Abstract
Two Arabidopsis thaliana ABC transporter genes linked to auxin transport by various previous results were studied in a reverse-genetic fashion. Mutations in Multidrug Resistance-Like1 (MDR1) reduced acropetal auxin transport in roots by 80% without affecting basipetal transport. Conversely, mutations in MDR4 blocked 50% of basipetal transport without affecting acropetal transport. Developmental and auxin distribution phenotypes associated with these altered auxin flows were studied with a high-resolution morphometric system and confocal microscopy, respectively. Vertically grown mdr1 roots produced positive and negative curvatures threefold greater than the wild type, possibly due to abnormal auxin distribution observed in the elongation zone. However, upon 90° reorientation, mdr1 gravitropism was inseparable from the wild type. Thus, acropetal auxin transport maintains straight growth but contributes surprisingly little to gravitropism. Conversely, vertically maintained mdr4 roots grew as straight as the wild type, but their gravitropism was enhanced. Upon reorientation, curvature in this mutant developed faster, was distributed more basally, and produced a greater total angle than the wild type. An amplified auxin asymmetry may explain the mdr4 hypertropism. Double mutant analysis indicated that the two auxin transport streams are more independent than interdependent. The hypothesis that flavanols regulate MDR-dependent auxin transport was supported by the epistatic relationship of mdr4 to the tt4 phenylpropanoid pathway mutation.
INTRODUCTION
The hormone auxin is an important regulator of root growth and development. The mechanisms responsible for distributing auxin from sites of synthesis and their relationship to auxin-mediated development have been major subjects of study since metabolically driven, polar auxin transport was first established (Goldsmith, 1977; Muday and DeLong, 2001; Leyser, 2006). Auxin entering the root from the shoot is transported through the central tissues of the root toward the tip, where it is presumably combined with apically produced auxin (Ljung et al., 2005), redistributed toward the flanks, and then transported basipetally through the lateral root cap and epidermis (Swarup and Bennett, 2003). The strong bias in the direction of transport within a tissue results from asymmetry in the cellular localization of an efflux apparatus that contains PIN-type efflux facilitators (Friml, 2003). For example, localization of PIN1 at the apical ends of cells in the stele of the root is thought to promote net movement of auxin toward the root tip (Blilou et al., 2005). Laterally symmetric PIN3 in the columella cells of a vertically growing root facilitates a uniform centrifugal flow of auxin toward the flanks. When the root is rotated by 90°, PIN3 distribution becomes asymmetric, accumulating along the lower sidewall (Friml et al., 2002), which shifts the lateral auxin stream to the lower flank of the root. The auxin is presumed to enter the basipetal stream, which depends on the basally localized PIN2 protein (Müller et al., 1998; Abas et al., 2006) for its directionality. This results in a higher concentration of auxin on the lower side of the root in the zone 50 to 800 μm from the tip where cells are rapidly elongating. Because auxin concentrations above the nanomolar range inhibit cell elongation in this region of the root, which can be separated into distal and central elongation zones (Evans et al., 1994; Wolverton et al., 2002), expansion of cells on the lower side slows relative to the upper side and downward curvature results. Mutations in PIN2 (Chen et al., 1998; Müller et al., 1998) or auxin transport inhibitors such as naphthylphthalamic acid (NPA) impair basipetal auxin transport and gravitropism (Muday, 2001), consistent with the above explanation.
In addition to the PIN proteins, the multidrug resistance/P-glycoprotein (MDR/PGP)-type ABC transporters also function in the process of auxin transport and distribution (Noh et al., 2001; Geisler and Murphy, 2006). These ATP binding, large, glycosylated membrane proteins were first identified in plants by Dudler and Hertig (1992) and shown by overexpression and antisense manipulations to affect hypocotyl elongation by Sidler et al. (1998). A connection to auxin and tropisms for two of the 22 members of the family was shown by studies of the mdr1 and pgp1 mutants. (MDR1 [At3g28860] has also been referred to as PGP19 [Martinoia et al., 2001] and MDR11 [Sánchez-Fernández et al., 2001].) Basipetal transport of auxin in the hypocotyl and inflorescence stems of mdr1 mutants was impaired by ∼80% and nearly completely blocked in mdr1 pgp1 double mutants (Noh et al., 2001). Surprisingly, tropisms were enhanced in mdr1 hypocotyls relative to the wild type and even more so in mdr1 pgp1 double mutants (Noh et al., 2003). The explanation offered to reconcile a block in auxin transport with enhanced tropisms was that reduced polar transport increased the potential for lateral auxin gradients to form in response to tropic stimuli (Noh et al., 2003).
