Abstract
Endogenous small RNAs, including microRNAs (miRNAs) and short interfering RNAs (siRNAs), are critical components of plant gene regulation. Some abundant miRNAs involved in developmental control are conserved between anciently diverged plants, while many other less-abundant miRNAs appear to have recently emerged in the Arabidopsis thaliana lineage. Using large-scale sequencing of small RNAs, we extended the known diversity of miRNAs in basal plants to include 88 confidently annotated miRNA families in the moss Physcomitrella patens and 44 in the lycopod Selaginella moellendorffii. Cleavage of 29 targets directed by 14 distinct P. patens miRNA families and a trans-acting siRNA (ta-siRNA) was experimentally confirmed. Despite a core set of 12 miRNA families also expressed in angiosperms, weakly expressed and apparently lineage-specific miRNAs accounted for the majority of miRNA diversity in both species. Nevertheless, the molecular functions of several of these lineage-specific small RNAs matched those of angiosperms, despite dissimilarities in the small RNA sequences themselves, including small RNAs that mediated negative feedback regulation of the miRNA pathway and miR390-dependent ta-siRNAs that guided the cleavage of AUXIN RESPONSE FACTOR mRNAs. Diverse, lineage-specific, small RNAs can therefore perform common biological functions in plants.
INTRODUCTION
Most eukaryotic organisms are capable of producing small RNAs, typically between 21 and 24 nucleotides in length, that serve as sequence-specific posttranscriptional regulators. Many short interfering RNAs (siRNAs) are initially produced as short duplexes containing two-nucleotide, 3′ overhangs via the enzymatic activity of endonucleases of the Dicer family acting upon long double-stranded RNA (dsRNA) (Baulcombe, 2005). One of the two siRNA strands is stably incorporated into an ARGONAUTE (AGO) protein that then functions to repress the expression of target RNAs, which are selected via base-pairing interactions with the siRNA (Hammond, 2005). This repression can take several forms, including AGO-catalyzed target cleavage (Liu et al., 2004; Meister et al., 2004) and translational repression (Doench et al., 2003). Some endogenous siRNAs also direct DNA and histone modifications to homologous genomic loci (Zilberman et al., 2003). Such siRNAs are especially numerous and diverse in flowering plants (Lu et al., 2005; Rajagopalan et al., 2006; Kasschau et al., 2007).
MicroRNAs (miRNAs) are a distinct type of small RNA that also function as negative regulators of gene expression. Primary miRNA transcripts (pri-miRNAs) adopt stem-loop RNA secondary structures from which a specific ∼21-nucleotide duplex is excised by a Dicer endonuclease (Bartel, 2004). Similar to siRNA duplexes, the resulting miRNA/miRNA* duplex possesses characteristic two-nucleotide 3′ overhangs and is unwound coincident with the association of the mature, single-stranded miRNA with an AGO protein. The mature miRNA then serves to guide the bound AGO protein to target mRNAs based upon complementarity. In both plants and animals, the regulation of target gene expression via miRNAs is critical for numerous biological processes (Wienholds and Plasterk, 2005; Jones-Rhoades et al., 2006; Mallory and Vaucheret, 2006). In plants, miRNA-directed target cleavage can sometimes stimulate the production of dsRNA using the cleaved product as a template; the resulting dsRNA is subsequently processed by a Dicer protein to produce siRNAs referred to as trans-acting siRNAs (ta-siRNAs) because of their miRNA-like ability to repress mRNAs from loci distinct from the originating locus (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005; Axtell et al., 2006).
Although the miRNAs characterized in angiosperms have numerous distinct biological functions, there is a striking tendency for many of them to regulate developmental processes (Jones-Rhoades et al., 2006). Previous studies have shown that some miRNA–target interactions first identified in Arabidopsis thaliana are deeply conserved among land plants (Floyd and Bowman, 2004; Arazi et al., 2005; Axtell and Bartel, 2005). Interestingly, the ancient miRNA regulatory interactions are disproportionately those thought to be involved in development, suggesting that these units of posttranscriptional gene control have been indispensable during the diversification of land plants.
The sequencing of a moss (Physcomitrella patens) genome and a lycopod (Selaginella moellendorffii) genome provides an opportunity to explore the diversity of small RNA functions in nonseed plants and to more rigorously address the trajectories of small RNA evolution in three widely separated plant lineages. To date, >40 distinct miRNA families have been annotated from P. patens based upon analysis of sequenced small RNAs or bioinformatic predictions (Arazi et al., 2005; Talmor-Neiman et al., 2006a; Fattash et al., 2007). In lycopods, the expression of miR166 has been confirmed in S. moellendorffii (Floyd and Bowman, 2004), and the expression of two additional families proposed based upon microarray experiments using RNA from Selaginella uncinata (Axtell and Bartel, 2005).
In this study, we applied high-throughput small RNA sequencing to the discovery and analysis of miRNAs from the moss P. patens and the lycopod S. moellendorffii. Our results directly demonstrated the expression of a much large number of distinct miRNA families in P. patens and S. moellendorffii then has previously been reported. Only a few of these miRNAs were expressed in common between mosses, lycopods, and angiosperms; these core conserved plant miRNAs were abundantly expressed in both angiosperms and basal pants, and all, save one, appeared to be involved on the regulation of developmental processes. The majority of miRNA diversity in both lycopods and mosses appeared lineage specific and generally expressed at lower levels than were the core conserved miRNAs. The experimental isolation of 29 cleavage products directed by 14 distinct P. patens miRNAs and a ta-siRNA demonstrated that the core conserved plant miRNAs have generally retained homologous target interactions during the diversification of land plants. Surprisingly, experimentally validated targets also included several instances in which lineage-specific P. patens miRNAs and ta-siRNAs shared common functions with sequence-distinct Arabidopsis small RNAs.
RESULTS
Identification of miRNAs from P. patens and S. moellendorffii
Direct sequencing of small RNAs and computational predictions have resulted in the annotation of 44 distinct P. patens miRNA families (Arazi et al., 2005; Talmor-Neiman et al., 2006a; Fattash et al., 2007). We have recently reported a data set of 127,135 unique P. patens small RNAs from three libraries, represented by a total of 561,102 reads, which provided insights into siRNA biogenesis in plants (Axtell et al., 2006). Because of the >100-fold increase in sequencing depth over previous P. patens small RNA sequencing efforts, we reasoned that additional less-abundant miRNAs might be found by analysis of these data. In addition, a single small RNA library prepared from the above-ground tissues of S. moellendorffii was sequenced to yield 36,582 unique small RNAs, represented by 149,586 reads that matched the genome (whole-genome shotgun traces).
To produce a list of possible miRNA loci, consensus genomic sequences surrounding each small RNA were derived by assembly of the relevant whole-genome shotgun (WGS) traces, followed by RNA secondary structure prediction. As a preliminary screen, the minimum free energy structure for each locus was analyzed using MIRcheck (Jones-Rhoades and Bartel, 2004), which evaluates paired regions of a predicted RNA secondary structure for characteristic features of miRNA stem loops. The candidate miRNA loci that passed MIRcheck were then analyzed for evidence of miRNA* accumulation. A miRNA/miRNA* duplex, with two-nucleotide 3′ overhangs indicative of DICER-LIKE1 (DCL1) cleavage, would be expected if a moss DCL1 ortholog excised the miRNA from the hairpin precursor as indicated by a previous study (Talmor-Neiman et al., 2006a). Indeed, the sequencing of both a miRNA and a miRNA* constitutes evidence of DCL1-like processing from a hairpin precursor, which is evidence that can supplement, or even substitute for, other criteria for confidently distinguishing miRNAs from endogenous siRNAs and other RNAs that should not be annotated as miRNAs (Rajagopalan et al., 2006; Ruby et al., 2006; Fahlgren et al., 2007).
In total, 88 distinct P. patens miRNA families, arising from 205 miRNA genes, were recovered by this analysis (Tables 1 and 3; see Supplemental Table 1 online). This set included all previously recognized P. patens miRNA families except for 13 of the 22 families uniquely reported by Fattash et al. (2007). Analysis of the deep sequencing data from P. patens also suggested refinements to a small number of previous miRNA annotations; these suggestions included reversals of miRNA/miRNA* designations, changes in the annotated positions of the dominant mature miRNA within stem-loop precursors, and consolidations of redundantly reported families (see Supplemental Table 1 online). Our resulting roster of confidently identified P. patens miRNAs (see Supplemental Table 1 online) included 12 families conserved with angiosperms and/or lycopods (Table 3). An additional six families of P. patens miRNAs annotated on the basis of computational comparisons to angiosperm miRNAs (miR167, miR172, miR395, miR414, miR418, and miR419; Fattash et al., 2007) were not observed in our sequence libraries. The expression of the remaining 76 sequence-distinct families was, to the best of our current knowledge, bryophyte specific. These included 57 novel P. patens miRNA families that have not previously been reported (Table 1).
