similar phenotypes to that of P35S:WSEP-A (data not shown).
Figure 7.
Phenotype Analysis of Transgenic Arabidopsis Plants.
(A) A plant overexpressing GUS was used as the control. The typical phenotype of a P35S:WSEP-A transgenic plant. The transgenic plant produced a terminal flower and showed extremely early flowering. Bar = 1 cm.
(B) The typical phenotypes of plants carrying the transgene P35S:WLHS1-A, P35S:WLHS1-B, or P35S:WLHS1-D. No morphological changes were observed in the P35S:WLHS1-A plant, but the P35S:WLHS1-B and P35S:WLHS1-D plants showed early flowering and late production of a terminal flower. Morphological changes were not present in the floral organs of all transgenic plants. Bar = 1 cm.
Arabidopsis plants transformed with P35S:WLHS1 exhibited a dramatically different phenotype from those of WSEP-transformed plants. A total of 30 independent P35S:WLHS1-A transformants were produced; none of these showed any indication of a change in morphology or in flowering time compared with control P35S:GUS plants (Figure 7B). However, the P35S:WLHS1-B and P35S:WLHS1-D transformants showed early flowering and late production of terminal flowers (Figure 7B). Transformants with ectopic expression of WLHS1 genes were fertile and showed no morphological changes in any organ.
WLHS1 Protein Accumulation in Wheat Inflorescences
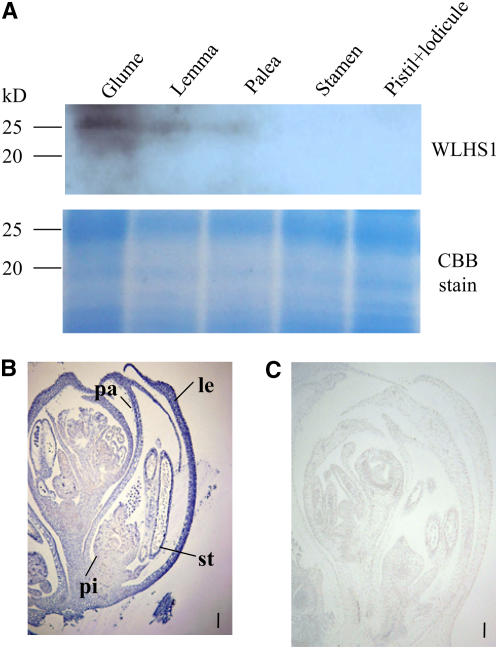
The accumulation of WLHS1-A, WLHS1-B, or WLHS1-D proteins in wheat inflorescences was examined by protein gel blot analysis using a WLHS1-specific antibody against a synthetic oligopeptide, HPEHDTSMQIGYPQ, which corresponds to the C terminus of WLHS1. We could not detect an 18.7-kD band, the predicted size of the product of the WLHS1-A gene in floral organs (Figure 8A). The products of WLHS1-B and WLHS1-D are 27.2 and 26.1 kD, respectively. We found a signal of ∼26 kD in the glume, palea, and lemma (Figure 8A), indicating that WLHS1-D protein accumulated in these organs. No signal was found for WLHS1-B, which is silenced at the transcription level.
Figure 8.
Protein Gel Blotting Analysis and Distribution of WLHS1 Proteins in Wheat Inflorescences.
(A) Protein gel blotting of individual parts of floral organs at the prebooting stage. Total proteins were extracted from each tissue and separated by SDS-PAGE. The proteins were stained either with Coomassie blue or, after membrane transfer, with antibodies raised against the C-terminal peptide of WLHS1. The antibody produced a band at ∼26 kD, corresponding to the WLHS1-B and/or WLHS1-D protein. No bands were seen at 19 kD, which corresponds to the expected size of the WLHS1-A protein.
(B) Immunolocalization of WLHS1. Strong staining is seen in the palea and lemma. Bar = 100 μm. pi, pistil; st, stamen; pa, palea; le, lemma.
(C) Control section. No staining was seen using serum from a preimmunized rabbit.
Bar = 100 μm.
To investigate the distribution of WLHS1-D protein, we performed immunolocalization studies on sections of immature wheat spikelets. During late flower development, we found heavy staining in the lemma and palea (Figure 8B), which is in good agreement with the results of the protein gel blot analysis. As a negative control, we performed the same procedure on similar tissues using a serum from a preimmunized rabbit and found no signals (Figure 8C).
DISCUSSION
Functional Diversification of Two Class E Genes, WSEP and WLHS1
Arabidopsis has four class E MADS box genes for floral organ identity, SEP1, SEP2, SEP3, and SEP4, which exhibit functional redundancy. These SEP genes can form heterochimeric protein complexes with the products of the class A, B, C, and D genes that regulate identity acquirement of sepals, petals, stamens, carpels, and ovules (Honma and Goto, 2001; Pelaz et al., 2001; Favaro et al., 2003). In rice, five class E MADS box genes have been identified: Os MADS1, Os MADS5, Os MADS24 (identical to Os MADS8), Os MADS34 (identical to Os MADS19), and Os MADS45 (identical to Os MADS7) (Kater et al., 2006; Yamaguchi and Hirano, 2006). These rice class E genes can be separated into two groups: one consists of Os MADS1, Os MADS5, and Os MADS34, and the other consists of Os MADS24 and Os MADS45. Two of the genes in the latter group, Os MADS24 and Os MADS45, show high sequence similarities to SEP genes and also display similar expression and interaction profiles as SEP proteins, indicating that they are orthologs of the Arabidopsis SEP genes (Favaro et al., 2002; Pelucchi et al., 2002; Malcomber and Kellogg, 2004; Prasad et al., 2005). Phylogenetic analysis showed that the WSEP homoeologous genes clustered in the same group as Os MADS24 and Os MADS45 (Figure 1). In situ hybridization experiments clearly showed that WSEP was expressed in the inner three whorls (lodicules, stamens, and pistils) at the floral organ differentiation stage (Figures 2B and 2C). Interestingly, after floral organ identities had been determined, strong expression of WSEP was observed in the palea (Figure 2D). The high expression level of WSEP in the palea was also confirmed by real-time PCR (Figure 4A). The expression patterns suggest that WSEP genes are not only involved in floral organ differentiation but also in their subsequent development.