Biochemical evidence of a role for these ABC transporters in auxin transport has also been obtained. The MDR1 and PGP1 proteins when extracted from plant membranes bind to an NPA affinity chromatography column, and yeast expressing MDR1 also bind NPA (Noh et al., 2001). Expression of PGP1 in cultured HeLa cells conferred auxin efflux activity upon the heterologous system, and auxin efflux from pgp1 protoplasts was reduced relative to the wild type (Geisler et al., 2005). These and related results (Petrášek et al., 2006) can be interpreted as evidence that MDR/PGP proteins are auxin efflux transporters (Geisler and Murphy, 2006), but they may also affect the mechanism responsible for the subcellular localization of PIN proteins (Noh et al., 2003), which has at least one NPA-sensitive step (Geldner et al., 2003). Recent experiments indicated that a direct, synergistic interaction between MDR1 and PIN1 affects the rate and substrate specificity of auxin transport relative to the transport properties of either single molecule measured separately (Blakeslee et al., 2007).
MDR4/PGP4 (At2g47000) is 46% identical to MDR1 at the overall amino acid level and resides in a different subclade within the MDR family. However, restricting comparisons to only the C termini shows MDR4 to be the family member most similar to MDR1. The C terminus is of particular interest because in the case of MDR1 it has been shown to interact with the TWD1 immunophilin-like protein (Geisler et al., 2003). The wavy inflorescence stems and hypocotyls and epinastic cotyledons of twd1 seedlings create a phenotype very similar to that of mdr1 pgp1 mutants (Geisler et al., 2003), and TWD1 affects auxin efflux activities attributable to MDR/PGP proteins in heterologous systems (Bouchard et al., 2006), indicating that the TWD1 interaction at the C terminus is relevant to MDR1 function. Therefore, the similarity between MDR1 and MDR4 C termini may be evidence of functional similarity, even though one study shows MDR4 mediates auxin accumulation, not efflux, in heterologous systems (Terasaka et al., 2005). Particularly relevant to this work is that the expression patterns of MDR4 and MDR1 in the root are more complementary than overlapping. MDR4 is expressed primarily in the outer cell layers of the root (Birnbaum et al., 2003; Terasaka et al., 2005; www.arexdb.org), whereas MDR1 is present primarily in cells of the central cylinder (Wu et al., 2007). These two mutants were used here as genetic tools to dissect the roles of the two antiparallel auxin streams in root growth and development, which was quantified with unprecedented spatiotemporal resolution by a novel morphometric platform based on computer vision techniques (Miller et al., 2007). An accompanying article (Wu et al., 2007) used the same mutants to investigate the role of the auxin transport streams in lateral root growth and development.
RESULTS
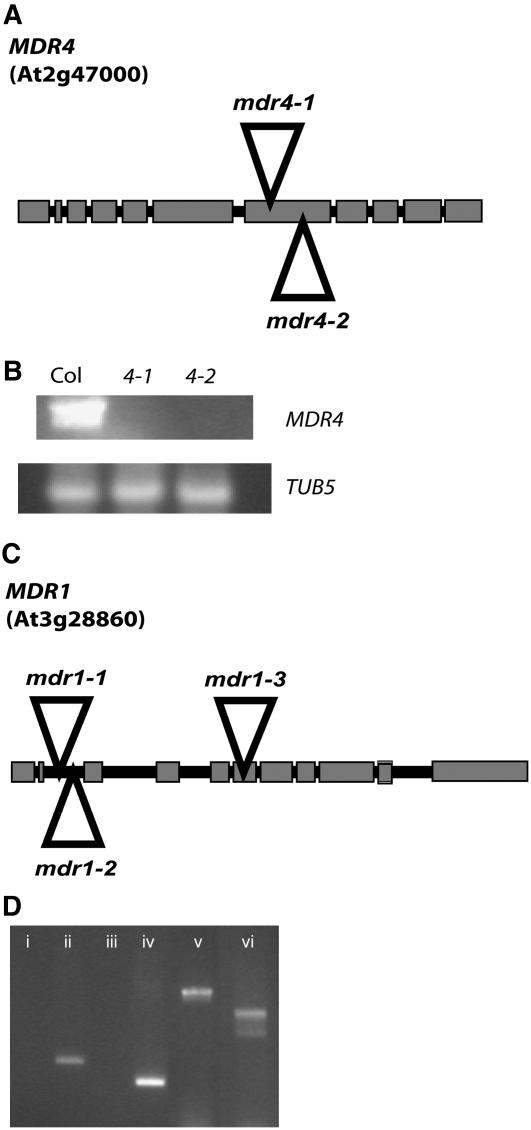
The experiments described here were performed with T-DNA insertion mutants of two genes in the MDR family of ABC transporters. The mdr1-1 and mdr1-2 null alleles (Wassilewskija [Ws] ecotype) were isolated from the University of Wisconsin collection and described in detail by Noh et al. (2001). The mdr4 alleles used here are in the Columbia-0 (Col-0) ecotype, and their characterization is presented in Figures 1A and 1B. To generate mdr1 mdr4 double mutants without mixing ecotypes, the mdr1-3 knockout allele in the Col-0 background was isolated. Description of the mdr1-3 allele and PCR results to document the double mutant genotype are presented in Figures 1C and 1D.