Table 1.
Novel P. patens miRNAs
| Normalized Abundance (TPQ)a
|
|||||||
|---|---|---|---|---|---|---|---|
| miRNA | Sequence (5′–3′)b | Loci | Length | P | PG | GS | Armc |
| ppt-miR1023(5′) | aCACUCUCUCCaUUUCuCUaC | 4 | 21 | 513 (1696) | 4341 (12373) | 392 (929) | Amb |
| ppt-miR1023(3′) | AGaGAAUuGgAgAGAGUGcAu | 21 | 983 (1696) | 6666 (12373) | 328 (929) | Amb | |
| ppt-miR1024 | UCUGGUUGGAUUGUAGGCCUC | 2 | 21 | 1102 (1258) | 1236 (1744) | 841 (1157) | 3′ |
| ppt-miR1025 | UGCCACAACAAAGCUAAUAAC | 1 | 21 | 8486 (9486) | 8690 (10624) | 9370 (10954) | 3′ |
| ppt-miR1026 | UGAGAAAGACUUGAGAGGACA | 2 | 21 | 480 (534) | 174 (187) | 42 (44) | 5′ |
| ppt-miR1027 | UUUCUAUCUUCUCUUCCAAUC | 2 | 21 | 102 (383) | 129 (503) | 156 (406) | 5′ |
| ppt-miR1028(5′) | UCUUAGAUCUACAAUGucaCa | 3 | 21 | 934 (1744) | 235 (582) | 933 (3607) | Amb |
| ppt-miR1028(3′) | uGgCAUUGUaGguuUaAGagC | 21 | 491 (1744) | 216 (582) | 2437 (3607) | Amb | |
| ppt-miR1029 | UCUCUCUCAACCAACCAUAC | 1 | 20 | 16 (113) | 31 (158) | 104 (234) | 3′ |
| ppt-miR1030 | uCUGCAUCUGCACCUGCACCa | 10 | 21 | 64 (367) | 35 (220) | 140 (638) | 5′ |
| ppt-miR1031 | UCAAUGCUCUCUGGAGCUUCU | 2 | 21 | 421 (491) | 233 (264) | 458 (490) | 5′ |
| ppt-miR1032 | AGGUGACUGCCUGGAAUUGGG | 1 | 21 | 86 (118) | 147 (176) | 174 (222) | 5′ |
| ppt-miR1033 | UGACGGGUCGUGAUGGGCACU | 5 | 21 | 5 (10) | 24 (239) | 276 (767) | 3′ |
| ppt-miR1034 | UUACUUUGGCAGCGCUGUGCU | 1 | 21 | 0 (5) | 22 (37) | 98 (188) | 3′ |
| ppt-miR1035 | CGUCUUAGCCCACAAAACGAA | 1 | 21 | 54 (91) | 74 (143) | 94 (98) | 5′ |
| ppt-miR1036(5′) | UGUGGAGUCCGUAAUUAGCUG | 1 | 21 | 16 (54) | 18 (49) | 14 (42) | Amb |
| ppt-miR1036(3′) | AGCUAAUUAAGGAUUCUACAC | 21 | 21 (54) | 12 (49) | 4 (42) | Amb | |
| ppt-miR1037 | AGCCUUAUUAGGAUUUGAUGG | 1 | 21 | 0 (0) | 4 (12) | 14 (24) | 3′ |
| ppt-miR1038(5′) | UAGGUGCGUUUCCACCAAAG | 1 | 20 | 37 (86) | 22 (70) | 40 (94) | Amb |
| ppt-miR1038(3′) | CAUGGUGGAAUCGCAUCCAGG | 21 | 27 (86) | 8 (70) | 30 (94) | Amb | |
| ppt-miR1039(5′) | UCUUUGGGUCUUUCUCUCCUG | 1 | 21 | 0 (5) | 10 (20) | 2 (4) | Amb |
| ppt-miR1039(3′) | GGGGAGACGGGCUCAAGGAUC | 21 | 0 (5) | 6 (20) | 0 (4) | Amb | |
| ppt-miR1040 | UGAACGCAAAUGAACAUGUUC | 1 | 21 | 21 (48) | 37 (52) | 40 (50) | 3′ |
| ppt-miR1041 | UUUUCGGGUGAGAGUGGUCCU | 1 | 21 | 16 (54) | 2 (4) | 6 (22) | 5′ |
| ppt-miR1042(5′) | GCUGUGCAGGAACUAGAUAGG | 1 | 21 | 0 (0) | 4 (8) | 0 (4) | Amb |
| ppt-miR1042(3′) | UGUCUAGUCUCUCCACGGCCCG | 22 | 0 (0) | 2 (8) | 2 (4) | Amb | |
| ppt-miR1043(5′) | CCUUCAACUUCAUGCGUGCA | 1 | 20 | 0 (5) | 8 (14) | 0 (18) | Amb |
| ppt-miR1043(3′) | UUGCGCGUGAAUUUGAAGGCU | 21 | 0 (5) | 4 (14) | 10 (18) | Amb | |
| ppt-miR1044(5′) | GCACAAAUAUGCACUACAAAC | 1 | 21 | 0 (0) | 16 (47) | 0 (8) | Amb |
| ppt-miR1044(3′) | UUGUAGUGCAUAUUUGUUUU | 20 | 0 (0) | 16 (47) | 0 (8) | Amb | |
| ppt-miR1045 | AAGUGCUGGCUUUUUGACGUU | 1 | 21 | 0 (0) | 20 (37) | 64 (196) | 5′ |
| ppt-miR1046(5′) | UGGAUUUCAUAUUUUUCACG | 1 | 20 | 16 (54) | 8 (14) | 6 (46) | Amb |
| ppt-miR1046(3′) | UGGUGAAAAAUAUGAAAAAUC | 21 | 5 (54) | 2 (14) | 12 (46) | Amb | |
| ppt-miR1047(5′) | CUUGAUUACAAAGGCCUUAAU | 1 | 21 | 5 (27) | 12 (62) | 14 (72) | Amb |
| ppt-miR1047(3′) | UGAUCAGCGAUGACCUAGUUG | 21 | 10 (27) | 22 (62) | 42 (72) | Amb | |
| ppt-miR1048(5′) | UAGAACAUGAGUGUAGACGAC | 1 | 21 | 27 (59) | 14 (29) | 6 (14) | Amb |
| ppt-miR1048(3′) | UGUCUACACUCAUGUUCUAGA | 21 | 16 (59) | 12 (29) | 2 (14) | Amb | |
| ppt-miR1049 | UCUCUCUUAGCCAAACAGUCU | 1 | 21 | 108 (124) | 10 (18) | 2 (2) | 3′ |
| ppt-miR1050 | UGACCACCUUGAUUCCGGCCU | 1 | 21 | 10 (59) | 60 (114) | 474 (661) | 5′ |
| ppt-miR1051(5′) | GGUUCAAGUGAACAGGAAGA | 1 | 20 | 0 (5) | 27 (47) | 2 (24) | Amb |
| ppt-miR1051(3′) | UUCCUGUCUACUUGAAGCCAC | 21 | 0 (5) | 14 (47) | 20 (24) | Amb | |
| ppt-miR1052 | UUCCUUUUAUUGAUUGUGGUA | 1 | 21 | 189 (216) | 647 (907) | 372 (470) | 3′ |
| ppt-miR1053(5′) | GAUGGGUUAUCUCAAAGUGG | 1 | 20 | 0 (0) | 2 (4) | 0 (2) | Amb |
| ppt-miR1053(3′) | CUCUACUUUGACAUAGCCCACU | 22 | 0 (0) | 0 (4) | 2 (2) | Amb | |
| ppt-miR1054 | UAAACCCUCUCUCUAUUCCUG | 1 | 21 | 43 (59) | 79 (118) | 288 (326) | 3′ |
| ppt-miR1055 | UUAGGGGUGUUUUCCAGUGACU | 1 | 22 | 5 (5) | 27 (56) | 0 (6) | 3′ |
| ppt-miR1056 | UGGAUCUUUGCAUCAUAACCC | 1 | 21 | 37 (102) | 27 (52) | 10 (50) | 3′ |
| ppt-miR1057 | UUCUUGUGUUCUAGCUGGGGGU | 1 | 22 | 0 (0) | 0 (2) | 4 (8) | 3′ |
| ppt-miR1058 | AGAGAUUCCAUCACGAAGCAC | 1 | 21 | 10 (32) | 27 (93) | 174 (252) | 3′ |
| ppt-miR1059 | UGAAAGUCCUUCACAAACAAC | 1 | 21 | 81 (86) | 108 (120) | 62 (64) | 5′ |
| ppt-miR1060 | UUUGUCAUAGGAUUACACACG | 1 | 21 | 16 (21) | 4 (8) | 8 (10) | 5′ |
| ppt-miR1061(5′) | UUAUUUCAUAGACUACUCAAA | 1 | 21 | 5 (5) | 2 (4) | 2 (6) | Amb |
| ppt-miR1061(3′) | UGAGUAGUCAAUGGAAUAAUG | 21 | 0 (5) | 2 (4) | 2 (6) | Amb | |
| ppt-miR1062 | UCCCUCACGUGGUGUUUGCAGC | 1 | 22 | 5 (5) | 10 (18) | 16 (18) | 5′ |
| ppt-miR1063 | CAUCUUGGAGUACUGCAUCUU | 8 | 21 | 378 (615) | 235 (345) | 34 (62) | 5′ |
| ppt-miR1064(5′) | CAUUCUGAGAAUAAUCAUCCG | 1 | 21 | 0 (10) | 0 (4) | 4 (4) | Amb |
| ppt-miR1064(3′) | GGGUGAUUAUUCUCAGAGUUC | 21 | 5 (10) | 2 (4) | 0 (4) | Amb | |
| ppt-miR1065 | ACAGUCUCUGACUUCUCGCAG | 1 | 21 | 0 (10) | 8 (18) | 218 (296) | 5′ |
| ppt-miR1066 | ACAUGUUGCAGAGCGGGGUAC | 1 | 21 | 16 (64) | 22 (58) | 46 (112) | 3′ |
| ppt-miR1067 | ACAUACUGAAGUUUGAUGCCA | 1 | 21 | 27 (32) | 16 (29) | 2 (6) | 5′ |
| ppt-miR1068 | UAGCCAUUUGCUUGAAGGUCA | 1 | 21 | 21 (27) | 10 (14) | 76 (92) | 5′ |
| ppt-miR1069(5′) | UCUCAAGUCUUAUCAUUGGAU | 1 | 21 | 5 (10) | 4 (14) | 0 (4) | Amb |
| ppt-miR1069(3′) | UGAUAAAUCAAAGUGCUCACU | 21 | 5 (10) | 2 (14) | 4 (4) | Amb | |
| ppt-miR1070 | UCGGUUUCUAGUAAAACUUGC | 1 | 21 | 21 (21) | 10 (16) | 0 (0) | 5′ |
| ppt-miR1071(5′) | GCGCAAUUCCCGAACGGGUAG | 1 | 21 | 5 (10) | 0 (0) | 0 (0) | Amb |
| ppt-miR1071(3′) | ACCCGUUCUGGAAUUGCGCGG | 21 | 5 (10) | 0 (0) | 0 (0) | Amb | |
| ppt-miR1072 | UGCAUUGUGUUAUUUGAAGCUUGA | 1 | 24 | 43 (54) | 37 (77) | 56 (108) | 3′ |
| ppt-miR1073(5′) | UGAAUGAUAUUAACGUCCACG | 1 | 21 | 189 (659) | 239 (1041) | 138 (418) | Amb |
| ppt-miR1073(3′) | UGGGCGUUAAUACAUUUCAUA | 21 | 383 (659) | 587 (1041) | 232 (418) | Amb | |
| ppt-miR1074 | AGGGUUGUUAGUUGUGUUGAU | 1 | 21 | 70 (232) | 66 (154) | 20 (106) | 5′ |
| ppt-miR1075 | UGUUUCAGUCAUGGUUUCUAC | 1 | 21 | 48 (81) | 20 (49) | 18 (24) | 3′ |
| ppt-miR1076(5′) | UAUCACAAGGUGCGAUAAUUG | 1 | 21 | 27 (172) | 56 (331) | 20 (224) | Amb |
| ppt-miR1076(3′) | CCAAGCACUUAUCGCACCCUG | 21 | 21 (172) | 45 (331) | 60 (224) | Amb | |
| ppt-miR1077(5′) | UUCAAAUGUUCGGAUGCGUGC | 1 | 21 | 0 (0) | 0 (6) | 8 (24) | Amb |
| ppt-miR1077(3′) | AGGGAUGGACGUAUACGAGCC | 21 | 0 (0) | 4 (6) | 10 (24) | Amb | |
| ppt-miR1078 | UUUGGAUGAUUCAAUUGUGAU | 1 | 21 | 91 (124) | 160 (195) | 28 (36) | 5′ |
| ppt-miR1079(5′) | CACCGAGCUCUUCUUUUUGGAGG | 1 | 23 | 0 (5) | 2 (6) | 0 (0) | Amb |
| ppt-miR1079(3′) | ACCAAAAGAAUAGCUAUGUGGU | 22 | 0 (5) | 4 (6) | 0 (0) | Amb | |
To enable comparison between samples, the number of reads corresponding to the mature miRNA and all reads corresponding to the miRNA hairpin (in parentheses) were converted into transcripts per quarter million (TPQ) by dividing the raw number of reads by the total number of reads that matched the Phypa1_1 genome assembly, multiplying by 250,000, and rounding to the nearest integer. P, protonemata library (GSM115095); PG, protonemata and young gametophores library (GSM115096); GS, mature gametophores and sporophytes library (GSM115097).
Sequence of the most abundant mature miRNA is shown, with lowercase letters indicating positions where the sequence of less-abundant paralogs differ.
Arm of stem loop where the mature miRNA resides. Amb, ambiguous.
Table 3.
Deeply Conserved Plant miRNA Families
| Family | At Locia | Pt Locia | Os Locia | Sm Locib | Pp Locic |
|---|---|---|---|---|---|
| miR156/157 | 12 | 11 | 12 | 4 | 3 |
| miR160 | 3 | 8 | 6 | 2 | 9 |
| miR165/166 | 9 | 17 | 14 | 3 | 13 |
| miR170/171 | 4 | 11 | 9 | 4 | 2 |
| miR159/319 | 6 | 15 | 8 | 2 | 5 |
| miR390 | 2 | 4 | 1 | 3 | |
| miR396 | 2 | 7 | 5 | 1 | |
| miR408 | 1 | 1 | 1 | 1 | 2 |
| miR473/477 | 2 | 8 | |||
| miR529(5′)d | 1 | 7 | |||
| miR535 | 1 | 4 | |||
| miR536 | 1 | 6 |
Data from miRBase version 9.2.
Data from this work.
Data from miRBase version 9.2 and this work.
Based upon the sequencing frequencies obtained in P. patens, we suggest the previously annotated miR529 from O. sativa actually represents the miRNA*, and the sequence from the 5′ arm represents the miRNA. miR529(5′) may also be related to miR156.
In S. moellendorffii, 44 distinct miRNA families were found to originate from 58 miRNA genes (see Supplemental Table 2 online), including miR156, miR159/319, and miR166, as expected (Floyd and Bowman, 2004; Axtell and Bartel, 2005), as well as five additional deeply conserved miRNA families (miR160, miR171, miR396, miR408, and miR536; Table 3). However, the majority of S. moellendorffii miRNA diversity, comprising 36 novel families, was to the best of our present knowledge restricted to the lycopod lineage (Table 2). The deeply conserved S. moellendorffii miRNAs were often encoded by fewer paralogs than in other species (Table 3), and most of the novel S. moellendorffii miRNAs appeared to arise from only a single locus (Table 2). This reduction in paralogs was consistent with the reduced genome size of S. moellendorffii compared with most land plants with fully sequenced genomes (Wang et al., 2005). Collectively, the results from P. patens and S. moellendorffii were consistent with recent results from angiosperms, which demonstrated a large diversity of recently evolved miRNAs expressed at low levels that only became apparent after deep sequencing of small RNA libraries (Rajagopalan et al., 2006; Fahlgren et al., 2007).
Table 2.