Protein–protein interactions between class E and class A, B, C, or D proteins have been observed in a number of plant species (Egea-Cortines et al., 1999; Honma and Goto, 2001; Favaro et al., 2002, 2003; Ferrario et al., 2003). Yeast two- and three-hybrid experiments showed that WSEP formed a complex with wheat class B and C genes (Table 2). Furthermore, overexpression of WSEP in Arabidopsis caused early flowering and terminal flower formation (Figure 7A). These characteristics are similar to phenotypes caused by ectopic expression of Arabidopsis SEP3, or its counterparts, in petunia (Petunia hybrida) and lily (Lilium longiflorum; Pelaz et al., 2001; Ferrario et al., 2003; Tzeng et al., 2003). In addition to WSEP, Ta MADS1 has been identified and characterized as a wheat class E gene (Zhao et al., 2006). Our phylogenetic study revealed that WSEP is an ortholog of rice Os MADS45 and that Ta MADS1 corresponds to Os MADS24, suggesting that SEP orthologs have diverged into two groups in monocot species. Transgenic Arabidopsis plants overexpressing Ta MADS1 showed early flowering and terminal flower formation. Although protein–protein interactions between Ta MADS1 and wheat class B or C genes have not yet been examined, WSEP and Ta MADS1 may share a similar function.
Recent studies revealed that Os MADS1, the gene affected in the lhs1 mutation, has an E function in floral organ specification in rice (Jeon et al., 2000; Lim et al., 2000; Prasad et al., 2001, 2005; Malcomber and Kellogg, 2004; Agrawal et al., 2005). Based on phylogenetic studies, WLHS1 is a wheat ortholog of Os MADS1 (Figure 1). However, the expression pattern of WLHS1 differed slightly from that of Os MADS1. In rice inflorescences, Os MADS1 expression occurs at the stage after formation of the panicle branches; expression occurs at high levels in the lemma and palea, at a low level in the carpel, and is not detectable in the glume (Jeon et al., 2000; Prasad et al., 2001, 2005). By contrast, transcripts of WLHS1 accumulated at high levels in the inflorescence stem, spikelet stem, glume, lemma, and palea, and at a low level in the pistil (Figures 2E to 2H and 4B). It has been reported that Os MADS1 expression in inflorescences varies among cereals, such as Chasmanthium latifolium, Pennisetum glaucum, and Sorghum bicolor (Malcomber and Kellogg, 2004). The differences in expression patterns of Os MADS1–like genes between rice and wheat may be associated with differences in the structures of their respective inflorescences.
The Effect of Structural Alteration of WLHS1-A on Expression and Function
In this study, we found two WLHSI transcripts of different sizes in wheat inflorescences (data not shown). Sequence analysis revealed that the shorter transcript corresponded to WLHS1-A, which encoded an in-frame 170–amino acid protein containing an internal deletion covering the K domain. In rice, a previous study showed that the K domain is essential for protein–protein interactions between Os MADS1 and other MADS box proteins (Lim et al., 2000). Yeast two-hybrid and three-hybrid analyses revealed that WLHS1-B and WLHS1-D interacted with class B genes and that they could form a homodimer (Table 2). By contrast, WLHS1-A did not interact with any wheat MADS box gene. Furthermore, overexpression of WLHS1-A did not induce any morphological changes in transgenic Arabidopsis plants, whereas transgenic plants that overexpressed WLHS1-B or WLHS1-D showed early flowering (Figure 7B). These observations suggest that the WLHS1-A protein, which lacks a K domain, has lost the normal MADS box function. Furthermore, the WLHS1-A protein was not detected in the protein gel blot experiment using a specific antibody (Figure 8), indicating that posttranscriptional silencing occurred in immature wheat floral organs.
It has been reported that polyploidization induces genetic and epigenetic modifications in the genomes of higher plants (reviewed in Comai, 2000; Chen and Ni, 2006). Elimination of noncoding and low-copy DNA sequences has been described in synthetic allopolyploids of Triticum and Aegilops species (Feldman et al., 1997; Liu et al., 1998). Sequence elimination was found to start earlier in the synthetic allopolyploids, generally during the first allopolyploid generation (Ozkan et al., 2001; Shaked et al., 2001). Furthermore, sequence elimination was a nonrandom and reproducible event whose direction was determined by the identities of the genomes present in the allopolyploid, suggesting that elimination in synthetic allopolyploids resembles the process in naturally occurring allopolyploids (Ozkan et al., 2001). Contrary to the expectation of simple sequence elimination, WLHS1-A is an instance where polyploidization has resulted in a gene-specific alteration of a homoeolog, through replacement of an intragenic region to a novel sequence (Figure 3).
Evolutionary Origin of the Variant WLHS1-A
To obtain further insight of the origin of the sequence change, we compared the WLHS1-A locus in diploid, tetraploid, and hexaploid species of Triticum. We detected the variant WLHS1-A (WLHS1-Anovel) in T. dicoccum (AABB), T. macha (AABBDD), and T. aestivum (AABBDD) (Table 1). Interestingly, they showed the same alteration (data not shown), suggesting that a variant arose once during allopolyploid evolution in wheat species. Among the lines of tetraploid species examined, only T. dicoccum carried WLHS1-Anovel. Furthermore, only some of the hexaploid wheat lines had WLHS1-Anovel. These findings indicate that the sequence change in WLHS1-A occurred in a lineage of T. dicoccum (domesticated tetraploid) and that hexaploid species originated on multiple occasions from crosses of the domesticated tetraploid species and the D genome donor Ae. tauschii. Multiple origin of hexaploid wheat has also been suggested by sequence comparisons of low-copy DNA (Talbert et al., 1998) and the high molecular weight glutenin gene Glu-l (Gu et al., 2006). Therefore, it seems a reasonable hypothesis. However, we cannot exclude the following hypothesis as yet. An alternative explanation for the origin of WLHS1-Anovel is that it arose in a lineage of hexaploid T. aestivum and that T. macha originated from the T. aestivum lineage carrying this sequence change. Then, the WLHS1-Anovel locus may have passed into T. dicoccum by introgression from T. aestivum. It could occur because spontaneous DNA introgression from T. aestivum into wild tetraploid species has been known (Weissmann et al., 2005). A third possible explanation is that WLHS1-A has a hot spot in its intron region and that a replacement event has occurred independently in T. dicoccum, T. macha, and T. aestivum. However, no transposon-related sequences have been detected in the WLHS1-A locus (data not shown).