Figure 1.
Genomic Structure of T-DNA Insertion Alleles.
(A) Structure of the MDR4 gene. Boxes represent exons, and lines represent introns. The positions of the T-DNA insertions in mdr4-1 (SALK_010207) and mdr4-2 (SALK_072038) are represented by triangles.
(B) RT-PCR analysis of MDR4 transcript levels in Col-0 and the two independent mdr4 T-DNA alleles used in this study.
(C) Gene diagram of mdr1 T-DNA insertion alleles used in this study. Ws is the genetic background of the previously described mdr1-1 and mdr1-2 alleles. To obtain an allele in the Col-0 background, mdr1-3 (Salk_033455) was isolated.
(D) PCR results showing interruption of MDR1 and MDR4 in the mdr1-3 mdr4-1 double mutant (i to iv) and proper function of the gene-specific primers on wild-type DNA (v and vi). (i) MDR1 5′ primer plus MDR1 3′ primer gave no product. (ii) MDR1 5′ primer plus T-DNA Lb1a primer gave a product with the expected size. (iii) MDR4 5′ primer plus MDR4 3′ primer gave no product. (iv) MDR4 3′ primer plus T-DNA Lb1a primer gave a product with the expected size. (v) Wild-type DNA: product of the MDR1 5′ primer plus the MDR1 3′ primer was the expected size. (vi) Wild-type DNA: product of the MDR4 5′ primer plus the MDR4 3′ primer was the expected size.
Genetic Dissection of Basipetal and Acropetal Auxin Transport Streams
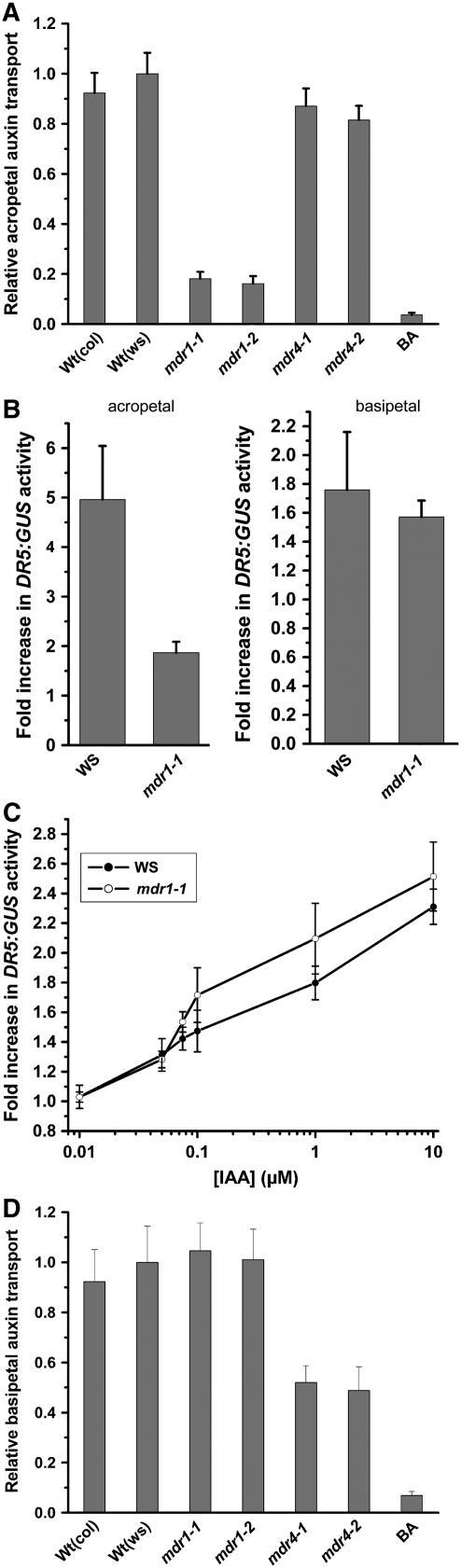
The extent to which polar auxin transport depends on MDR1 in both axial directions was examined by measuring the effects of mdr1 knockout mutations on the movement of locally applied 3H-indole-3-acetic acid (3H-IAA). The data in Figure 2A show that acropetal transport of auxin placed near the primary root-shoot junction of mdr1-1 and mdr1-2 seedlings was only ∼20% of the wild type. A ProDR5:β-glucuronidase (GUS)–based assay was designed to independently test the conclusion that acropetal auxin transport is highly MDR1 dependent in root. Endogenous levels of ProDR5-driven GUS activity in apical portions of roots quantified with a 4-methylumbelliferyl-β-d-glucuronide (MUG) assay were 30% lower in mdr1 than the wild type, indicating that impaired acropetal transport resulted in less auxin in general in the mdr1 root. Application of IAA to the root-shoot junction raised the GUS activity in the apical half of the root approximately fivefold in the wild type but less than twofold in mdr1-1 (Figure 1B). The interpretation that much less IAA was transported acropetally in mdr1-1 compared with the wild type is valid only if sensitivity of the signaling mechanism linking auxin to the DR5 promoter is similar in the two genotypes. Figure 1C shows that ProDR5:GUS was induced by a wide range of exogenous auxin concentrations similarly in the roots of mdr1-1 and the wild type. Thus, the 3H-IAA and ProDR5:GUS results both demonstrate that loss of MDR1 greatly impairs acropetal IAA transport. The 3H-IAA assay is more sensitive than an assay of GUS activity, but it relies on the IAA not being metabolized during the course of the experiment. The ProDR5:GUS-based assay is less sensitive but may have more physiological relevance because it measures the output of an auxin signal transduction chain. A difference between two genotypes in the ProDR5:GUS assay means that the difference in transport is sufficiently large to affect an auxin