Novel S. moellendorffii miRNAs
| Abundancea
|
||||||
|---|---|---|---|---|---|---|
| miRNA | Sequence (5′–3′)b | No. of Loci | Length | miRNA | Total | Armc |
| smo-miR1080 | UUCACUAUCUGCAAACACCUCU | 1 | 22 | 79 | 110 | 3′ |
| smo-miR1081 | UGAGGCUUGCCUUUGAUUCUC | 1 | 21 | 5001 | 5469 | 5′ |
| smo-miR1082 | GcGUUGGCCUGCUGGCCGGuG | 2 | 21 | 6188 | 8320 | 5′ |
| smo-miR1083 | UAGCCUGGAACGAAGCACGGU | 1 | 21 | 5 | 8 | 5′ |
| smo-miR1084 | UAACUCAGGUGGUAUGUUCCCA | 1 | 22 | 1346 | 1430 | 5′ |
| smo-miR1085(5′) | CCCCUACAAGAGAUAUCAAUC | 1 | 21 | 1 | 3 | Amb |
| smo-miR1085(3′) | UUGAUAUGUCUUGUAGGGAC | 20 | 1 | 3 | Amb | |
| smo-miR1086 | UAGUGCCGUGGUCCUUUUGGC | 1 | 21 | 19 | 22 | 5′ |
| smo-miR1087 | UACAGAUUGGUGUUAGUGCUU | 1 | 21 | 5 | 11 | 3′ |
| smo-miR1088(5′) | CAGAAGAAAGAGAGCACGCAU | 1 | 21 | 22 | 56 | Amb |
| smo-miR1088(3′) | GCGUGCUCUUUUUCUUCUGUC | 21 | 29 | 56 | Amb | |
| smo-miR1089 | AGAUCAUCUAGGAUUGUUUGC | 1 | 21 | 12 | 18 | 3′ |
| smo-miR1090 | UGAAAGCCAUUCUCAACAAAA | 1 | 21 | 7 | 11 | 5′ |
| smo-miR1091 | CGGCAGUGAGGGAGGAUUUGC | 1 | 21 | 191 | 294 | 5′ |
| smo-miR1092 | UGACAGGAAUGCAUUGGUGUU | 1 | 21 | 10 | 23 | 5′ |
| smo-miR1093 | UGGAGGUGUCGUUGCCAAGGA | 1 | 21 | 17 | 30 | 5′ |
| smo-miR1094 | UACUUCuCcGUgCCACAGUA | 3 | 20 | 785 | 1406 | 3′ |
| smo-miR1095 | UAGGUUCCCUUGUUUCCAAAU | 2 | 21 | 1694 | 2381 | 3′ |
| smo-miR1096 | CUGCCUCUUUGCUUCAGGAAU | 1 | 21 | 11 | 37 | 3′ |
| smo-miR1097 | UAGCCAUUGUUGUUGUUGGAA | 1 | 21 | 11 | 19 | 5′ |
| smo-miR1098 | UGAUGACGUUUGUGCUGAAAU | 1 | 21 | 2415 | 2835 | 5′ |
| smo-miR1099 | UAUAGCAAUGGUGUUUUUGUC | 1 | 21 | 23 | 41 | 5′ |
| smo-miR1100 | UGUCACGGACAGAACCCCACUC | 1 | 22 | 41 | 68 | 5′ |
| smo-miR1101(5′) | GGCAUUCUCUCGAAACAAGUG | 1 | 21 | 1 | 2 | Amb |
| smo-miR1101(3′) | UUGUUUGCAGAGAAUGCCCAA | 21 | 1 | 2 | Amb | |
| smo-miR1102 | AGCUAUUGCCAGCGUCAAGCA | 1 | 21 | 196 | 210 | 5′ |
| smo-miR1103(5′) | AAGAGUGCACCCCCUUUCCAAU | 1 | 22 | 1 | 4 | Amb |
| smo-miR1103(3′) | UGGAAAAAGGAGGUGCAUUCUUGU | 24 | 1 | 4 | Amb | |
| smo-miR1104 | CGCAGCUGUUCUUUUUCCUUC | 1 | 21 | 692 | 862 | 3′ |
| smo-miR1105 | UUCGGAUGUGAAGAUCGCUGC | 1 | 21 | 48 | 87 | 3′ |
| smo-miR1106 | UUUAAAGGUGUUAAUGUGUGA | 1 | 21 | 30 | 38 | 3′ |
| smo-miR1107 | GGUGCUGGUUCCAAUUCAGgU | 1 | 21 | 2405 | 2556 | 5′ |
| smo-miR1108 | UGAGCUGAGAGACAUAACCCC | 1 | 21 | 10 | 32 | 3′ |
| smo-miR1109 | UAGUGGGAGAUUUUGUGUAAC | 1 | 21 | 46 | 129 | 5′ |
| smo-miR1110 | GCUAGGGGCAGUGGUCAAGGA | 1 | 21 | 12 | 22 | 5′ |
| smo-miR1111 | GCCAUGACUAAGCAGGACCAG | 1 | 21 | 4 | 10 | 3′ |
| smo-miR1112(5′) | ACUUUGUUAUGGCAUUCCUCG | 1 | 21 | 33 | 61 | Amb |
| smo-miR1112(3′) | AGGAAUGCUAUAACAAAGUCA | 21 | 26 | 61 | Amb | |
| smo-miR1113 | UGAGCAGUCAUAAGGUAGCCU | 1 | 21 | 5443 | 5919 | 5′ |
| smo-miR1114 | AAGGGACAAAACAUUGAGCAG | 1 | 21 | 6 | 14 | 3′ |
| smo-miR1115(5′) | UGAGCUCAGGCACUUUGGUGG | 1 | 21 | 1 | 3 | Amb |
| smo-miR1115(3′) | UCACCAAAGUGCCUGAGCUCA | 21 | 1 | 3 | Amb | |
Number of times the mature miRNA was sequenced (miRNA) or any small RNA matching the hairpin was sequenced. A total of 149,586 S. moellendorffii small RNAs that matched at least one WGS trace were sequenced from a single library.
Sequence of the most abundant mature miRNA is shown, with lowercase letters indicating positions where the sequence of less-abundant paralogs differ.
Arm of stem loop where the mature miRNA resides. Amb, ambiguous.
A few plant pri-miRNA transcripts have been observed to produce more than one miRNA/miRNA* duplex, including Arabidopsis miR161 (Allen et al., 2004) and miR163 (Kurihara and Watanabe, 2004) as well as P. patens miR319d and miR1219a (Talmor-Neiman et al., 2006a; Fattash et al., 2007). Certain miRNAs from the unicellular green alga Chlamydomonas reinhardtii also produce multiple miRNA/miRNA* duplexes from a single pri-miRNA transcript (Molnár et al., 2007; Zhao et al., 2007). We found that two novel S. moellendorffii pri-miRNAs produced two distinct miRNA/miRNA* duplexes, in both cases separated from each other by distances that were rough multiples of 21 (see Supplemental Figure 1A and Supplemental Table 2 online). This spatial separation was consistent with sequential DCL1-mediated processing of the pri-miRNA, as reported for Arabidopsis miR163 (Kurihara and Watanabe, 2004), although independent recognition and processing of the two miRNA/miRNA* duplexes could not be excluded. Production of three sequential miRNA/miRNA* duplexes is a conserved feature of plant miR159 and miR319 loci (see Supplemental Figure 1B and Supplemental Tables 1 and 2 online; Rajagopalan et al., 2006; Talmor-Neiman et al., 2006a; Fattash et al., 2007). The previously annotated mature miR159 and miR319 miRNAs are the best conserved of these sequences, likely reflecting their conserved functions in targeting messages in trans. However, as noted previously by comparing Arabidopsis and Oryza sativa miR159 and miR319 pri-miRNAs (Li et al., 2005), the loop-proximal alternative miRNA/miRNA* duplex was also relatively well conserved among land plants (see Supplemental Figure 1B online). Although some potential targets could be predicted for the Arabidopsis and P. patens alternative miR319 species, we were not able to verify them using RNA ligase–mediated 5′ rapid amplification of cDNA ends (RLM 5′-RACE; Llave et al., 2002a). Thus, these data are inconclusive as to whether any of these conserved alternative miR319 miRNAs are in fact incorporated into a functional silencing complex.
P. patens miRNA Loci Are Often Clustered and Frequently Overlap with Protein-Coding Genes
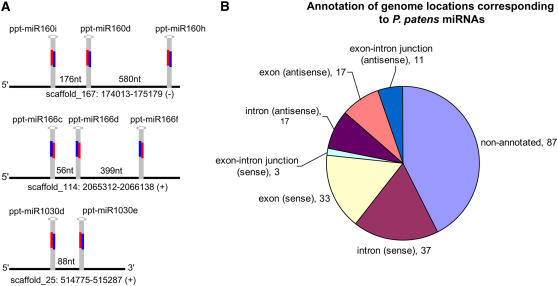
The availability of a draft assembly and transcript annotations of the P. patens genome allowed the detailed examination of miRNA loci. Most miRNA loci characterized in angiosperms appear to arise from independent transcription units, each encoding a single miRNA-producing stem loop that does not overlap with annotated protein-coding loci (Jones-Rhoades et al., 2006). Four P. patens miR1219 loci are contained in two clusters, strongly suggesting that they are expressed from two polycistronic precursors (Talmor-Neiman et al., 2006a). We found that clustered miRNA expression was common for P. patens miRNAs (Figure 1A; see Supplemental Table 1 online). A total of 48 of the 205 P. patens miRNA loci were arranged in closely linked clusters of two or three miRNA stem loops. Clustered miRNA stem loops were observed for both ancient miRNAs, such as miR160 and miR166, and for novel bryophyte-specific miRNAs (Figure 1A). With only a few exceptions, these clusters contained paralogs encoding mature miRNA families in the same family, rather than encoding distinct mature miRNAs unrelated in sequence as is frequently the case for animal miRNA clusters (see Supplemental Table 1 online). Similarly, recent results in maize (Zea mays) have demonstrated the expression of a polycistronic miRNA precursor for miR156 (Chuck et al., 2007), and a miRNA from the green alga C. reinhardtii is also arranged in a tandem cluster (Zhao et al., 2007).
Figure 1.
Properties of miRNA Expression in P. patens.
(A) Schematic depictions of representative clustered P. patens miRNAs that are potentially processed from polycistronic pri-miRNAs, indicated by black lines. In total, 48 out of the 205 P. patens miRNA loci were contained in 21 clusters containing two or three miRNA stem loops each (see Supplemental Table 1 online). Distances refer to the number of nucleotides separating the regions annotated as miRNA foldbacks.
(B) Mapping of mature P. patens miRNAs with respect to annotated protein-coding genes. The numbers of mature miRNAs that overlapped various annotations of the P. patens Phypa1_1 genome assembly are indicated.
Strikingly, the majority of the P. patens miRNA hairpins overlapped with the positions of annotated protein-coding loci in the P. patens version 1_1 (Phypa1_1) draft genome assembly (Figure 1B). Of the miRNAs that overlapped with protein-coding annotations, approximately two-thirds were in the sense orientation with respect to the annotated gene, and approximately equal numbers overlapped exonic compared with intronic sequences (Figure 1B). The preponderance of P. patens miRNA loci that overlapped with annotated protein-coding loci was more reminiscent of miRNA expression in the unicellular green alga C. reinhardtii than that of angiosperms (Zhao et al., 2007). The Phypa1_1 genome release contains 35,938 annotated protein coding genes, the majority of which were annotated using a combination of 5′ and 3′ EST sequences and de novo gene-finding algorithms. We therefore considered the possibility that many of the miRNA-overlapping protein-coding loci represented erroneous annotations triggered by the fact that, except for protein-coding potential, miRNA loci have typical features of RNA polymerase II–transcribed regions that might be recognized by automated gene annotation software (Lee et al., 2004; Xie et al., 2005). If this were the case, then the lengths of the miRNA-overlapping annotated proteins would be predicted to be much shorter than the typical length of all of the annotated proteins. However, this was not the case: The median length for all annotated proteins in the draft P. patens Phypa1_1 assembly was 277 amino acids compared with a median of 267 for the subset of annotated protein-coding genes that overlapped mature miRNAs. The median lengths of proteins that overlapped with miRNA loci in introns and exons was similar (medians of 269.5 and 256, respectively), suggesting that there is no consistent bias for the overlapping genes of either intronic or exonic miRNAs to be misannotated. We conclude that many P. patens miRNAs are indeed expressed from loci that also have protein-coding potential.