Gene-Specific Silencing of the WLHS1-B Homoeolog Is Caused by Epigenetic Regulation
Real-time PCR analysis using gene-specific primers showed that the expression levels of the three WSEP homoeologs were almost identical (Figure 4A). By contrast, real-time PCR analysis for WLHS1 indicated that expression of WLHS1-B was downregulated compared with its homoeologs (Figure 4B). The transient promoter assay demonstrated that the promoter regions of WLHS1-B possessed transcriptional activation activities (Figure 6A), indicating that alteration of the cis-element was not the cause of WLHS1-B silencing. Using a bisulfite genome sequencing analysis, we examined the methylation levels of the 5′ CpG and CpNpG islands of the WLHS1 homoeologs. This analysis indicated that gene-specific hypermethylation was present in exon 1 of WLHS1-B, which seems to be associated with silencing of this gene (Figure 6B). Other studies have shown that epigenetic alterations accompany polyploidization and can occasionally lead to gene silencing: for example, in Arabidopsis (Chen et al., 1998; Lee and Chen, 2001; Wang et al., 2004), Triticum (Shaked et al., 2001; Kashkush et al., 2002; He et al., 2003), and Brassica (Lukens et al., 2006). This study found that silencing of WLHS1-B did not occur in tetraploid species having the B genome nor in diploid Ae. speltoides with the S genome, which is possibly a modified B genome (Figure 4C). This suggests that the epigenetic downregulation of WLHS1-B occurred at the origin of the hexaploid genome, in which the AB genome was combined with the D genome. However, a synthetic hexaploid wheat that is the F2 generation of the cross between T. durum and Ae. tauschii contains nonsilenced WLHS1-B (Figure 4C). This suggests that the silencing of WLSH1-B does not occur soon after the formation of the hexaploid. The molecular mechanism of the effect of interaction between homoeologous genes on epigenetic regulation during allopolyploid formation is an interesting subject for future study.
The Three WLHS1 Homoeologs Show a Differential Contribution to Flower Development in Hexaploid Wheat
In this study, we showed that the three homoeologous genes for WLHS1 have genetic and epigenetic alterations. WLHS1-A has a structural alteration and contains a large novel sequence instead of the K domain (Figure 3), and WLHS1-B was silenced by cytosine methylation (Figures 4B and 6B). Consequently, of the three WLSH1 homoeologous genes present in hexaploid wheat, only WLHS1-D is functional. Differential contribution of three homoeologous genes has been reported for several hexaploid wheat genes. In Ta Hd1, a gene that controls the photoperiodic flowering pathway, the homoeolog on the B genome is silenced by a deletion within the promoter region (Nemoto et al., 2003). In Ha, a locus that influences grain hardness, a large genomic deletion has occurred independently in the A and B genomes, and only the Ha locus genes present on the D genome (Pina, Pinb, and Gsp-1) are functional (Chantret et al., 2005). In Ta Bx, a gene responsible for biosynthesis of benzoxazinones (Bx), expression analysis and the determination of the proteins' catalytic properties revealed that a homoeolog from the B genome is generally the largest contributor to Bx biosynthesis (Nomura et al., 2005). These studies indicated that expression of homoeologous genes is regulated by genetic or epigenetic mechanisms. Here, we have provided a description of a situation where three homoeologous genes are differentially regulated by genetic and epigenetic mechanisms.
Feldman and Levy (2005) have proposed a classification system for genome evolution induced by allopolyploidization: the first is rapid genome change (revolutionary change) through generation of genetic and epigenetic alterations in a short period; and the second is slow genome change (evolutionary change) during polyploid speciation by allopolyploidization. Revolutionary changes comprise (1) nonrandom elimination of coding and noncoding DNA, (2) epigenetic changes of coding and noncoding DNA, and (3) activation of genes and retroelements induced by alteration of adjacent DNA sequences. Evolutionary changes comprise (1) intergenomic transfer of DNA segments between the constituent genomes, (2) production of recombinant genomes through hybridization or introgression to different polyploid or diploid species, and (3) mutation. In this study, we showed that WLHS1-A has a structural alteration that includes a large novel sequence instead of the K domain (Figure 3). The variant WLHS1-A was not always detected in tetraploid and hexaploid species having the A genome (Table 1), indicating that this alteration is a type of evolutionary change. Furthermore, epigenetic alteration in WLHS1-B seems to be a type of evolutionary change because WLHS1-B is not silenced in a newly synthesized synthetic hexaploid (Figure 4C). WLHS1 genes could be useful models to investigate the triggers of revolutionary or evolutionary changes in homoeologs and to determine which of three homoeologs is functional.