response. The two independent methods of measuring polar auxin transport both showed a major role for MDR1 in moving physiologically relevant amounts of auxin toward the tip of roots. The two assays were also used to test basipetal transport. Figure 1D shows that neither knockout allele of mdr1 differed from the wild type with respect to basipetal auxin movement in the primary root. The results of the radioactive assays were again confirmed by experiments with ProDR5:GUS auxin reporter plants (Figure 1B). Thus, MDR1 plays a critical role in acropetal but not basipetal auxin transport. A complementary set of experiments was performed with mdr4 knockout mutants. Movement of 3H-IAA in the roots of two separate mdr4 knockout alleles was measured. Both mdr4 alleles displayed normal acropetal IAA transport (Figure 2A), but basipetal auxin transport was reduced by ∼50% (Figure 2D), consistent with previous findings by Terasaka et al. (2005) who used a different allele and different methods. Thus, ∼80% of the acropetal auxin transport stream depends on MDR1, and 50% of the basipetal stream depends on MDR4.
Figure 2.
Contribution of MDR1 and MDR4 to Acropetal and Basipetal Auxin Transport in Roots.
(A) Acropetal auxin transport measured by applying 3H-IAA to the root-shoot junction zone and later determining the amount of radioactivity in an apical portion of the root. Values shown are mean ± se of five independent trials, each involving eight roots per genotype. BA, benzoic acid, a molecule not acted on by the polar transport stream that was used as a control.
(B) Auxin transport assayed by induction of ProDR5:GUS. Acropetal: auxin applied at the junction zone activated GUS expression and quantified by MUG assay in more apical regions of the root. Baseline GUS activity was 860 ± 150 relative fluorescence units h−1 in the wild type and 590 ± 95 relative fluorescence units h−1 in mdr1. Basipetal: auxin applied at the root tip induced GUS expression in more basal portions of the root. Values are mean fold induction over mock treatment ± se, and n = 6 trials of 10 roots per genotype.
(C) Dose–response curve for ProDR5:GUS induction shows that roots of mdr1 and the wild type were similarly sensitive to auxin; mean ± se, n = 6 trials for each point, with 10 roots per measurement.
(D) Basipetal auxin transport measured by applying 3H-IAA to the root apex and later determining the amount of radioactivity in a basal segment of the root. Values shown are mean ± se of seven independent trials, each involving eight roots per genotype.
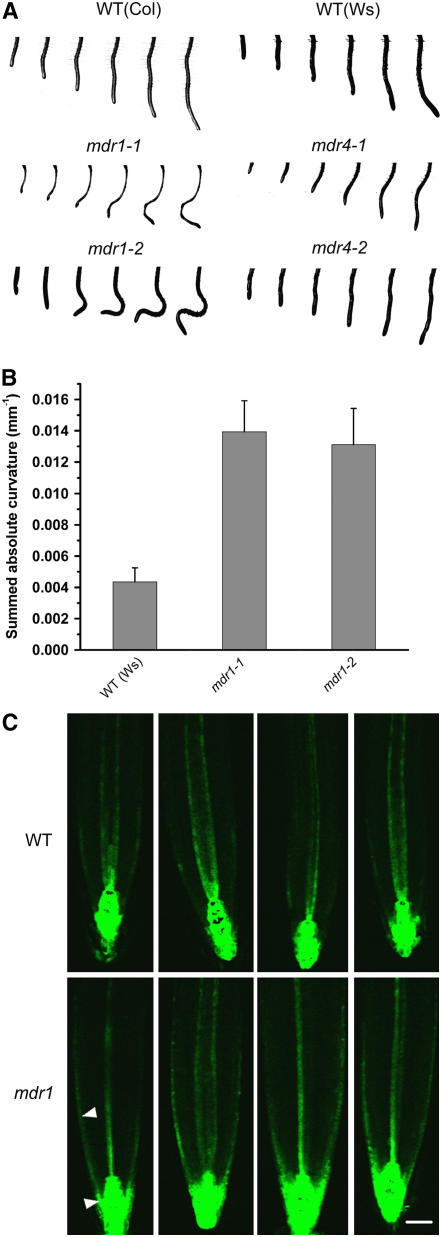
Spurious Curvature and Altered Auxin Distribution in Vertical Roots of mdr1
An observable mdr1 phenotype having a plausible connection with its defect in acropetal auxin transport is a wavy root (Figure 3A). A newly developed computer vision tool was used to quantify this phenotype. The technique is described in detail in Miller et al. (2007). Briefly, electronic images of roots were acquired at 7.5-min intervals by a CCD camera equipped with a macro lens. Custom software developed by Miller et al. (2007) extracted a smoothed set of root midline points from each image in the time series and then fit polynomials to the family of midline point sets. Mathematical analysis quantified curvature (K, in