Some lineage-specific miRNA hairpins expressed in Arabidopsis possess extended complementarity to protein-coding transcripts that are frequently, though not always, the targets of such miRNAs (Allen et al., 2004; Rajagopalan et al., 2006; Fahlgren et al., 2007). This suggests that the initiation of novel miRNA–target interactions at least sometimes involves the inverted duplication of transcribed regions. Similar analyses for P. patens miRNA loci were complicated by the fact that more than half of the miRNA hairpins overlapped annotated protein-coding regions; thus, any extended homology found between such hairpins and other protein-coding transcripts would be more likely due to conserved protein domains rather than indicative of the origin of the miRNA locus. Nonetheless, of the 87 loci that did not overlap with annotated protein-coding loci, five were found to have extended complementarity with annotated transcripts (FASTA search E < 0.05): miR1026a, miR1072, miR1073, miR1220a, and miR1220b. Interestingly, none of the presumed originating transcripts of these five loci were also predicted as targets of their mature miRNAs, suggesting that these loci might have been selected to have targets distinct from their originating gene families as appears to have happened for a few Arabidopsis miRNAs (Fahlgren et al., 2007). However, the majority of P. patens miRNA loci that don't themselves overlap with annotated protein-coding transcripts did not appear to have extended complementarity to annotated transcripts.
P. patens miRNA Targets
Several targets of P. patens miRNAs and ta-siRNAs have been predicted and/or validated using EST data, WGS trace data, or directed cDNA cloning (Floyd and Bowman, 2004; Arazi et al., 2005; Axtell et al., 2006; Floyd et al., 2006; Talmor-Neiman et al., 2006a, 2006b; Fattash et al., 2007). The availability of a large set of annotated mRNAs derived from the draft genome assembly coupled with the expanded number of P. patens miRNAs discovered in this study prompted us to revisit miRNA and ta-siRNA target prediction. Targets were predicted using the methodology described by Allen et al. (2005), except that conservation of targeting in other plant genomes was not considered (see Supplemental Table 3 online). This methodology performed reasonably well, as assessed by a signal-to-noise ratio of 2.78 to 1 when compared with a set of shuffled miRNA sequences controlled for di- and trinucleotide composition.
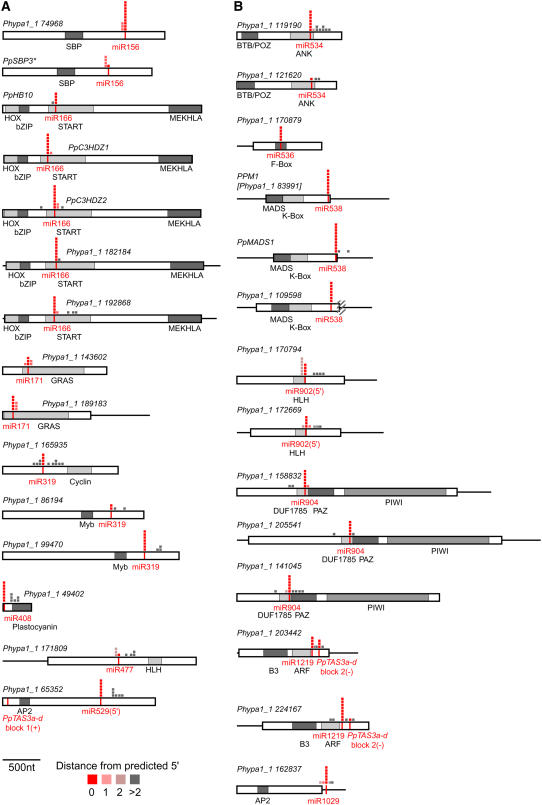
Computational target predictions were based upon the full contingent of annotated transcripts that accompanied the Phypa1_1 genome release and thus are expected to be more comprehensive than those based upon EST or WGS sequence data. However, we recognized that this database of P. patens transcripts was probably not as accurate as those derived from the current annotations of the more thoroughly studied Arabidopsis and O. sativa genomes; thus, failure to annotate P. patens transcripts could have led to false negatives (missed predictions), while the presence of inaccurately annotated transcripts and inability to consider conservation in a related genome could have led to false positive predictions. We therefore used RLM 5′-RACE (Llave et al., 2002a) to test many of these predictions by assaying for the existence of cDNAs corresponding to miRNA-mediated cleavage products. A total of 29 targets of 14 miRNA families and a ta-siRNA were validated (Figure 2). As expected based upon previous observations (Floyd and Bowman, 2004; Arazi et al., 2005; Axtell and Bartel, 2005; Floyd et al., 2006; Fattash et al., 2007), most of the miRNAs conserved between Arabidopsis and P. patens had similar targets in both organisms (Figure 2A; see Supplemental Table 3 online), supporting the hypothesis that these trans-regulatory interactions have remained constant during the diversification of land plants. For instance, miR156 directed the cleavage of two SBP box genes (including Pp SBP3, which had previously been demonstrated to be a miR156 target; Arazi et al., 2005), miR166 directed the cleavage of the five class III HD-ZIP genes that had been previously been observed to possess miR166 complementary sites (Floyd et al., 2006), miR171 directed the cleavage of two GRAS domain transcription factor genes, and miR408 directed the cleavage of a plastocyanin domain–containing gene. In flowering plants, the miR159/319 family targets subsets of the MYB and TCP transcription factor gene families (Rhoades et al., 2002; Palatnik et al., 2003); none of the initially predicted P. patens miR319 targets had recognizable homology to MYB or TCP genes (see Supplemental Table 3 online). However, a gene with a cyclin domain was among the initially predicted P. patens miR319 targets and was indeed cleaved in vivo at the predicted miR319 complementary site (Figure 2A). Reducing the stringency of the search for P. patens miR319 targets allowed the identification of two homologs of MYB transcription factors that were similar to the angiosperm targets of the sequence-related miR159 (see Supplemental Table 3 online); RLM 5′-RACE demonstrated that these two genes were also cleaved by a miR319-guided activity in P. patens (Figure 2A). Thus, the initial search parameters were too stringent to identify all bona fide cleavage targets. This sensitivity trade-off when predicting plant miRNA targets with acceptable specificity without recourse to evolutionary conservation was expected. For example, the target prediction score cutoffs would have been too stringent to identify the 3′ miR390 complementary site of Pp TAS3a and also would have failed to identify the 5′ miR390 complementary sites of angiosperm TAS3 loci, even though these have been confirmed by direct experimentation to be functional (Axtell et al., 2006; Talmor-Neiman et al., 2006b).
Figure 2.
Validated P. patens miRNA and ta-siRNA Targets.
(A) Cleaved targets of conserved P. patens miRNAs. Schematics of target mRNAs along with the relative positions of the 5′ residues of all sequenced uncapped cDNAs are shown. The positions of the miRNA complementary sites are indicated with red vertical lines. Gray boxes indicate the positions of regions coding for conserved protein domains, whose abbreviated names are listed below. Boxes correspond to open reading frames and horizontal black lines to untranslated regions. Shaded pixels indicate the positions of 5′ residues of individual cDNA clones, which are shaded as indicated in the key below to indicate their proximity to the site predicted by miRNA-mediated cleavage between the tenth and eleventh nucleotide of complementarity. An asterisk indicates that Pp SBP3 was included as a positive control (Arazi et al., 2005). Alignments of target sites with small RNAs are provided in Supplemental Table 3 online. nt, nucleotides.
(B) Cleaved targets of nonconserved P. patens miRNAs as in (A). Hatch marks in Phypa1_1 109598 indicate a region where the splicing of the observed cDNA fragments differed from the annotation.
The predicted and experimentally confirmed targets of the P. patens–specific miRNAs included many potential developmental regulatory genes and a set of genes whose sequences indicated more diverse functions (Figure 2B; see Supplemental Table 3 online). For instance, miR534 directed the cleavage of two mRNAs encoding ankyrin repeat proteins, while miR536 directed the cleavage of an F-box encoding mRNA. The P. patens–specific miR538 was demonstrated to target three MADS box transcription factors: PPM1, Pp MADS1, and a close relative of PPM2, Phypa1_1 109598 (Krogan and Ashton, 2000; Henschel et al., 2002). Among these miR538 targets, PPM1 has been demonstrated to play a role in moss development; antisense knockdown of PPM1 results in delayed gametangia development and aberrant leaf development (Singer et al., 2007). Interestingly, the Arabidopsis-specific miR824 also directs the cleavage of a MADS box mRNA (Fahlgren et al., 2007) as does the O. sativa–specific miR444 (Sunkar et al., 2005). There is no detectable sequence similarity among miR538, miR824, and miR444 despite the fact that all three regulate MADS box–containing genes in their respective lineages. The two small RNAs comprising the miRNA/miRNA* duplex of miR902 were roughly equivalent in sequencing abundance, and both had several predicted targets with favorable scores (see Supplemental Table 3 online). miR902(5′) was predicted to target several potential helix-loop-helix (HLH) transcription factor genes, while the targets of miR902(3′) were predicted to include PHD/SET domain genes potentially involved in the regulation of chromatin structure. Although the predictions suggested that miR902(5′) and miR902(3′) might both have posttranscriptional regulatory functions, we were only able to experimentally validate the HLH targets of miR902(5′) (Figure 2B).
Recurrence of miRNA-Mediated Negative Feedback Loops in Plants
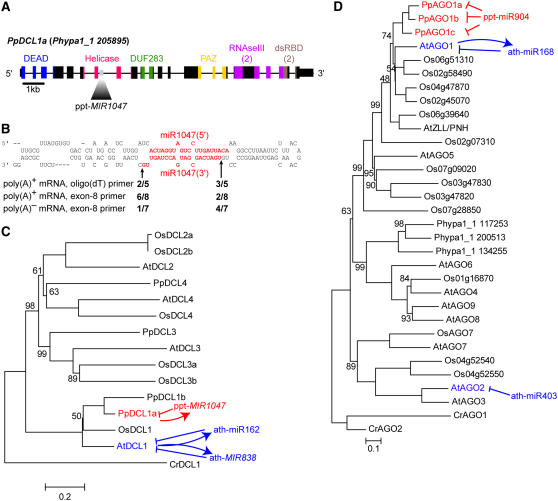
The gene encoding the DCL protein chiefly responsible for miRNA biogenesis in Arabidopsis, DCL1, is itself regulated by miRNAs: miR162 is expressed from two distinct loci and can target DCL1 mRNA via a complementary site formed by the splicing of exon 12 to exon 13 (Xie et al., 2003), while miR838 is excised from a stem loop present in intron 14 of the DCL1 pre-mRNA (Rajagopalan et al., 2006). Target cleavage mediated by miR162 and excision of the miR838/miR838* duplex, both of which would lead to repression of DCL1 mRNA expression, have been hypothesized to require the activity of the DCL1 protein and thereby constitute negative feedback circuits. We found that the P. patens MIR1047 locus resided in the sense orientation relative to intron seven of a gene whose predicted protein product had strong overall similarities to DCL proteins (Figure 3A). RLM 5′-RACE experiments detected RNA fragments terminating at positions consistent with the excision of the miR1047/miR1047* duplex from this pre-mRNA (Figure 3B). Evidence for miR1047 excision from pre-mRNA in which intron seven was not spliced was found from both poly(A)+ and poly(A)− RNA samples. We also noted that approximately half of these truncated pre-mRNAs terminated at the loop-proximal cleavage position, indicating that the first DCL-mediated cleavage of the stem loop must have occurred at the loop-proximal position (Figure 3B). This was in contrast with the processing of animal pri-miRNAs (Lee et al., 2003) and certain other plant pri-miRNAs (Kurihara and Watanabe, 2004; Vaucheret et al., 2006), for which the first cleavage events can occur proximal to the base of the hairpins. Nevertheless, similar results for Arabidopsis miR838 processing (Rajagopalan et al., 2006) support the conclusion that the loop-proximal cleavage event can sometimes occur first during the biogenesis of some plant miRNAs.
Figure 3.
Analogous miRNA-Mediated Feedback Loops Despite Divergent Small RNA Sequences.
(A) Schematic of the Pp DCL1a pre-mRNA, which contains an expressed miRNA within intron seven. Thick rectangles represent predicted coding exons, thin rectangles represent untranslated regions, and thin lines represent predicted introns. Colored exonic regions correspond to conserved protein domains as indicated. Gray indicates the location of the MIR1047 hairpin within intron seven.
(B) Cleavage at miR1047 in intron seven of the Pp DCL1a pre-mRNA. The predicted secondary structure of MIR1047 is shown, with the miRNA/miRNA* duplex highlighted in red. Fractions indicate the number of 5′-RACE products observed to terminate at the indicated positions over the total number of 5′-RACE clones sequenced from three different experiments from the indicated combinations of RNA samples and RT primers.
(C) An unrooted phylogenetic reconstruction of relationships among Arabidopsis, O. sativa, and P. patens DCL proteins using C. reinhardtii DCL1 as an outgroup. The known miRNA-mediated feedback loops between At DCL1 pre-mRNA, mRNA, protein, and DCL1-dependent miRNAs are indicated in blue. The hypothesized feedback loop between Pp DCL1a pre-mRNA, protein, and ppt-MIR1047 is indicated in red. Nodes with <100% bootstrap support are noted with the percentage of supporting replicates. Accession numbers are given in Methods.