Is epigenetic silencing of a homoeologous gene stable in different organs and at different growth stages? Expression of 40 homoeologous gene pairs was assayed in natural and tetraploid cotton (Gossypium) (Adams et al., 2003). It was found that silencing of some gene pairs is reciprocal and developmentally regulated, with one homoeolog showing silencing in some organs and the other silencing in other organs. Additionally, Adams et al. (2004) showed that allopolyploidization in cotton induces an immediate (in the first generation of a newly created allotetraploid) and widespread effect on the expression of duplicate homoeologs, which varies between different organs. These findings suggest that in allopolyploids there is rapid and differential specialization of homoeologs in the various organs. In wheat, expression profiles of 90 genes were estimated using the frequencies of ESTs (Mochida et al., 2003). The expression patterns of homoeologs were classified into two major groups: (1) genes almost equally expressed from all three genomes and (2) genes expressed with a significant preference, which varied between tissues. Using the same method for estimating expression patterns of genes for the seed storage proteins gliadin and glutenin, Kawaura et al. (2005) reported that the homoeologs of the D genome are preferentially expressed during the seed maturation process, but those from the A genome are strongly expressed at later stages. Moreover, using the single-strand conformation polymorphism technique with 70 single-copy loci, it was found that there is a modest bias toward silencing of the homoeolog on the D genome in the leaf but not in the root (Bottley et al., 2006). In this study, we observed silencing of the WLHS1-B gene in all reproductive organs tested: young spike, glume, lemma, palea, stamen, pistil, and lodicule (Figure 4B). However, the expression level of WLHS1-A was higher than that of WLHS1-D in young spikes, but WLHS1-D was preferentially expressed in the glume, lemma, and palea. Interestingly, WLHS1-A does not produce a functional protein (Table 2, Figures 7B and 8), suggesting that organ/tissue-specific expression of homoeologous genes is not associated with gene products.
METHODS
Plant Materials
The common wheat (Triticum aestivum) cv CS was used in the cDNA cloning, RT-PCR, in situ hybridization, and immunolocalization analyses. To examine phylogenetic origin of the variant WLHS1-A, 90 lines of diploid, tetraploid, and hexaploid species and of synthetic hexaploids were used. The plants were grown in soil-filled pots in a greenhouse. Arabidopsis thaliana ecotype Columbia was used for the experiments on ectopic expression of WLHS1-A, WLHS1-B, WLHS1-D, WSEP-A, WSEP-B, and WSEP-D. The Arabidopsis plants were cultivated in growth chambers at 22°C under 12-h-light/12-h-dark conditions.
Cloning of Wheat Class E MADS Box Genes
The full-length cDNAs of WSEP and WLHS1 homoeologous genes were obtained from the wheat EST database (Ogihara et al., 2003) and by PCR-based cloning. Clone numbers used here were as follows: WSEP-A, whoh6p21; WSEP-B, whms14i23; WSEP-D, whh21o05; WLHS1-A, wh5b06; WLHS1-D, wh17d02. Full-length cDNAs were amplified by PCR with the following primer pairs: WSEP full-L (5′-ATGGGGAGGGGGAGGGTGGAG-3′) and WSEP full-R (5′-TCAAGGCAACCACGGGGGCAT-3′); WLHS1 full-L (5′-GACATCGCTCTGCTGGCTG-3′) and WLHS1 full-R, (5′-CAACAATAGCGACCTCAGCACAC-3′). PCR products were subcloned and sequenced. The upstream regions of the WLHS1 homoeologous genes were cloned using the Universal GenomeWalker kit (BD Biosciences Clontech) according to the manufacturer's instructions using the following primers: WLHS1-proGW-1R (5′-CTCCACCTTCCCCCGACCCATCTC-3′) and WLHS1-proGW-2R (5′-CCTTTTATTTTCCGCAGCCAGC-3′).
Phylogenetic Analysis
Multiple amino acid sequence alignment was performed using the computer program ClustalW (Thompson et al., 1994) with matrix blosum (gap open penalty, 10; gap extension penalty, 0.2; gap distance, 8), and a phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987). Support values for nodes on the tree were estimated with 1000 bootstrap replicates (Felsenstein, 1985). Programs used here were provided by the DDBJ (http://www.ddbj.nig.ac.jp/search/clustalw-e.html).
Characterization of the Variant WLHS1-A
Genomic PCRs were conducted to search for the origin of the altered sequence in WLHS1-A in 90 lines of diploid, tetraploid, and hexaploid Triticum species, including synthetic hexaploid. Primers were designed using the DNA sequences of intron 2, the novel region of WLHS1-A, and exon 6. These primers amplify sequences that span the 3′ and 5′ ends of the novel DNA that has replaced the K domain in WLHSI-A; the primer pair for the 3′ sequence was WLHS1Aint 2L (5′-CTGTTGATGGGCATAAGG-3′) and WLHS1Ains 1R (5′-TGTTTTCATCCTGCAATTCT-3′) and that for the 5′ sequence was WLHS1Ains 1L (5′-GCAAGACCCCACACATTAG-3′) and WLHS1Aexon 6R (5′-GCAGAACATCATTTTAGGCAG-3′). The resulting PCR fragments were sequenced, and recombination points were determined.
In Situ Hybridization Analysis
In situ hybridization was performed using the method described previously (Shitsukawa et al., 2006). Spikes at different developmental stages (from prespikelet initiation to preheading) were sampled from CS plants and fixed with FAA solution (3.7% paraformaldehyde, 5% acetic acid, and 50% ethanol) at 4°C overnight. The fixed tissues were dehydrated and embedded in Paraplast Plus (Oxford Labware). The tissues were cut into 20-μm sections and dried overnight. Hybridization was performed overnight at 55°C. DIG-labeled RNA probes were synthesized by T3 or T7 RNA polymerase in vitro transcription using a DIG RNA labeling kit (Roche Diagnostics). After hybridization, the sections were washed twice with 0.5× SSC at 52°C. Immunological detection of the hybridized probe was performed as described by Hama et al. (2004). As controls for specificity, consecutive sections were hybridized with sense and antisense probes of the same region of the WSEP or WLHS1 genes.
Binary Constructs and Arabidopsis Transformation
To examine ectopic expression of WSEP and WLHS1 homoeologous genes, cDNA fragments containing the coding region of the genes were cloned downstream of the cauliflower mosaic virus 35S promoter of pIG121 vectors. Binary vectors were introduced into Agrobacterium tumefaciens GV3101. Arabidopsis plants were transformed using the floral dip method described by Clough and Bent (1998).