units of mm−1) at ∼12-μm intervals along the root axis. K is either positive or negative (curvature is either concave or convex) relative to a reference axis, which is the vertical in this case. To quantify the waviness of the mdr1 roots, the absolute curvature at each point along the midline was summed. A perfectly straight root would give a sum of zero, and a wavy root would give a positive value even if it had equal amounts of positive and negative (convex and concave) curvature. Figure 3B shows that both alleles of mdr1 displayed threefold more absolute curvature than the wild type. The example time series of images in Figure 3A show that neither allele of mdr4 nor either wild-type ecotype displayed the spurious curves of mdr1 roots. These data may be taken as evidence that acropetal auxin transport, which is only 20% of wild-type levels in mdr1 roots (Figure 2), is required for balancing rates of cell elongation across vertical roots, but basipetal auxin transport, which is only 50% of wild-type levels in mdr4 roots, plays no detectable role in this process.
Figure 3.
Sporadic Curvature and Altered Auxin Distribution in mdr1 Roots.
(A) Image series showing sporadic root curvature in two alleles of mdr1. Wild-type and mdr4 alleles display relatively straight root growth.
(B) Summation of absolute curvature in wild-type and mdr1 roots quantifies the phenotype displayed in (A). The absolute value of curvature at each point along the midline of the root was summed to create a measure of deviation from straight growth. On average, mdr1 roots display approximately threefold more curvature when growing vertically than the wild type. Plotted are mean values ± se of eight trials.
(C) Auxin distribution indicated by ProDR5:GFP and measured by laser scanning confocal microscopy in vertically grown wild-type and mdr1-3 roots. Four representative mdr1-3 examples show more GFP signal in the epidermis and portions of the root cap (arrowheads) than the wild type. Bar = 50 μm.
Root waving in wild-type plants is exaggerated when the plants are grown on back-tilted, stiff agar plates (Okada and Shimura, 1990; Rutherford and Masson, 1996; Rutherford et al., 1998; Thompson and Holbrook, 2004). The mdr1 phenotype shown here was similar when seedlings were grown on 0.8% agar or 1.5% agar plates (data not shown), indicating that it may be mechanistically distinct from conventional root waving.
The spurious curvature may result from altered auxin distribution across mdr1 roots that somehow results from impaired acropetal transport. To explore this possibility, the ProDR5:GFP auxin reporter was crossed into mdr1-3 seedlings, and green fluorescent protein (GFP) was visualized in mutant and wild-type root apices by confocal microscopy. The results show that in almost all cases, GFP signal extended back (basally) from the tip substantially farther in mdr1 root epidermal and lateral root cap cells compared with the wild type (Figure 3C). In mdr1, the signal extended into the elongation zone. It is possible that higher auxin levels in this part of the root, if not distributed symmetrically, would cause a large imbalance in cell elongation rates, leading to the observed curvatures. How loss of MDR1 results in higher levels of auxin in the epidermis is considered in the Discussion.