(D) An unrooted phylogenetic reconstruction of relationships among Arabidopsis, O. sativa, and P. patens AGO proteins using C. reinhardtii AGO1 and AGO2 as outgroups as displayed in (B). miRNA-mediated control of Arabidopsis AGO genes is shown in blue, and miRNA-mediated control of P. patens AGO genes by ppt-miR904 is shown in red. Accession numbers are given in Methods.
Phylogenetic reconstruction of the relationships between Arabidopsis, O. sativa, C. reinhardtii, and P. patens DCL proteins clearly demonstrated that Pp DCL1a (Phypa1_1 205895), within whose pre-mRNA MIR1047 is embedded, was part of the land plant DCL1 clade (Figure 3C). Neither miR1047(5′), miR1047(3′), nor the entire Pp DCL1a intron seven sequence possessed detectable similarity with miR838 or with the entire At DCL1 intron 14 sequence, which encodes miR838. MIR838 and MIR1047 also differed with respect to their positions within their host genes, in that the intron encoding MIR838 is flanked by exons coding for the PAZ region of DCL1 (Rajagopalan et al., 2006), whereas the MIR1047-encoding intron is flanked by exons coding for the helicase region (Figure 3A). Because the number and positions of introns were identical between At DCL1 and Pp DCL1a, this difference suggested that miR838 and miR1047 arose independently rather than having diverged from a common ancestral miRNA. Nonetheless, despite their sequence and positional differences, miR838 and miR1047 appeared to have analogous functions in the negative feedback regulation of DCL1 pre-mRNAs. We did not find any O. sativa small RNAs that corresponded to the Os DCL1 locus among two large data sets of sequenced rice small RNAs (Johnson et al., 2007; Nobuta et al., 2007).
The founding member of the AGO gene family, Arabidopsis AGO1, encodes a miRNA-guided endonuclease required for the trans-regulatory effects of many miRNAs (Vaucheret et al., 2004; Baumberger and Baulcombe, 2005; Qi et al., 2005). AGO1 is required for the stability and function of miR168, which targets the AGO1 mRNA; this homeostatic feedback loop is critical for proper Arabidopsis development and the normal functioning of many other Arabidopsis miRNAs (Vaucheret et al., 2004, 2006). We found that the three validated targets of the P. patens–specific miR904 were homologous to AGO genes (see Supplemental Table 3 online; Figures 2B and Figure 3D). The three P. patens AGO proteins whose mRNAs were validated targets of miR904 were clearly embedded within the AGO1 clade of land plant AGO proteins (Figure 3D), suggesting that one or more of these miR904 targets might be the major miRNA-guided endonucleases in P. patens; to reflect this similarity, we named these genes Pp AGO1a (Phypa1_1 205541), Pp AGO1b (Phypa1_1 158832), and Pp AGO1c (Phypa1_1 141045). Despite having homologous targets, no similarity between the sequences of miR168 and miR904 was detected. This suggested that the miRNA-AGO feedback loops either appeared independently in angiosperms and bryophytes by convergent evolution or that the sequences of the miRNA and AGO target complementary sites have diverged from those of a common ancestor. Both miR168 and miR904 are released from the 5′ arms of their respective precursor RNAs, and their cleavage sites in both Arabidopsis and P. patens AGO mRNAs lie just upstream of the region encoding the conserved PAZ domain. These observations leave open the possibility that the miR168-AGO and miR904-AGO regulatory interactions could have descended from a common ancestor. The observation that negative feedback interactions between miRNAs, DCL genes, and AGO genes exist in both bryophytes and angiosperms suggests that these interactions are critical for control of miRNA functions in both lineages.
The phylogenetic comparison of the DCL and AGO families in Arabidopsis, O. sativa, P. patens, and C. reinhardtii provided the opportunity to consider the evolution of these families during the divergence of land plants. The DCL family appeared to have diverged and specialized early, with at least three members present in the last common ancestor of these land plants and rather modest duplication since then. We failed to observe a DCL2 homolog in P. patens, though the node supporting the placement of Pp DCL4 in the DCL4 clade was weak (Figure 3C). Whether this reflected the first appearance of DCL2 in the seed plant lineage after the divergence of the bryophytes, selective loss of DCL2 in the bryophyte lineage, or simply a failure to annotate the Pp DCL2 transcript in the Phypa1_1 draft genome was unclear. Because the genome of S. moellendorffii has not yet been assembled and annotated, we were unable to include lycopod DCL proteins in this analysis. Evolution of the AGO family appears to have been more dynamic, with much more expansion, and perhaps loss, of paralogs since the last common ancestor. None of the six identified P. patens AGO proteins fell in the AGO2/3/7-like clade (Figure 3D); whether this reflected selective loss in the bryophyte lineage, diversification of the AGO2/3/7 clade of AGO proteins in the seed plant lineage after divergence from the bryophytes, or a failure to annotate some P. patens AGO loci in the Phypa1_1 assembly was unclear. It is also possible that some of the apparent duplications and extinctions in the AGO genes were artifacts derived from errors in phylogenetic reconstruction; as for the DCL proteins, the S. moellendorffii AGO proteins were not available to assist in resolving this issue.
Pp TAS3a-d Targets Include Genes Similar to Arabidopsis ETT/ARF3 and ARF4
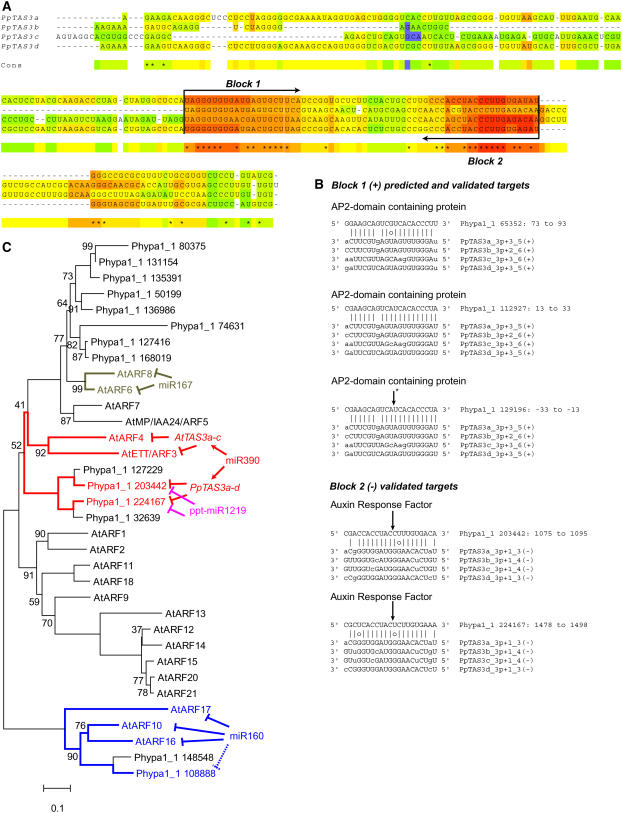
We previously described four ta-siRNA loci targeted by miR390 in P. patens, which we referred to as Pp TAS1-4 (Axtell et al., 2006). The existence of what we called Pp TAS1 was first suggested by Arazi et al. (2005) and was functionally characterized by Talmor-Neiman et al. (2006b), who named it TAS4. We suggest that these four P. patens ta-siRNA loci would be better referred to as Pp TAS3a-d for three reasons: All four loci possess dual miR390 complementary sites, similar to angiosperm TAS3 loci (Axtell et al., 2006; Howell et al., 2007), an unrelated Arabidopsis ta-siRNA locus has also recently been named TAS4 (Rajagopalan et al., 2006), and targets identified and validated in this study (see below) demonstrate that ta-siRNAs derived from these moss loci function to regulate AUXIN RESPONSE FACTOR (ARF) genes similar to those regulated by the angiosperm TAS3 loci.
Pp TAS3a produces abundant phased siRNAs that are dependent upon the RNA-dependent RNA polymerase Pp RDR6 (Talmor-Neiman et al., 2006b). Interestingly, Pp rdr6 mutants also display accelerated differentiation of leafy gametophores from protonemata, suggesting that the Pp TAS3 loci are important for the timing of gametophyte development (Talmor-Neiman et al., 2006b); in Arabidopsis, rdr6 mutants display an At TAS3–dependent acceleration of sporophytic vegetative phase change (Peragine et al., 2004; Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006). Alignment of angiosperm TAS3 genes from multiple species allowed the identification of two conserved, nearly identical ta-siRNAs that function in Arabidopsis to regulate ETT/ARF3 and ARF4 (Allen et al., 2005; Fahlgren et al., 2006; Hunter et al., 2006), but these ta-siRNAs did not have any recognizable homology to the Pp TAS3a-d loci (Axtell et al., 2006).
Given their similar complementarity to miR390, we reasoned that Pp TAS3a-d might be paralogs derived from duplication and divergence of an ancestral locus, and perhaps the areas producing biologically relevant ta-siRNAs may have been preferentially conserved. This appears to have been the case, as alignment of the ta-siRNA–producing regions of Pp TAS3a-d revealed two ∼21-nucleotide blocks of strong conservation (Figure 4A). Sense ta-siRNAs whose 5′ residues were coincident with the beginning of block 1 were two to three nucleotides offset from the phasing register set by cleavage at the 3′ miR390 complementary site, while antisense ta-siRNAs whose 5′ ends were coincident with the beginning of block 2 were exactly coincident with the phasing register set by the 3′ miR390 complementary site (Figure 4A). We found no evidence for miR390 expression in S. moellendorffii, suggesting that the miR390-TAS3 cascade of small RNA production might have been lost in that lineage.
Figure 4.
Similar ARF Targets for Diverse ta-siRNAs and miRNAs.
(A) T-COFFEE alignment of the regions of Pp TAS3a-d between the two miR390 complementary sites. Two ∼21-nucleotide blocks conserved between the four paralogs are indicated.
(B) Alignments of the block 1 (+) siRNAs and the block 2 (-) RNAs with their predicted and validated targets. Lowercase letters indicate positions that cannot form either Watson-Crick or G-U pairs with the target. Arrows indicate cleavage-validated target sites. The asterisk indicates the cleavage site of the AP2 domain–containing gene Phypa1_1 129196 by Pp TAS3a-d block 1 (+)–derived siRNAs as demonstrated by Talmor-Neiman et al. (2006b).
(C) An unrooted phylogenetic reconstruction of the relationships between Arabidopsis and P. patens ARF proteins. Nodes with <100% bootstrap support are noted with the percentage of supporting replicates. ARF genes known or predicted to be under small RNA–mediated regulation are indicated. Accession numbers are given in Methods.
Target predictions using the block 1(+) ta-siRNAs recovered three predicted targets of Pp TAS3a-d. These included Phypa1_1 129196, an AP2 domain–containing gene that has been directly demonstrated to be a target of a block 1(+) ta-siRNA by RLM 5′-RACE (Talmor-Neiman et al., 2006b), and two other AP2 domain–containing genes (Figure 4B). Interestingly, the Pp TAS3a-d, block 1(+) ta-siRNAs that could optimally pair with these predicted targets were all offset by two or three nucleotides from the phasing register predicted by the 3′ miR390 cleavage site and were closer to the register predicted by the 5′ miR390 cleavage site. The experimentally determined cleavage site of Phypa1_1 129196 is consistent with the offset ta-siRNAs that are optimal for interaction with these targets (Talmor-Neiman et al., 2006b; Figure 4B). One of these predicted block 1(+) targets was also a validated target of miR529(5′) (Phypa1_1 65352; Figure 2A). Our failure to detect cleavage products corresponding to the upstream Pp TAS3a-d block 1(+) complementary site in Phypa1_1 65352 could have been due to highly efficient cleavage at the 3′ miR529(5′) site. No secondary siRNAs that corresponded to the region of Phypa1_1 65352 between the two sites were sequenced in our study.
The block 2(-) ta-siRNAs were predicted to target four distinct transcripts (see Supplemental Table 3 online), two of which encoded proteins homologous to angiosperm ARFs (Figures 4B and 4C). Cleavage products were detected for both ARF messages, thereby demonstrating that these P. patens ARF mRNAs were indeed regulated by Pp TAS3–derived ta-siRNAs (Figure 2B). Unexpectedly, these RLM 5′-RACE experiments revealed a second, longer species of uncapped mRNA for both of the P. patens ARF genes, leading us to investigate whether the longer products also corresponded to small RNA-mediated cleavage products. The putative upstream small RNA complementary sites did not appear to correspond to any ta-siRNA produced from Pp TAS3a-d nor were these two ARF genes predicted as targets of any P. patens miRNAs using established protocols (Allen et al., 2005; Rajagopalan et al., 2006). However, relaxing the stringency of the target prediction parameters revealed that the upstream cleavage sites corresponded with the bryophyte-specific miR1219 such that the observed cleavage product termini aligned exactly with the tenth nucleotide of complementarity (Figure 2B). We concluded that the two P. patens ARF genes were targeted at distinct sites by miR390-initiated, Pp TAS3a-d–derived ta-siRNAs and by the bryophyte-specific miR1219. We did not sequence any small RNAs that corresponded to the regions of the two P. patens ARF genes between the Pp TAS3a-d block 2(-) and miR1219 cleavage sites.