Transient Promoter Activity Assay Using a Microbombardment
The 5′ upstream regions of WLHS1-A, -B, and -D were amplified with the linker-added primer sets WLHS1pro-XbaI L (5′-CCTCTAGACTTCTTGAGCAGCCCATTCC-3′) and WLHS1pro-HindIII R (5′-CCAAGCTTCGCAGAAGCATTTGGGTACA-3′). The PCR products were digested with XbaI and HindIII and inserted into pIG121 to produce GUS proteins controlled by each WLHS1 promoter. These GUS constructs were introduced into young spikes of CS plants by particle bombardment using the conditions described by Takumi et al. (1994). After introducing these plasmids, immature spikes were incubated on hormone-free MS medium at 22°C for 1 d. GUS expression was assayed using a staining solution containing 1.9 mM 5-bromo-4 chloro-3 indoryl-β-d-glucuronic acid (X-gluc) and 0.5 mM potassium ferrocyanide. The transgenic young spikes were incubated with the staining solution at 37°C for 24 h and decolorized by 70% ethanol.
Quantification of Transcripts by Real-Time PCR
Total RNA was isolated from spikes at various developmental stages (<3 mm, 3 to 10 mm, 10 to 15 mm, and 15 to 25 mm in length; flag leaf unfolding, before booting, booting, and heading stages) from CS plants using ISOGEN (Nippon-gene). Total RNA was also isolated from separate parts of the floral organs (glume, lemma, palea, stamen, and pistil) of CS plants at the booting stage. First-strand cDNA synthesis was performed using 5 μg of DNase-digested total RNA, derived from the above organs, with oligo(dT) primer according to the protocol for RT-PCR first-strand synthesis (GE Healthcare Bio-Sciences). We tested the amplification efficiency of each primer set using four twofold gene-specific plasmid dilutions (5 ng/μL to 625 pg/μL) and compared their amplification efficiency relative to primers for CS wheat ACTIN (actin361-L, 5′-TATGCCAGCGGTCGAACAAC-3′; actin361-R, 5′-GGAACAGCACCTCAGGGCAC-3′). Based on single nucleotide polymorphisms in the cDNA sequence, homoeologous gene-specific primers for quantitative PCR were designed as follows: WSEP-A-specific L (5′-TTGCTTGGTGAAGATCTTGATTCC-3′) and WSEP-A-specific R (5′-GTGAAGTGTGGGTTCACCAGCT-3′); WSEP-B-specific L (5′-GGAACAAATGTTTTCGGAGGCA-3′) and WSEP-B-specific R (5′-TGCATGAGTTACTCAGGGACTCG-3′); WSEP-D-specific L (5′-CTGGGAGCACAACAACAATGTACTGG-3′) and WSEP-D-specific R (5′-AGCAGCATCAAGGGGGTGGAAA-3′); WLHS1-A-specific L (5′-GCACCTCCGCTAGAAAATGAAGA-3′) and WLHS1-A-specific R (5′-TATCATGCTCCGGGTGTTGG-3′); WLHS1-B-specific L (5′-AGGAAGCAACACCTCCGCTAGAAAG-3′) and WLHS1-B-specific R (5′-ACCACGCAGCTTAGCACACACA-3′); WLHS1-D-specific L (5′-TCAAGCATATCAGGTCAAAAAAGAATCAA-3′) and WLHS1-D-specific R (5′-GCTGTCAAACTTTTGGGCCTTCT-3′). Quantitative PCR experiments were performed using a LightCycler 2.0 (Roche Diagnostics), and in all cases quantity was determined by SYBR GREEN fluorescence and CS wheat ACTIN as endogenous controls.
Yeast Two- and Three-Hybrid Assays
In the yeast two-hybrid assay, the Gal4 Two-hybrid Phagemid Vector kit (Stratagene) was used to investigate protein–protein interactions among wheat MADS box genes. The vector pBD-Gal4 was used to clone the entire open reading frame sequences of WAP1, WAG-1, WAG-2, WSEP-A, WSEP-B, WSEP-D, WLHS1-A, WLHS1-B, and WLHS1-D into EcoRI and SalI sites. The entire open reading frame sequences of WSEP and WLHS1 orthologs were cloned into EcoRI and SalI sites of pAD-GAL4-2.1. Ternary complex formation was studied with a pBridge vector (BD Biosciences Clontech), which expresses a DNA binding domain fusion and an additional protein. The complete WAP3 coding sequence was cloned into MCS1 to generate a hybrid protein that contains the sequences for the GAL4 DNA binding domain; WPI-1 or WPI-2 was cloned into MCS2 to generate as third protein. All constructs were sequenced and then transformed into the yeast strain YRG2, which has His3 and LacZ reporter genes, using the Saccharomyces cerevisiae direct transformation kit (Wako). Double transformants were grown on selective medium and tested by histidine prototrophy.
DNA Isolation and Bisulfite Sequencing
Genomic DNA (1 μg) was isolated from leaves of CS plants at the floret differentiation stage and treated with sodium bisulfite for use as the PCR template. Briefly, a 50-μL solution containing the DNA was denatured with 0.2 M NaOH and then treated with hydroquinone and sodium bisulfite at 52°C for 18 h. Bisulfite-treated DNA was purified using the QIAquick gel extraction kit (Qiagen). Modification was completed by treatment with 0.3 M NaOH for 5 min at room temperature. The bisulfite-treated DNA was precipitated with ammonium acetate and ethanol, and the pellets were washed once with 70% ethanol and dissolved in 20 μL of water. PCR analysis was performed at 48°C using the primer set WLHS-bis #3L (5′-GAAAATAAAAGGAGATTTAGAGA-3′) and WLHS-bis #4R (5′-CCTTCTTAAACAACCCATTCC-3′); this primer set is common to all three homoeologous genes. PCR products were cloned into the pCR4 Blunt-TOPO vector using the Zero Blunt TOPO PCR cloning kit for sequencing (Invitrogen) and sequenced.