Normal Gravitropism in mdr1 Despite Large Defects in Acropetal Auxin Transport
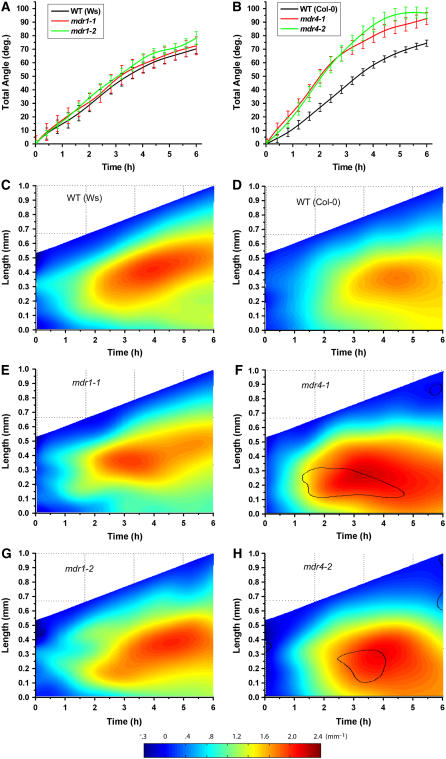
The effect of disrupted acropetal auxin transport on gravitropism was determined by subjecting mdr1 and wild-type roots undergoing gravitropism to the morphometric analysis developed by Miller et al. (2007). Electronic images of roots were acquired at 2-min intervals following reorientation. Midline point sets computationally extracted from these images were operated on by the newly developed analytical algorithms to obtain the tip (total) angle. This is the angle the root tip takes with respect to the vertical, which conventionally would be measured by a protractor or its equivalent. Figure 4A shows, surprisingly, that total angle accruement by mdr1-1 and mdr1-2 roots was similar in extent and time course to the wild type. All three genoptypes began to curve within 60 min of reorientation and reached an angle of 70° to 80° within 6 h. The Miller et al. (2007) method also determined the distribution of curvature (K) along the root axis over time, with spatial and temporal resolution of ∼5 μm and 2 min, respectively. These results are shown in two-dimensional plots in which K is color-coded for the z-dimension. Moving from left to right across the plots, in the direction of time, the increasing length of the midline is shown. Superimposed are the color-coded values of K, with blue indicating low curvature and red indicating high curvature. The preponderance of blue along the left edge of the plots indicates that the roots were mostly straight at the onset of the experiment. Moving rightward, with time, redder colors appear first ∼0.25 mm from the tip within the first hour. Curvature in the wild type continued to develop over time, concentrating in a zone centered ∼0.3 mm behind the tip. Both alleles of mdr1 developed curvature very similarly to each other and to the Ws wild type (Figures 4C, 4E, and 4G), consistent with the total angle determinations in Figure 4A. Thus, a major reduction in acropetal auxin transport does not measurably affect gravitropic curvature development.
Figure 4.
Gravitropism of Wild-Type, mdr1, and mdr4 Roots Quantified with Computational Morphometrics.
(A) Total angle of the root is plotted versus time after reorientation for the wild type (n = 6) and two alleles of mdr1 (n = 7). There is no effect of the mutation.
(B) Total angle of the root is plotted versus time after reorientation for the wild type (n = 8) and two alleles of mdr4 (n = 10) ± se. The mutants respond faster and to a greater extent than the wild type.
(C) Spatiotemporal distribution of gravitropic curvature (K) in the Ws wild type. Length of the root axis is plotted in the y-dimension, and time is plotted along the x-dimension. K is color-coded and plotted in the z-dimension. Straight areas of the root are shown in cool colors, and curvature is shown as warm colors, as shown by the horizontal color scale bar. Shown is the average of six [0] individual roots.
(D) Spatiotemporal distribution of gravitropic curvature of Col wild-type roots, with an average of eight individuals.
(E) and (G) Spatiotemporal distribution of gravitropic curvature of mdr1 roots, with an average of seven individuals. The mdr1 mutations did not affect the response.
(F) and (H) Spatiotemporal distribution of gravitropic curvature of mdr4 roots, with an average of 10 individuals. The area of main curvature is shifted basally relative to the wild type. The black contour lines demark areas where the difference between the mutant and the wild type is significant to a level of P = 0.05.
Enhanced Gravitropism in mdr4 Mutants Deficient in Basipetal Auxin Transport
The mdr4 mutants were evaluated with the same method but with notably different results. Total angle accrued faster in both alleles of mdr4 compared with the wild type over the entire 6-h period monitored, resulting in a complete 90° reorientation over a period during which the wild type achieved only ∼70° (Figure 4B). Also, the area of curvature concentration was localized more basally than the wild type (Figures 4F to 4H). The black contour lines in the mdr4 plots indicate the regions of the mutant response that differ from the wild type to a statistically significant extent (P = 0.05) as determined by two-sample two-tailed Student's t tests executed at each point within the plot. Both alleles of mdr4 displayed a region of significantly higher curvature than the wild type 0.4 to 0.8 mm behind the tip within 2 h of reorientation (area bounded by the contour). These data indicate that reduced basipetal auxin transport through the elongation zone of the root alters the location, persistence, and/or magnitude of the gravitationally induced lateral auxin gradient, an interpretation that was experimentally tested as reported below. Both alleles of mdr4 grew on average 20 to 25% faster than the wild type during the measurement period, all with a standard error of ∼7.5%. To produce the fairest comparison of wild-type and mdr4 curvature distributions, eight wild-type and 10 mdr4 individuals having similar growth rates (0.125 to 0.2 mm h−1) were used to produce the results shown in Figures 4D, 4F, and 4H. Pooling all the trials regardless of growth rate increased the difference between both mdr4 alleles and the wild type.