Phylogenetic comparison of P. patens ARF proteins to Arabidopsis ARF proteins revealed that the Pp TAS3a-d block 2(-) and miR1219-targeted ARFs were closely related to At ETT/ARF3 and At ARF4 (Figure 4C). At ETT/ARF3 and At ARF4 are targeted by miR390-phased, At TAS3–derived ta-siRNAs at two distinct complementary sites located downstream of the region coding for the ARF domain (Allen et al., 2005). Our data thus demonstrated that the arrangement of dual small RNA target sites within ARF mRNAs was shared between bryophytes and angiosperms despite divergence of both the sequence composition and of the biogenesis of the small RNAs themselves, providing a striking example of common regulatory functions being fulfilled by lineage-specific small RNAs. Moreover, the Pp TAS3a-d genes appear to transmute the expression of one miRNA (miR390) into two biologically relevant sets of related small RNAs that are capable of the coordinated regulation of a diverse set of regulatory transcripts. One set of Pp TAS3a-d ta-siRNAs regulates AP2 domain genes in concert with miR529(5′), while another set acts in concert with a lineage-specific miRNA, miR1219, to regulate the expression of ETT/ARF3 and ARF4-like genes.
Other Small RNAs in Basal Plants
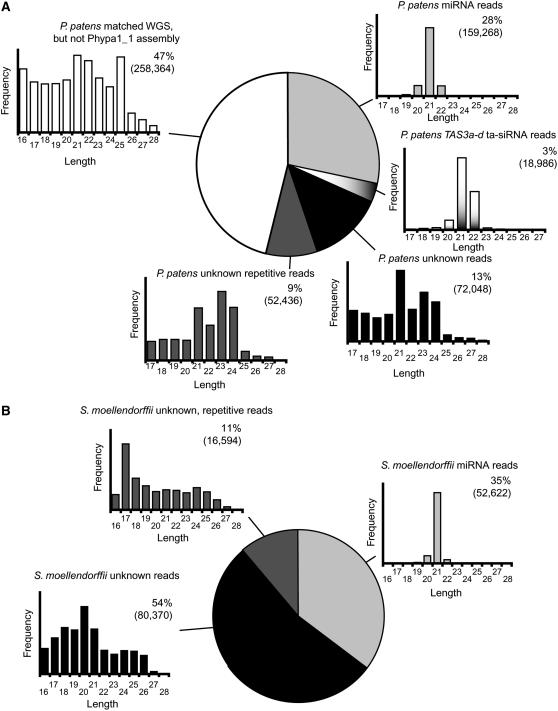
The entire set of 561,102 P. patens small RNA reads that matched at least one WGS trace (Axtell et al., 2006) were categorized based upon genomic and length characteristics (Figure 5A). miRNAs and ta-siRNAs accounted for 28 and 3% of the reads, respectively. Both subpopulations were dominated by 21-nucleotide sequences, with a considerable minority of 22-nucleotide sequences deriving from the Pp TAS3a-d ta-siRNAs (Figure 5A); these properties were consistent with those of angiosperm miRNAs and ta-siRNAs (Reinhart et al., 2002; Rajagopalan et al., 2006). Strikingly, 47% of the reads that initially matched at least one raw WGS trace did not match anywhere along the Phypa1_1 draft genome assembly. This subpopulation of reads included many short sequences of sizes inconsistent with the activities of any known Dicer endonuclease and some small RNAs between 20 and 24 nucleotides (Figure 5A). A substantial number of these reads were only sequenced once and disproportionately tended to match only a single WGS trace (data not shown), a statistically unlikely occurrence considering that shotgun coverage of the P. patens genome is ∼8×. This suggested that this subpopulation of reads contains some convergent sequencing errors (i.e., a sequencing error in a single small RNA and a single WGS trace that causes a spurious match). It might also contain breakdown products of RNAs and Dicer-derived small RNAs originating from genomic regions that are difficult to sample and assemble using shotgun sequencing and thus were left out of the draft genome assembly.
Figure 5.
Small RNA Populations in P. patens and S. moellendorffii.
(A) Categorization of 561,102 P. patens small RNA reads that matched at least one WGS trace. Reads were categorized as miRNAs (matching one or more of the 205 annotated miRNA hairpins), ta-siRNAs (from PpTAS3a-d), unknown (corresponding to 10 or fewer loci in the Phypa1_1 genome assembly), unknown-repetitive (corresponding to 11 or more loci in the Phypa1_1 genome assembly), or those not matching any loci in the Phypa1_1 genome assembly. Histograms display the length distributions of each of these small RNA populations. Reads that corresponded to the nuclear rRNAs and the chloroplast genome were not included.
(B) Categorization of 149,586 S. moellendorffii small RNA reads that matched at least one WGS trace. Reads are categorized as miRNAs (matching one or more of the 58 annotated miRNA hairpins), unknown (matching 100 or fewer WGS traces), or unknown-repetitive (corresponding to 101 or more WGS traces). Histograms display the length distributions of each of these small RNA populations. Reads that corresponded to nuclear rRNA were not included.
The remaining 22% of the P. patens reads that matched the version 1_1 genome assembly, but did not appear to originate from recognizable miRNA or ta-siRNA loci, were classified as unknown reads (Figure 5A). Repetitive unknown reads were those that corresponded to >10 loci in the draft genome assembly. Both repetitive and nonrepetitive subpopulations of unknown small RNA reads had clear size peaks at 21, 23, and 24 nucleotides, consistent with the hypothesis that many corresponded to endogenous siRNAs produced by DCL activities. In Arabidopsis, many endogenous 24-nucleotide siRNAs are dependent on the DCL3, AGO4, and AGO6 proteins (Qi et al., 2006; Kasschau et al., 2007; Zheng et al., 2007). Phylogenetic reconstructions of DCL proteins indicated that the four recognizable P. patens DCL proteins fell into three clades: a DCL1-like clade, a DCL3-like clade, and a DCL4-like clade (Figure 3C). Similarly, P. patens AGO proteins also fell into two distinct groups: an AGO1-like clade and an AGO4/6-like clade (Figure 3D). Taken together, these results suggest that, similar to angiosperms, P. patens produces at least some miRNA-independent (i.e., non-TAS) siRNAs that might participate in DNA silencing.
A similar analysis of the S. moellendorffii small RNA reads found that 35% of them corresponded to miRNA hairpins, most of which were 21 nucleotides in length (Figure 5B). The remaining 65% of the reads that matched at least one S. moellendorffii WGS trace were binned into repetitive and nonrepetitive subpopulations, which comprised 11 and 54% of the total, respectively (Figure 5B). The length distributions of the S. moellendorffii unknown reads were dispersed and overall were not consistent with the small RNA sizes produced by the biochemically or genetically characterized DCL proteins from other species. However, based upon the experience with P. patens small RNAs, we expect that approximately one-half of these unknown small RNAs will not be matched to the forthcoming S. moellendorffii draft genome sequence. As with P. patens, subtracting these reads may reveal the signature of a small population of potential miRNA-independent siRNAs in S. moellendorffii.
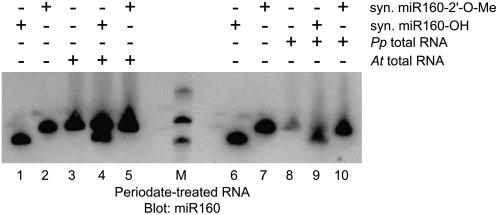
The proportion of P. patens and S. moellendorffii potential endogenous siRNAs was lower than that observed in similar deep sequencing projects that examined Arabidopsis small RNAs (Lu et al., 2005; Rajagopalan et al., 2006; Kasschau et al., 2007), raising the possibility that endogenous siRNAs play a less prominent role in genome regulation in basal plants. However, it is also possible that basal plant endogenous siRNAs possess biochemical characteristics that render them recalcitrant to the sequencing strategy employed in this study, which is highly selective for molecules with both a 5′ monophosphate and a 3′ hydroxyl. This would not be without precedent. In Caenorhabditis elegans, secondary siRNAs from an exogenous trigger and a subset of endogenous siRNAs are recalcitrant to this procedure because they possess 5′-triphosphates, reflecting their likely origins as short unprimed RdRp transcripts (Ruby et al., 2006; Pak and Fire, 2007; Sijen et al., 2007). Angiosperm siRNA and miRNA/miRNA* duplexes are methylated by the HEN1 protein on the 2′ hydroxyl of the 3′-most residue (Yu et al., 2005; Yang et al., 2006). Periodate treatment of P. patens total RNA demonstrated that the 3′-most residue of miR160 does not possess both free 2′ and 3′ hydroxyls, which was strongly suggestive of a conservation of HEN1-mediated small RNA methylation in bryophytes (Figure 6). Although methylation of the terminal 2′ hydroxyl would not have had a major effect on sequencing frequency, other modifications of this terminal ribose might have, especially if different classes of basal plant small RNAs possess different modifications.
Figure 6.
The 3′-Most Residue of P. patens miR160 Is Modified.
The indicated RNA samples were oxidized with periodate, blotted, and probed for miR160. Markers (M) from top to bottom are 24, 21, and 18 nucleotides, respectively.
DISCUSSION
Recent Diversification of Many Weakly Expressed miRNAs in Three Major Land Plant Lineages
The first small RNA sequencing efforts in both animals (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001) and plants (Llave et al., 2002b; Park et al., 2002; Reinhart et al., 2002) were performed at relatively low sequencing depths comprising hundreds of reads and therefore captured only the most abundant of the miRNAs expressed in the sampled tissues. Within both lineages, these initially discovered and most abundant miRNAs were very well conserved, leading to the hypothesis that miRNAs are generally ancient posttranscriptional regulators. The use of high-throughput DNA sequencing for small RNA discovery has allowed a 100- to 1000-fold increase in sequencing depth for various Arabidopsis small RNA samples (Lu et al., 2005, 2006; Henderson et al., 2006; Rajagopalan et al., 2006; Kasschau et al., 2007; Zhang et al., 2007). Analyses of these large data sets have revealed that the majority of Arabidopsis miRNA diversity (but not numerical abundance) is accounted for by weakly expressed miRNAs that lack identifiable homologs in other species. These nonconserved miRNAs tend to be encoded by only a single locus and also tend to have few, if any, confidently predicted targets. These properties have led to the suggestion that many of the weakly expressed, less well-conserved miRNAs represent a pool of evolutionarily transient regulators with minimal fitness consequences that may be occasionally recruited into biologically relevant circuits of posttranscriptional control but are more likely to decay and die due to mutational drift of the DCL1 recognized stem loops of their pri-miRNAs (Rajagopalan et al., 2006; Fahlgren et al., 2007). Many of the miRNAs discovered from the lycopod S. moellendorffii and P. patens in this study are reminiscent of the recently evolved miRNAs of Arabidopsis. Weakly expressed miRNAs that tend to originate from a single genomic locus and have few predicted mRNA targets comprise the majority of miRNA diversity in all three lineages. Thus, our data strongly support the hypothesis that miRNA-producing loci are frequently born and can rapidly die over relatively short evolutionary time scales, with only a select few being stabilized during their otherwise brief existence by recruitment into a beneficial regulatory interaction (Rajagopalan et al., 2006; Fahlgren et al., 2007). This process clearly occurs in all three deeply branching lineages of land plants (angiosperms, lycopods, and bryophytes), whose small RNAs have been sampled by deep sequencing, and might also occur in animals (Bentwich et al., 2005; Berezikov et al., 2006).
Common Functions for Plant Small RNAs, with and without Conservation of Sequence Identity
The targets of the deeply conserved P. patens miRNAs were clearly homologous with the known targets of the same miRNAs in Arabidopsis, as observed previously for deeply conserved miRNAs (Floyd and Bowman, 2004; Arazi et al., 2005; Axtell and Bartel, 2005; Floyd et al., 2006). The sequences of these targets strongly suggest that most of these deeply conserved regulatory circuits function to control transcriptional regulators that influence multicellular development and morphology. It will be fascinating to experimentally dissect how these conserved molecular modules have been recruited to control the very different developmental programs of angiosperms and bryophytes.