Protein Extraction and Protein Gel Blot Analysis
Proteins were extracted from individual parts of florets before the booting stage using the P-PER plant protein extraction kit (Pierce) in accordance with the manufacturer's instructions. An anti-WLHS1 polyclonal antibody was raised against the peptide HPEHDTSMQIGYPQ. This peptide sequence was completely identical to those of WLHS1-A and WLHS1-D and differed by one residue from that of WLHS1-B. The specificity of this antibody was confirmed using recombinant antigens that express each WLHS1 protein fused with a 220–amino acid Tag protein from the pET-41a-c(+) vector (Novagen). The antibody was purified on an affinity column, and its antigen specificity was checked by protein gel blotting using the WLHS1-A, WLHS1-B, and WLHS1-D recombinant proteins. Proteins were resolved on 12.5% polyacrylamide gels and transferred to polyvinylidene fluoride membranes by semidry blotting (Bio-Rad Laboratories) or stained with Coomassie Brilliant Blue to confirm protein quantity and assess protein integrity. For protein gel blots, polyvinylidene fluoride membranes were blocked with 5% (w/v) nonfat dry milk in Tris-buffered saline (TBS; 20 mM Tris and 137 mM NaCl, pH 7.7) with 0.1% (v/v) Tween-20. Affinity-purified anti-WLHS1 antibody was used at 1/300 to 1/500 dilution in TBS-Tween/5% milk solution and incubated for 1 h at room temperature. Blots were washed with TBS-Tween 6 times for 10 min each and then incubated with horseradish peroxidase–conjugated goat anti-rabbit antibody (GE Healthcare Bio-Sciences) at 1/250,0000 dilution and then washed as above. Bound antibodies were detected with ECL Plus (GE Healthcare Bio-Sciences) and exposure to XAR x-ray film (Fuji Photo Film).
Immunolocalization Analysis
Spikelets from prebooting stage plants were fixed and embedded, and sections were cut as described above for the in situ hybridization technique. Tissues sections were deparaffinized with xylene and hydrated through an ethanol series. WLHS1 antigens were activated by autoclaving tissues in citrate buffer (0.5 M, pH 6.0) at 121°C for 60 min. The sections were blocked by 1.5% BSA and incubated in primary antibody against WLHS1 diluted in Can Get Signal Solution A (TOYOBO) and rinsed and soaked in goat anti-rabbit F′ab fragments conjugated to alkaline phosphatase. After the slides were washed, signals were detected with NBT/BCIP (Roche Diagnostics). As controls for specificity, consecutive sections were incubated with serum from a preimmunized rabbit.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL/DDBJ data libraries under the following accession numbers: WSEP-A, AB295659; WSEP-B, AB295660; WSEP-D, AB295661; WLHS1-A, AB295662; WLHS1-B, AB295663; WLHS1-D, AB295664. The accession numbers of the other genes in the phylogenetic tree (Figure 1) are as follows; BM7 (AJ249145), BM9 (AJ249147), Lp MADS9 (AY198334), As LHS1 (AY597512), ZMM6 (AJ430692), Os MADS24 (U78892), Os MADS45 (U78891), Ta MADS1 (AF543316), Al LHS1 (AY597511), Cl LHS1 (AY597513), Ds LHS1 (AY597514), ZMM14 (AJ005338), ZMM8 (Y09303), Os MADS1 (L34271), Os MADS5 (AAB71434), WAP1 (AB007504), Ee LHS1 (AY597515), Ec LHS1 (AY597516), Lv LHS1 (AY597517), Lh LHS1 (AY597518), Si LHS1 (AY597521), Lp MADS6 (AY198331), Pm LHS1 (AY597519), Pg LHS1 (AY597520), Sb LHS1 (AY597522), and ZMM3 (Y09301). The accession number of CS wheat ACTIN is AB181991.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Deduced Amino Acid Sequences of Class E MADS Box Genes in Monocots.
Supplemental Table 1. Lines Used for Screening of the Variant WLHS1-A.
Supplementary Material
Acknowledgments
We thank Hiro-Yuki Hirano, Shoji Ohta, and Yoshihiro Matsuoka for valuable suggestions and Yoshito Ishida for the gift of two-hybrid vector for WPI. We also thank the National Bioresource Project–Wheat (NBRP-KOMUGI) for providing wheat materials and the EST database. This work was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology specific area research GROUP 2: Comparative Genomics B03: Comparative genomics of model organisms and their close relatives, and by the Fukui Prefectural Government (to K.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Koji Murai (murai@fpu.ac.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adams, K.L., Cronn, R., Percifield, R., and Wendel, J.F. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K.L., Percifield, R., and Wendel, J.F. (2004). Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal, G.K., Abe, K., Yamazaki, M., Miyao, A., and Hirochika, H. (2005). Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol. Biol. 59 125–135. [DOI] [PubMed] [Google Scholar]
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5 569–579. [DOI] [PubMed] [Google Scholar]
- Bottley, A., Xia, G.M., and Koebner, R.M. (2006). Homoeologous gene silencing in hexaploid wheat. Plant J. 47 897–906. [DOI] [PubMed] [Google Scholar]
- Cacharron, J., Saedler, H., and Theissen, G. (1999). Expression of MADS box genes ZMM8 and ZMM14 during inflorescence development of Zea mays discriminates between the upper and the lower floret of each spikelet. Dev. Genes Evol. 209 411–420. [DOI] [PubMed] [Google Scholar]
- Chantret, N., et al. (2005). Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.J., Comai, L., and Pikaard, C.S. (1998). Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc. Natl. Acad. Sci. USA 95 14891–14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.J., and Ni, Z. (2006). Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. Bioessays 28 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, Y.Y., Kim, S.R., Finkel, D., Yanofsky, M.F., and An, G. (1994). Early flowering and reduced apical dominance result from ectopic expression of a rice MADS box gene. Plant Mol. Biol. 26 657–665. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Comai, L. (2000). Genetic and epigenetic interactions in allopolyploid plants. Plant Mol. Biol. 43 387–399. [DOI] [PubMed] [Google Scholar]
- Danyluk, J., Kane, N.A., Breton, G., Limin, A.E., Fowler, D.B., and Sarhan, F. (2003). TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 132 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, B., Egea-Cortines, M., de Andrade Silva, E., Saedler, H., and Sommer, H. (1996). Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 15 4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., and Yanofsky, M.F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14 1935–1940. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro, R., Immink, R.G., Ferioli, V., Bernasconi, B., Byzova, M., Angenent, G.C., Kater, M., and Colombo, L. (2002). Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol. Genet. Genomics 268 152–159. [DOI] [PubMed] [Google Scholar]
- Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., Yanofsky, M.F., Kater, M.M., and Colombo, L. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15 2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M. (2001). The origin of cultivated wheat. In The Wheat Book, A. Bonjean and W. Angus, eds (Paris: Lavoisier), pp. 1–56.