Visualizing the Auxin Gradient across Wild-Type and mdr4 Roots during Gravitropism
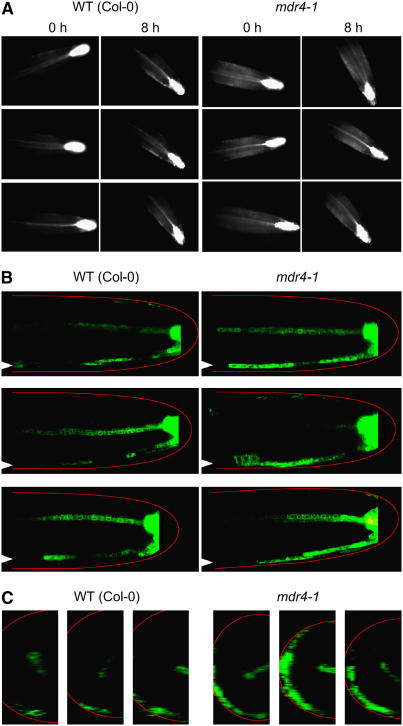
The ProDR5:GFP lines were used to visualize the change in distribution of auxin signaling activity during gravitropism using a horizontally mounted fluorescence microscope that permitted the simultaneous monitoring of the curvature response and the fluorescent auxin reporter. The results were somewhat variable, with six out of 11 wild-type seedlings showing a clearly discernible auxin gradient following reorientation. Three representatives of these six are shown in Figure 5A as before-and-after pairs of images. A distinct increase in ProDR5:GFP signal was observed along the lower edge of the root. With mdr4 roots, 10 out of 12 trials showed an obvious auxin asymmetry, and the pattern, in most cases, was more diffuse and against a somewhat higher background of signal (Figure 5A). The general impression was that mdr4 roots had more auxin signaling activity than the wild type, and the gradient across the root resulting from reorientation was not as tightly focused as the wild type. As an independent test of this tentative conclusion, confocal microscopy was used to examine GFP levels and distribution in serial longitudinal optical sections through roots that had been gravistimulated for 6 h (Figure 5B). These results supported the conclusion that mdr4 roots produced a more robust gravity-induced auxin asymmetry following gravistimulation than the wild type. Figure 5B displays this result in two manners. First, a representative medial optical slice is shown for three representative wild-type and three representative mdr4 individuals. These optical slices show that the ProDR5:GFP signal in mdr4 was brighter, more continuous, and extended further basally than in the wild type. Second, a z-stack of 16 optical slices was used to compute a cross-sectional view of half of each root in the approximate region of the distal elongation zone. The prominent crescent-shaped, continuous green signal along the lower flank of mdr4 roots indicated that a robust auxin asymmetry had developed across the root. This signal was less extensive in the wild type, indicating that less auxin was redistributed to the lower half of the root compared with mdr4. Another view of the larger, broader auxin asymmetry induced by gravity across the mdr4 root apex compared with the wild type is presented in the form of rotating three-dimensional reconstructions created from the series of z-sections (see Supplemental Movies 1 and 2 online). Collectively, the data indicate that slowing basipetal auxin transport (Figure 2D) by mutation of MDR4 permits a larger, broader auxin asymmetry to develop in response to root reorientation (Figure 5), which affects the time course of gravitropism (Figure 4B) and the distribution of curvature along the root axis (Figures 4F and 4H).
Figure 5.
Gravity-Induced Auxin Asymmetry in Wild-Type and mdr4 Roots.
(A) ProDR5:GFP signal in wild-type and mdr4 roots before and after 8 h of reorientation. Images were obtained with a horizontally mounted epifluorescence microscope. Three example experiments are shown. The background auxin-dependent GFP signal in the elongation zone was generally higher in mdr4 roots, and the accumulation on the lower flank after gravitropism was more diffuse than in the wild type.
(B) Optical slices (longitudinal medial) through wild-type or mdr4 root apices expressing ProDR5:GFP obtained by laser scanning confocal microscopy after 6 h of gravitropism. A generalized root outline in red is superimposed for orientation because the intense ProDR5:GFP signal landmark at the root tip is omitted. Arrowheads at the left edge of each image point to the ProDR5:GFP signal along the lower flank of the root, which differs between the mutant and the wild type. Shown are three individuals that are representative of the six examined for each genotype.
(C) Cross sections computationally constructed from the z-series of confocal images from which the images in (B) were selected show greater ProDR5:GFP signal in the lower part of mdr4 roots compared with the wild type.