Analysis of the P. patens miRNA loci and their targets strongly suggests that miRNA-mediated feedback control on the miRNA pathway is also shared between bryophytes and angiosperms through the action of diverse miRNA loci generated through convergent and perhaps divergent evolutionary pathways. P. patens MIR1047 is expressed from the intron of a DCL1 homolog, and just as for Arabidopsis MIR838, processing of the stem loop by DCL1 cleaves the DCL1 pre-mRNA, implying a negative feedback loop. These two miRNAs appear to have arisen independently yet serve a common function and thus appear to constitute an example of convergent evolution for miRNA functions. Considering that large reservoirs of newly emergent and continuously evolving miRNAs have now been described in an angiosperm, a lycopod, and a moss, we expect that many additional examples of convergent miRNA functions will be found in land plants. Whether miR904 targeting of AGO1-like genes in P. patens and miR168 targeting of AGO1 in angiosperms was achieved through convergent or divergent emergence of these diverse miRNAs was less clear. Biogenesis from the same arm of the miRNA hairpins and cleavage at similar locations within the homologous AGO1 messages were both consistent with divergence from a common ancestor, but in either evolutionary scenario, our data demonstrated that analogous regulatory specificity of plant miRNAs does not always entail conservation of the mature miRNA sequences. Similar observations have been made for animal miRNAs, most notably the vertebrate miR-196 and insect miR-iab-4, which both target Ubx orthologs in their respective lineages (Yekta et al., 2004; Ronshaugen et al., 2005).
Just as for the miRNAs that appeared to mediate feedback regulation upon DCL and AGO genes in plants, the biologically relevant ta-siRNAs emanating from plant TAS3 loci differed in sequence but were found to perform analogous molecular functions in mosses and seed plants. The two P. patens ARF homologs possessed dual cleavage sites just downstream of the conserved ARF domain (Figure 2B), a location and arrangement closely mirroring that of the cleavage sites within Arabidopsis ETT/ARF3 and ARF4 (Allen et al., 2005). These data involving miR390-triggered siRNA regulation of similar angiosperm and bryophyte ARF transcripts could thus be regarded as evidence that the At TAS3 and Pp TAS3a-d genes might have descended from a common molecular ancestor. However, Arabidopsis ETT/ARF3 and ARF4 both possessed dual complementary sites to identical At TAS3–derived ta-siRNAs, whereas the P. patens ARFs possessed only one site complementary to Pp TAS3 ta-siRNAs. Cleavage at the other site in the P. patens ARF mRNAs was guided by the lineage-specific miR1219, which did not have any detectable sequence similarity to the ARF-targeting At TAS3 ta-siRNAs nor to any Pp TAS3a-d–derived ta-siRNAs. These observations add to the examples of convergent evolutionary origins for similarly functioning small RNAs, while illustrating that cleavage of a homologous target at an analogous location does not always indicate descent from a common ancestor. Moreover, the ARF-targeting ta-siRNAs are derived from the (+) strand of the angiosperm TAS3 loci, whereas they derived from the (–) strands of the moss TAS3 loci, suggesting a more complex evolutionary path for plant TAS3 loci than simple sequence divergence from a common ancestor. Data from additional plant lineages might provide important clues for distinguishing between these interesting possibilities.
The AGO1-targeting miRNAs and the miR390-phased ta-siRNAs might be the descendants of anciently derived small RNA–target interactions that have coordinately diverged from common molecular ancestors over long periods of time. This process would thus have hidden the ancestral relationships of the functionally homologous small RNA–target interactions in divergent contemporary organisms, making them appear to be nonconserved when considering primary sequence alone. In other cases, such as miRNAs excised from DCL1 pre-mRNAs, independently derived, nonconserved miRNAs appear to have converged upon an identical regulatory function. We conclude that the sequence diversity of miRNAs and ta-siRNAs expressed in various plants can sometimes obfuscate analogous regulatory functions.
METHODS
Small RNA sequencing
Sequencing of Physcomitrella patens small RNAs has been described (Axtell et al., 2006). Selaginella moellendorffii was purchased from Plant Delights Nursery. A total RNA sample derived from the above-ground parts of S. moellendorffii was isolated using the method of Chang et al. (1993). Small RNAs from this sample were sequenced as described (Axtell et al., 2006).
Annotation of miRNAs and Target Predictions
Sequenced small RNAs were filtered to remove contaminant sequences derived from rRNA or the synthetic small RNAs used as guides during the small RNA excision procedure. The remaining sequences were then compared against the relevant WGS traces available from the National Center for Biotechnology Information (NCBI) trace archive and the corresponding matches recorded. After discarding small RNAs that were repetitive (matched >1000 different WGS traces), genomic sequences surrounding small RNA hits were examined for their potential to form an RNA secondary structure typical of pri-miRNA stem loops. A 600-nucleotide window centered upon a given small RNA hit was analyzed using MIRcheck (Jones-Rhoades and Bartel, 2004). Consensus genomic sequences were derived for each potential miRNA locus that passed MIRcheck by use of BLAT (Kent, 2002) to capture redundant WGS traces followed by consensus assembly using CAP3 (Huang and Madan, 1999). All sequenced small RNAs that matched either polarity of the assembled consensus sequences of potential miRNA loci were captured and aligned. Candidate loci where two dominant sequences accumulated from only a single polarity and that, when analyzed in the context of a predicted RNA secondary structure, could form a typical miRNA/miRNA* duplex were annotated as miRNAs. In cases where no sequence corresponding to the presumed miRNA* was sequenced, a locus was still annotated as a miRNA if there was a paralogous locus from which a miRNA/miRNA* was isolated. After the P. patens draft genome Phypa1_1 assembly became available, the WGS-derived consensus miRNA loci were all successfully mapped.
The transcripts annotated from the draft P. patens genome assembly were used to predict the potential targets of P. patens miRNAs. The list of mature P. patens miRNAs (see Supplemental Table 1 online) was first collapsed to generate a nonredundant set of queries, and targets were predicted as described (Allen et al., 2005; Rajagopalan et al., 2006). To assess the signal-to-noise ratio of these predictions, a set of randomized sequences with mononucleotide compositions identical to the query and di- and trinucleotide compositions similar to the transcript database (Farh et al., 2005) was produced for each miRNA query and used to predict targets (see Supplemental Table 3 online).
Molecular Phylogenetic Analyses
P. patens proteins homologous to Arabidopsis thaliana DCL1 (At1g01040.1), AGO1 (At1g48410.1), and ETT/ARF3 (At2g33860.1) were found using BLASTP and manually examined to identify hits with the domain architectures expected of the respective protein families. Alignments were constructed using the T-COFFEE server (Poirot et al., 2003) under default settings (alignment computations using Mlalign_id_pair and Mslow_pair). Alignment scores were 50, 63, and 57 for the DCL, AGO, and ARF alignments, respectively. Phylogenetic analyses were performed within MEGA 3.1 (Kumar et al., 2004) using neighbor joining. Options common to all three analyses were as follows: phylogeny test and options = bootstrap, 1000 replicates; model = amino: Poisson correction; substitutions to include = all; pattern among lineages = same (homogenous); rates among sites = uniform rates. The DCL1 and AGO1 analyses used gaps/missing data = pairwise deletion and the ARF analyses used gaps/missing data = complete deletion. The sequence alignments used to construct these phylogenies are available as Supplemental Figures 2 to 4 online.
Isolation of Small RNA–Mediated Cleavage Products
Isolation of uncapped mRNAs using RLM 5′-RACE was performed essentially as described (Kasschau et al., 2003) using P. patens RNA from 17-d-old protonematal cultures with young leafy gametophores. Experiments used cDNA produced by reverse transcription of poly(A)+ RNA with the GeneRacer oligo(dT) oligo (5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTGTTTTTTTTTTTTTTTTTTTTTTTT-3′), except those indicated otherwise in Figure 3. All cleavage products were amplified using two rounds of nested PCR with the GeneRacer 5′ oligo (5′-CGACTGGAGCACGAGGACACTGA-3′) and the GeneRacer 5′ nested oligo (5′-GGACACTGACATGGACTGAAGGAGTA-3′) in conjunction with outer and nested gene-specific oligos. Products were then cloned and sequenced to determine the 5′ ends. Pp DCL1a oligos were as follows: exon 8 RT oligo, 5′-GGCTTCCGAGCACGACCACGAGACTGAA-3′; Pp DCL1a outer (spans intron seven/exon eight junction), 5′-GCAACGTCACCTAGCCCAACCAACTT-3′; and DCL1a nested (intron seven), 5′-GCTGAGCAAACCAGCTCGACAAGATG-3′. All other gene-specific oligos are listed in Supplemental Table 4 online.
Periodate treatments
Total RNA from wild-type 7-d-old Arabidopsis seedlings and 7-d-old P. patens protonemata was isolated using TRI-Reagent (Ambion). Synthetic miR160 RNAs (5′-UGCCUGGCUCCCUGUAUGCCA-3′) with a 5′ monophosphate and either 2′, 3′ hydroxyls or a 2′-O-methyl, 3′ hydroxyl on the 3′-most residue were purchased from Dharmacon. In separate reactions, 90 μg of Arabidopsis and P. patens total RNA and 10 fmols of the synthetic miR160 RNAs were incubated for 45 min in 10 mM NaIO4 at 20° in the dark, followed by addition of l-Lys, pH 8.5, to a concentration of 1 M and incubation at 45° for 90 min. Reactions were then phenol-chloroform extracted, desalted through Sephadex G-25 spin columns (GE Healthcare), and ethanol precipitated. Plant RNA reactions were resuspended in 15 μL water, while reactions with synthetic miR160 were resuspended in 4 μL water. The samples were then fractionated, blotted, and hybridized with an oligonucleotide probe against miR160 as described (Axtell and Bartel, 2005). Lanes containing plant RNA had 5 μL of the relevant sample, while lanes containing synthetic RNAs had 1 μL of the relevant sample.
Databases and Gene Family Annotations
All referenced Arabidopsis sequences were obtained from The Arabidopsis Information Resource version 6 genome annotation; in cases where multiple versions of a transcript and/or protein were annotated, version 1 was used (e.g., At1g01040.1). The nomenclature of the Arabidopsis ARF, DCL, and AGO gene families was derived from Okushima et al. (2005), Margis et al. (2006), and Fagard et al. (2000), respectively. All referenced Oryza sativa sequences were obtained from The Institute for Genomic Research (TIGR) version 4 genome annotation; in cases where multiple versions of a transcript and/or protein were annotated, version 1 was used (e.g., Os06g39640.1). The nomenclature of the O. sativa DCL and AGO gene families was derived from Margis et al. (2006) and Shi et al. (2007), respectively. Annotations of the S. moellendorffii small RNAs made use of the raw WGS traces available from the NCBI trace server. Analyses of P. patens small RNAs made use of the Phypa1_1 draft assembly of the genome from the Joint Genome Institute and the associated transcript and protein models, except where noted. C. reinhardtii sequences were derived from the Joint Genome Institute's genome version 2.0; the nomenclature of the Cr DCL1, Cr AGO1, and Cr AGO2 genes follows Schroda (2006).
Accession Numbers
P. patens and S. moellendorffii small RNA data sets have been deposited with the Gene Expression Omnibus under accession numbers GSE5103 and GSE7320, respectively. Partial cDNAs corresponding to P. patens miRNA-mediated cleavage products have been deposited with GenBank under accession numbers ES492157 to ES492185. All novel miRNAs have been registered with miRBase. The Arabidopsis Genome Initiative identifiers for the Arabidopsis genes/proteins mentioned by other names in this article are as follows: At RDR6, At3g49500; At TAS3a, At3g17185; At DCL1, At1g01040; At DCL2, At3g03300; At DCL3, At3g43920; At DCL4, At5g20320; At AGO1, At1g48410; At AGO2, At1g31280; At AGO3, At1g31290; At AGO4, At2g27040; At AGO5, At2g27880; At AGO6, At2g32940; At AGO7, At1g69440; At AGO8, At5g21030; At AGO9, At5g21150; At ZLL/PNH, At5g43810; At ARF1, At1g59750; At ARF2, At5g6200; At ETT/ARF3, At2g33860; At ARF4, At5g60450; At MP/IAA24/ARF5, At1g19850; At ARF6, At1g30330; At ARF7, At5g20730; At ARF8, At5g37020; At ARF9, At4g23980; At ARF10, At2g28350; At ARF11, At2g46520; At ARF12, At1g34310; At ARF13, At1g34170; At ARF14, At1g35540; At ARF15, At1g35520; At ARF16, At4g30080; At ARF17, At1g77850; At ARF18, At3g61830; At ARF19, At1g19220; At ARF20, At1G35240; At ARF21, At1g34410; At ARF22, At1g34390; At ARF23, At1g43950. The TIGR locus identifiers for the O. sativa genes/proteins mentioned by other names in this article are as follows: Os DCL1, Os03g02970; Os DCL2a, Os03g38740; Os DCL2b, Os09g14610; Os DCL3a, Os01g68120; Os DCL3b, Os10g34430; Os DCL4, Os04g43050; Os AGO7, Os03g33550. Based on the results of phylogenetic analysis (Figure 3), we have designated Phypa1_1 205895, Phypa1_1 163666, Phypa1_1 130425, and Phypa1_1 234670 as Pp DCL1a, Pp DCL1b, Pp DCL3, and Pp DCL4, respectively. Similarly, Phypa1_1 205541, Phypa1_1 158832, and Phypa1_1 141045 have been designated as Pp AGO1a, Pp AGO1b, and Pp AGO1c, respectively. Most P. patens genes mentioned or used for analyses were as annotated from the Phypa1_1 data set, except the following: Pp RDR6, DQ531710; Pp TAS3a, BK005825; Pp TAS3b, BK005826; Pp TAS3c, BK005827; Pp TAS3d, BK005828; PPM1, AF150932; Pp MADS1, AB067688; Pp SBP3, AJ968318; Pp HB10, DQ385518; Pp C3HDZ1, DQ385516; Pp C3HDZ2, DQ385517. The Joint Genome Institute gene model names for the C. reinhardtii proteins referenced in the text are as follows: Cr DCL1, C_130110; Cr AGO1, C_130206; Cr AGO2, C_1700017.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alternative miRNA Production.