- Feldman, M., and Levy, A.A. (2005). Allopolyploidy – A shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109 250–258. [DOI] [PubMed] [Google Scholar]
- Feldman, M., Liu, B., Segal, G., Abbo, S., Levy, A.A., and Vega, J.M. (1997). Rapid elimination of low-copy DNA sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 147 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution Int. J. Org. Evolution 39 783–791. [DOI] [PubMed] [Google Scholar]
- Ferrandiz, C., Gu, Q., Martienssen, R., and Yanofsky, M.F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1, and CAULIFLOWER. Development 127 725–734. [DOI] [PubMed] [Google Scholar]
- Ferrario, S., Immink, R.G., Shchennikova, A., Busscher-Lange, J., and Angenent, G.C. (2003). The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15 914–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara, F., Parenicova, L., Falasca, G., Pelucchi, N., Masiero, S., Ciannamea, S., Lopez-Dee, Z., Altamura, M.M., Colombo, L., and Kater, M.M. (2004). Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol. 135 2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco, R., Stagi, L., Colombo, L., Angenent, G.C., Sari-Gorla, M., and Pe, M.E. (1997). MADS box genes expressed in developing inflorescences of rice and sorghum. Mol. Gen. Genet. 253 615–623. [DOI] [PubMed] [Google Scholar]
- Gu, Y.G., et al. (2006). Types and rates of sequence evolution at the high-molecular-weight glutenin locus in hexaploid wheat and its ancestral genomes. Genetics 174 1493–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama, E., Takumi, S., Ogihara, Y., and Murai, K. (2004). Pistillody is caused by alterations to the class-B MADS-box gene expression pattern in alloplasmic wheats. Planta 218 712–720. [DOI] [PubMed] [Google Scholar]
- He, P., Friebe, B.R., Gill, B.S., and Zhou, J.M. (2003). Allopolyploidy alters gene expression in the highly stable hexaploid wheat. Plant Mol. Biol. 52 401–414. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529. [DOI] [PubMed] [Google Scholar]
- Irish, V.F., and Litt, A. (2005). Flower development and evolution: Gene duplication, diversification and redeployment. Curr. Opin. Genet. Dev. 15 454–460. [DOI] [PubMed] [Google Scholar]
- Jack, T. (2004). Molecular and genetic mechanisms of floral control. Plant Cell 16(Suppl): S1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, J.S., Jang, S., Lee, S., Nam, J., Kim, C., Lee, S.H., Chung, Y.Y., Kim, S.R., Lee, Y.H., Cho, Y.G., and An, G. (2000). leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H.-G., Jeon, J.-S., Lee, S., and An, G. (1998). Identification of class B and class C floral organ identity genes from rice plants. Plant Mol. Biol. 38 1021–1029. [DOI] [PubMed] [Google Scholar]
- Kashkush, K., Feldman, M., and Levy, A.A. (2002). Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater, M.M., Dreni, L., and Colombo, L. (2006). Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 57 3433–3444. [DOI] [PubMed] [Google Scholar]
- Kaufmann, K., Melzer, R., and Theissen, G. (2005). MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 347 183–198. [DOI] [PubMed] [Google Scholar]
- Kawaura, K., Mochida, K., and Ogihara, Y. (2005). Expression profile of two storage-protein gene families in hexaploid wheat revealed by large-scale analysis of expressed sequence tags. Plant Physiol. 139 1870–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka, J., Kobayashi, T., Morita, M., and Shimamoto, K. (2000). Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol. 41 710–718. [DOI] [PubMed] [Google Scholar]
- Lee, H.-S., and Chen, Z.J. (2001). Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc. Natl. Acad. Sci. USA 98 6753–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J., Moon, Y.H., An, G., and Jang, S.K. (2000). Two rice MADS domain proteins interact with OsMADS1. Plant Mol. Biol. 44 513–527. [DOI] [PubMed] [Google Scholar]
- Litt, A., and Irish, V.F. (2003). Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: Implications for the evolution of floral development. Genetics 165 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Vega, J.M., Segal, G., Abbo, S., Rodova, M., and Feldman, M. (1998). Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. I. Changes in low-copy non-coding DNA sequences. Genome 41 272–277. [DOI] [PubMed] [Google Scholar]
- Lopez-Dee, Z.P., Wittich, P., Pé, M.E., Rigola, D., Buono, I.D., Gorla, M.S., Kater, M.M., and Colombo, L. (1999). OsMADS13, a novel rice MADS box gene expressed during ovule development. Dev. Genet. 25 237–244. [DOI] [PubMed] [Google Scholar]
- Lukens, L.N., Pires, J.C., Leon, E., Vogelzang, R., Oslach, L., and Osborn, T. (2006). Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 140 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber, S.T., and Kellogg, E.A. (2004). Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16 1692–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meguro, A., Takumi, S., Ogihara, Y., and Murai, K. (2003). WAG, a wheat AGAMOUS homolog, is associated with development of pistil-like stamens in alloplasmic wheats. Sex. Plant Reprod. 15 221–230. [Google Scholar]
- Mochida, K., Yamazaki, Y., and Ogihara, Y. (2003). Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags. Mol. Genet. Genomics 270 371–377. [DOI] [PubMed] [Google Scholar]
- Murai, K., Miyamae, M., Kato, H., Takumi, S., and Ogihara, Y. (2003). WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiol. 44 1255–1265. [DOI] [PubMed] [Google Scholar]
- Murai, K., Murai, R., Takumi, S., and Ogihara, Y. (1998). Cloning and characterization of cDNAs corresponding to the wheat MADS box genes. In Proceedings of the 9th International Wheat Genetics Symposium, A.E. Slinkard, ed (Saskatchewan, Canada: University Extension Press), pp. 89–94.