Supplemental Figure 2. T-COFFEE Alignment of Arabidopsis, O. sativa, C. reinhardtii, and P. patens DCL Proteins.
Supplemental Figure 3. T-COFFEE Alignment of Arabidopsis, O. sativa, C. reinhardtii, and P. patens AGO Proteins.
Supplemental Figure 4. T-COFFEE Alignment of Arabidopsis and P. patens ARF Proteins.
Supplemental Table 1. P. patens miRNA Loci.
Supplemental Table 2. S. moellendorffii miRNA Loci.
Supplemental Table 3. Predicted and Validated Targets of P. patens miRNAs and ta-siRNAs.
Supplemental Table 4. RLM 5′-RACE Gene-Specific Oligo Sequences.
Supplementary Material
Acknowledgments
We thank the Joint Genome Institute and the Physcomitrella patens sequencing consortium members for the availability of the draft genome assembly. We thank Graham Ruby for many stimulating discussions, Ramya Rajagopalan for discussions and critical comments, and Nina Federoff and John Carlson for constructive comments on this manuscript. This work was supported by awards from the National Institutes of Health, the Prix Louis D. from the Institut de France (D.P.B.), the Helen Hay Whitney Foundation, and Pennsylvania State University (M.J.A.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Michael J. Axtell (mja18@psu.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouche, N., Gasciolli, V., and Vaucheret, H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16 927–932. [DOI] [PubMed] [Google Scholar]
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. [DOI] [PubMed] [Google Scholar]
- Allen, E., Xie, Z., Gustafson, A.M., Sung, G.H., Spatafora, J.W., and Carrington, J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36 1282–1290. [DOI] [PubMed] [Google Scholar]
- Arazi, T., Talmor-Neiman, M., Stav, R., Riese, M., Huijser, P., and Baulcombe, D.C. (2005). Cloning and characterization of micro-RNAs from moss. Plant J. 43 837–848. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., and Bartel, D.P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17 1658–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., Jan, C., Rajagopalan, R., and Bartel, D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127 565–577. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2005). RNA silencing. Trends Biochem. Sci. 30 290–293. [DOI] [PubMed] [Google Scholar]
- Baumberger, N., and Baulcombe, D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich, I., et al. (2005). Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37 766–770. [DOI] [PubMed] [Google Scholar]
- Berezikov, E., Thuemmler, F., van Laake, L.W., Kondova, I., Bontrop, R., Cuppen, E., and Plasterk, R.H. (2006). Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 38 1375–1377. [DOI] [PubMed] [Google Scholar]
- Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11 113–116. [Google Scholar]
- Chuck, G., Cigan, A.M., Saeteurn, K., and Hake, S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39 544–549. [DOI] [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., and Sharp, P.A. (2003). siRNAs can function as miRNAs. Genes Dev. 17 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., Boutet, S., Morel, J.B., Bellini, C., and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N., Howell, M.D., Kasschau, K.D., Chapman, E.J., Sullivan, C.M., Cumbie, J.S., Givan, S.A., Law, T.F., Grant, S.R., Dangl, J.L., and Carrington, J.C. (2007). High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE 2 e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N., Montgomery, T.A., Howell, M.D., Allen, E., Dvorak, S.K., Alexander, A.L., and Carrington, J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16 939–944. [DOI] [PubMed] [Google Scholar]
- Farh, K.K., Grimson, A., Jan, C., Lewis, B.P., Johnston, W.K., Lim, L.P., Burge, C.B., and Bartel, D.P. (2005). The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 310 1817–1821. [DOI] [PubMed] [Google Scholar]
- Fattash, I., Voss, B., Reski, R., Hess, W.R., and Frank, W. (2007). Evidence for the rapid expansion of microRNA-mediated regulation in early land plant evolution. BMC Plant Biol. 7 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd, S.K., and Bowman, J.L. (2004). Gene regulation: Ancient microRNA target sequences in plants. Nature 428 485–486. [DOI] [PubMed] [Google Scholar]
- Floyd, S.K., Zalewski, C.S., and Bowman, J.L. (2006). Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics 173 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D., Collier, S.A., Byrne, M.E., and Martienssen, R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16 933–938. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M. (2005). Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 579 5822–5829. [DOI] [PubMed] [Google Scholar]
- Henderson, I.R., Zhang, X., Lu, C., Johnson, L., Meyers, B.C., Green, P.J., and Jacobsen, S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38 721–725. [DOI] [PubMed] [Google Scholar]
- Henschel, K., Kofuji, R., Hasebe, M., Saedler, H., Munster, T., and Theissen, G. (2002). Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 19 801–814. [DOI] [PubMed] [Google Scholar]
- Howell, M.D., Fahlgren, N., Chapman, E.J., Cumbie, J.S., Sullivan, C.M., Givan, S.A., Kasschau, K.D., and Carrington, J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., and Madan, A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, C., Willmann, M.R., Wu, G., Yoshikawa, M., de la Luz Gutierrez-Nava, M., and Poethig, R.S. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133 2973–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C., Bowman, L., Adai, A.T., Vance, V., and Sundaresan, V. (2007). CSRDB: A small RNA integrated database and browser resource for cereals. Nucleic Acids Res. 35 D829–D833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., and Bartel, D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14 787–799. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., Bartel, D.P., and Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57 19–53. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Fahlgren, N., Chapman, E.J., Sullivan, C.M., Cumbie, J.S., Givan, S.A., and Carrington, J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5 e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4 205–217. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. (2002). BLAT–The BLAST-like alignment tool. Genome Res. 12 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.T., and Ashton, N.W. (2000). Ancestry of plant MADS-box genes revealed by bryophyte (Physcomitrella patens) homologues. New Phytol. 147 505–517. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294 853–858. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., and Ambros, V. (2001). An extensive class of small RNAs in Caenorhabditis elegans. Science 294 862–864. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., and Kim, V.N. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425 415–419. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Kim, M., Han, J., Yeom, K.H., Lee, S., Baek, S.H., and Kim, V.N. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Li, W., and Jin, Y.X. (2005). Computational identification of novel family members of microRNA genes in Arabidopsis thaliana and Oryza sativa. Acta Biochim. Biophys. Sin. (Shanghai) 37 75–87. [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., and Hannon, G.J. (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305 1437–1441. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. (2002. b). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002. a). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lu, C., Kulkarni, K., Souret, F.F., MuthuValliappan, R., Tej, S.S., Poethig, R.S., Henderson, I.R., Jacobsen, S.E., Wang, W., Green, P.J., and Meyers, B.C. (2006). MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 16 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Tej, S.S., Luo, S., Haudenschild, C.D., Meyers, B.C., and Green, P.J. (2005). Elucidation of the small RNA component of the transcriptome. Science 309 1567–1569. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., and Vaucheret, H. (2006). Functions of microRNAs and related small RNAs in plants. Nat. Genet. 38(Suppl): S31–S36. [DOI] [PubMed] [Google Scholar]
- Margis, R., Fusaro, A.F., Smith, N.A., Curtin, S.J., Watson, J.M., Finnegan, E.J., and Waterhouse, P.M. (2006). The evolution and diversification of Dicers in plants. FEBS Lett. 580 2442–2450. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., and Tuschl, T. (2004). Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15 185–197. [DOI] [PubMed] [Google Scholar]
- Molnár, A., Schwach, F., Studholme, D.J., Thuenemann, E.C., and Baulcombe, D.C. (2007). miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature http://dx.doi.org/10.1038/nature05903. [DOI] [PubMed]
- Nobuta, K., Venu, R.C., Lu, C., Belo, A., Vemaraju, K., Kulkarni, K., Wang, W., Pillay, M., Green, P.J., Wang, G.L., and Meyers, B.C. (2007). An expression atlas of rice mRNAs and small RNAs. Nat. Biotechnol. 25 473–477. [DOI] [PubMed] [Google Scholar]
- Okushima, Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak, J., and Fire, A. (2007). Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315 241–244. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 257–263. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirot, O., O'Toole, E., and Notredame, C. (2003). Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 31 3503–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y., Denli, A.M., and Hannon, G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19 421–428. [DOI] [PubMed] [Google Scholar]
- Qi, Y., He, X., Wang, X.J., Kohany, O., Jurka, J., and Hannon, G.J. (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443 1008–1012. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110 513–520. [DOI] [PubMed] [Google Scholar]
- Ronshaugen, M., Biemar, F., Piel, J., Levine, M., and Lai, E.C. (2005). The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 19 2947–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby, J.G., Jan, C., Player, C., Axtell, M.J., Lee, W., Nusbaum, C., Ge, H., and Bartel, D. (2006). Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in Caenorhabditis elegans. Cell 127 1193–1207. [DOI] [PubMed] [Google Scholar]
- Schroda, M. (2006). RNA silencing in Chlamydomonas: Mechanisms and tools. Curr. Genet. 49 69–84. [DOI] [PubMed] [Google Scholar]
- Shi, Z., Wang, J., Wan, X., Shen, G., Wang, X., and Zhang, J. (2007). Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226 99–108. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Steiner, F.A., Thijssen, K.L., and Plasterk, R.H. (2007). Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315 244–247. [DOI] [PubMed] [Google Scholar]
- Singer, S.D., Krogan, N.T., and Ashton, N.W. (2007). Clues about the ancestral roles of plant MADS-box genes from a functional analysis of moss homologues. Plant Cell Rep. http://dx.doi.org/10.1007/s00299-007-0312-0. [DOI] [PubMed]
- Sunkar, R., Girke, T., Jain, P.K., and Zhu, J.K. (2005). Cloning and characterization of microRNAs from rice. Plant Cell 17 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmor-Neiman, M., Stav, R., Frank, W., Voss, B., and Arazi, T. (2006. a). Novel micro-RNAs and intermediates of micro-RNA biogenesis from moss. Plant J. 47 25–37. [DOI] [PubMed] [Google Scholar]
- Talmor-Neiman, M., Stav, R., Klipcan, L., Buxdorf, K., Baulcombe, D.C., and Arazi, T. (2006. b). Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 48 511–521. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Mallory, A.C., and Bartel, D.P. (2006). AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 22 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., and Crete, P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16 69–79. [DOI] [PubMed] [Google Scholar]
- Wang, W., et al. (2005). Construction of a bacterial artificial chromosome library from the spikemoss Selaginella moellendorffii: A new resource for plant comparative genomics. BMC Plant Biol. 5 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds, E., and Plasterk, R.H. (2005). MicroRNA function in animal development. FEBS Lett. 579 5911–5922. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Fahlgren, N., Calamar, A., Givan, S.A., and Carrington, J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Kasschau, K.D., and Carrington, J.C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13 784–789. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Ebright, Y.W., Yu, B., and Chen, X. (2006). HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 34 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta, S., Shih, I.H., and Bartel, D.P. (2004). MicroRNA-directed cleavage of HOXB8 mRNA. Science 304 594–596. [DOI] [PubMed] [Google Scholar]
- Yoshikawa, M., Peragine, A., Park, M.Y., and Poethig, R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R.W., Steward, R., and Chen, X. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Henderson, I.R., Lu, C., Green, P.J., and Jacobsen, S.E. (2007). Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA 104 4536–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, T., Li, G., Mi, S., Li, S., Hannon, G.J., Wang, X.J., and Qi, Y. (2007). A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 21 1190–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X., Zhu, J., Kapoor, A., and Zhu, J.K. (2007). Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 26 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 716–719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.