- Murai, K., Takumi, S., Koga, H., and Ogihara, Y. (2002). Pistillody, homeotic transformation of stamens into pistil-like structures, caused by nuclear-cytoplasm interaction in wheat. Plant J. 29 169–181. [DOI] [PubMed] [Google Scholar]
- Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H.-Y., Sakai, H., and Nagato, Y. (2003). SUPERWOMAN 1 and DROOPING LEAF gene control floral organ identity in rice. Development 130 705–718. [DOI] [PubMed] [Google Scholar]
- Nemoto, Y., Kisaka, M., Fuse, T., Yano, M., and Ogihara, Y. (2003). Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J. 36 82–93. [DOI] [PubMed] [Google Scholar]
- Nomura, T., Ishihara, A., Yanagita, R.C., Endo, T.R., and Iwamura, H. (2005). Three genomes differentially contribute to the biosynthesis of benzoxazinones in hexaploid wheat. Proc. Natl. Acad. Sci. USA 102 16490–16495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara, Y., Mochida, K., Nemoto, Y., Murai, K., Yamazaki, Y., Shin-I, T., and Kohara, Y. (2003). Correlated clustering and virtual display of gene expression patterns in the wheat life cycle by large-scale statistical analyses of expressed sequence tags. Plant J. 33 1001–1011. [DOI] [PubMed] [Google Scholar]
- Ozkan, H., Levy, A.A., and Feldman, M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Tapia-Lopez, R., Alvarez-Buylla, E.R., and Yanofsky, M.F. (2001). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11 182–184. [DOI] [PubMed] [Google Scholar]
- Pelucchi, N., Fornara, F., Facalli, C., Masiero, S., Lago, C., Pe, M.E., Colombo, L., and Kater, M.M. (2002). Comparative analysis of rice MADS-box genes expressed during flower development. Sex. Plant Reprod. 15 113–122. [Google Scholar]
- Prasad, K., Parameswaran, S., and Vijayraghavan, U. (2005). OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 43 915–928. [DOI] [PubMed] [Google Scholar]
- Prasad, K., Sriram, P., Kumar, C.S., Kushalappa, K., and Vijayraghavan, U. (2001). Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev. Genes Evol. 211 281–290. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Schmitz, J., Franzen, R., Ngyuen, T.H., Garcia-Maroto, F., Pozzi, C., Salamini, F., and Rohde, W. (2000). Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol. Biol. 42 899–913. [DOI] [PubMed] [Google Scholar]
- Sears, E.R. (1966). Nullisomic-tetrasomic combinations in hexaploid wheat. In Chromosome Manipulations and Plant Genetics, R. Riley and K.R. Lewis, eds (Edinburgh, UK: Oliver and Boyd), pp. 29–45.
- Shaked, H., Kashkush, K., Ozkan, H., Feldman, M., and Levy, A.A. (2001). Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitsukawa, N., Takagishi, A., Ikari, C., Takumi, S., and Murai, K. (2006). WFL, a wheat FLORICAULA/LEAFY ortholog, is associated with spikelet formation as lateral branch of the inflorescence meristem. Genes Genet. Syst. 81 13–20. [DOI] [PubMed] [Google Scholar]
- Takumi, S., Otani, M., and Shimada, T. (1994). Effect of six promoter-intron combinations on transient reporter gene expression in einkorn, emmer and common wheat cells by particle bombardment. Plant Sci. 103 161–166. [Google Scholar]
- Talbert, L.E., Smith, L.Y., and Blake, N.K. (1998). More than one origin of hexaploid wheat is indicated by sequence comparison of low-copy DNA. Genome 41 402–407. [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis, B., Bagnall, D.J., Ellis, M.H., Peacock, W.J., and Dennis, E.S. (2003). MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. USA 100 13099–13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng, T.Y., Hsiao, C.C., Chi, P.J., and Yang, C.H. (2003). Two lily SEPALLATA-like genes cause different effects on floral formation and floral transition in Arabidopsis. Plant Physiol. 133 1091–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Tian, L., Madlung, A., Lee, H.-S., Chen, M., Lee, J.J., Watson, B., Kagochi, T., Comai, L., and Chen, Z.J. (2004). Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167 1961–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann, S., Feldman, M., and Gressel, J. (2005). Sequence evidence for sporadic intergeneric DNA introgression from wheat into a wild Aegilops species. Mol. Biol. Evol. 22 2055–2062. [DOI] [PubMed] [Google Scholar]
- Wendel, J.F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42 225–249. [PubMed] [Google Scholar]
- Whipple, C.J., Ciceri, P., Padilla, C.M., Ambrose, B.A., Bandong, S.L., and Schmidt, R.J. (2004). Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131 6083–6091. [DOI] [PubMed] [Google Scholar]
- Whipple, C.J., Zanis, M.J., Kellogg, E.A., and Schmidt, R.J. (2007). Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc. Natl. Acad. Sci. USA 104 1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T., and Hirano, H.-Y. (2006). Function and diversification of MADS-box genes in rice. ScientificWorldJournal 6 1923–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T., Lee, D.Y., Miyao, A., Hirochika, H., An, G., and Hirano, H.-Y. (2006). Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T., Nagasawa, N., Kawasaki, S., Matsuoka, M., Nagato, Y., and Hirano, H.-Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn, L.N., Feng, B., and Ma, H. (2006). Beyond the ABC-model: Regulation of floral homeotic genes. In Developmental Genetics of the Flower: Advances in Botanical Research, Vol. 44, D.E. Soltis, J.H. Leebensmack, and P.S. Soltis, eds (Amsterdam: Academic Press), pp. 163–207.
- Zhao, X.Y., Cheng, Z.J., and Zhang, X.S. (2006). Overexpression of TaMADS1, a SEPALLATA-like gene in wheat, causes early flowering and the abnormal development of floral organs in Arabidopsis. Planta 223 698–707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.