Morphometric Analysis of mdr1 mdr4 Double Mutants
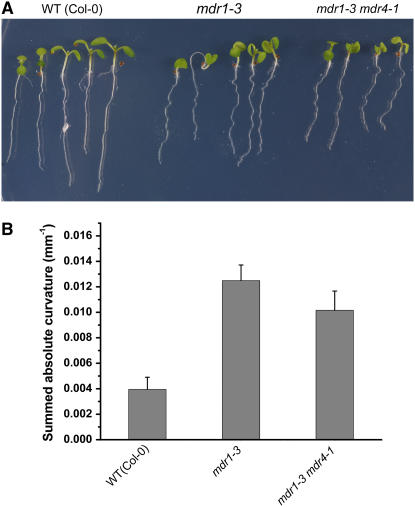
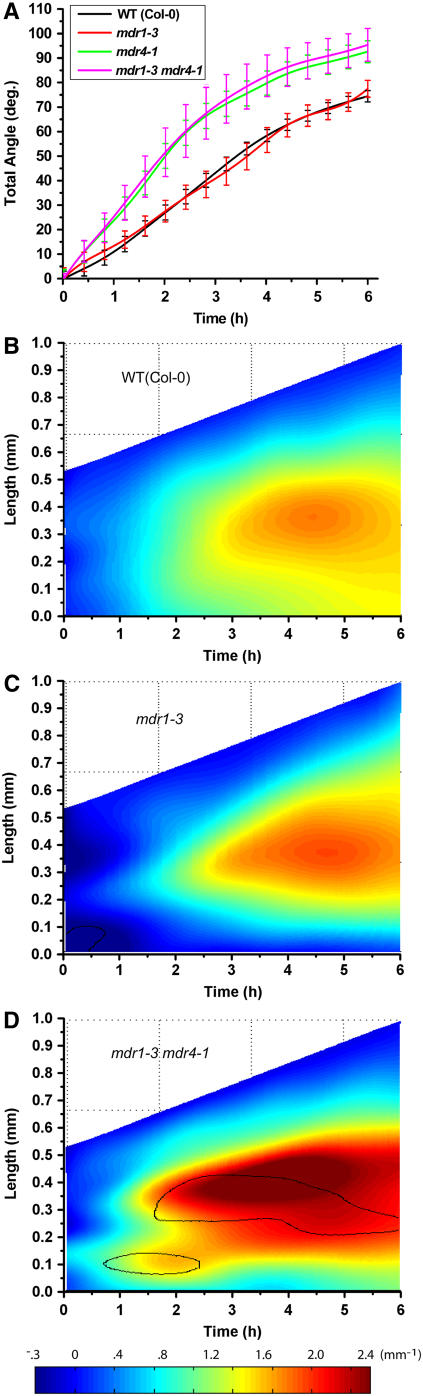
Seedlings homozygous for the mdr1-3 and mdr4-1 mutations were created by crossing and were identified by PCR genotyping (documentation in Figure 1D). Surprisingly, combining those mutations that individually impaired acropetal or basipetal auxin transport in the root produced a plant without any visible phenotype more severe than the epinastic cotyledons and wavy root characteristic of mdr1 mutants (Figure 6A). A detailed morphometric analysis of root curvature in vertically maintained plants demonstrated that the spurious curvature of mdr1-3 roots, which was similar to the mdr1-1 and mdr1-2 mutants in a different ecotype, was not affected by the additional loss of MDR4 (Figure 6B). Therefore, the phenotype is tightly correlated with defective acropetal transport but not affected by the addition of a major decrease in basipetal transport. Morphometric analysis of mdr1-3 (in Col-0) provided an independent test of the results presented in Figure 4 (Ws alleles). The results demonstrated that mdr1-3 did not differ from its wild type in total angle accruement (Figure 7A) or in the spatiotemporal distribution of gravitropic curvature (Figures 7B and 7C). The mdr1 mdr4 double mutant displayed a result much like mdr4 single mutants (Figure 7D). The effect of impaired basipetal transport on gravitropism was neither ameliorated nor exacerbated by additionally impairing acropetal transport. The fact that neither single mutation much affects the phenotype produced by the other indicates that the aspects of growth controlled by the acropetal auxin stream are independent of those controlled by the basipetal stream. Growth and development mediated by the two streams appear to be more independent than interdependent.
Figure 6.
Characterization of mdr1 mdr4 Double Mutant Phenotypes.
(A) Gross phenotype of mdr1-3 mdr4-1 double mutants showing the epinastic cotyledons and wavy roots of the mdr1 single mutant.
(B) Summation of absolute curvature ± se to quantify the wavy root phenotype. The mdr1 (n = 7) and mdr1 mdr4 double mutants (n = 8) were not different in this respect.
Figure 7.
Characterization of mdr1 mdr4 Gravitropism Using Computational Morphometrics.
(A) Total angle of mdr1 mdr4 (n = 6) roots accruing over time following reorientation ± se displayed the same hypertropic pattern as mdr4 single mutants, and like mdr1-1 and mdr1-2, mdr1-3 (n = 9) was indistinguishable from the wild type.
(B) Spatiotemporal distribution of gravitropic curvature of wild-type (Col) roots, with an average result of 16 individuals.
(C) Spatiotemporal distribution of gravitropic curvature of mdr1-3 roots, with an average of nine individuals.
(D) Spatiotemporal distribution of gravitropic curvature of mdr1-3 mdr4-1 roots, with an average of six individuals. The results demonstrate that mdr1-3 is indistinguishable from the wild type and that the double mutant behaves like mdr4. The black contour line shows where and when curvature is different from the wild type at a significance level of P = 0.05. In all phenotypes, no synergistic effects of the mutations were observed.
Genetic Evidence That Flavonols Regulate MDR4
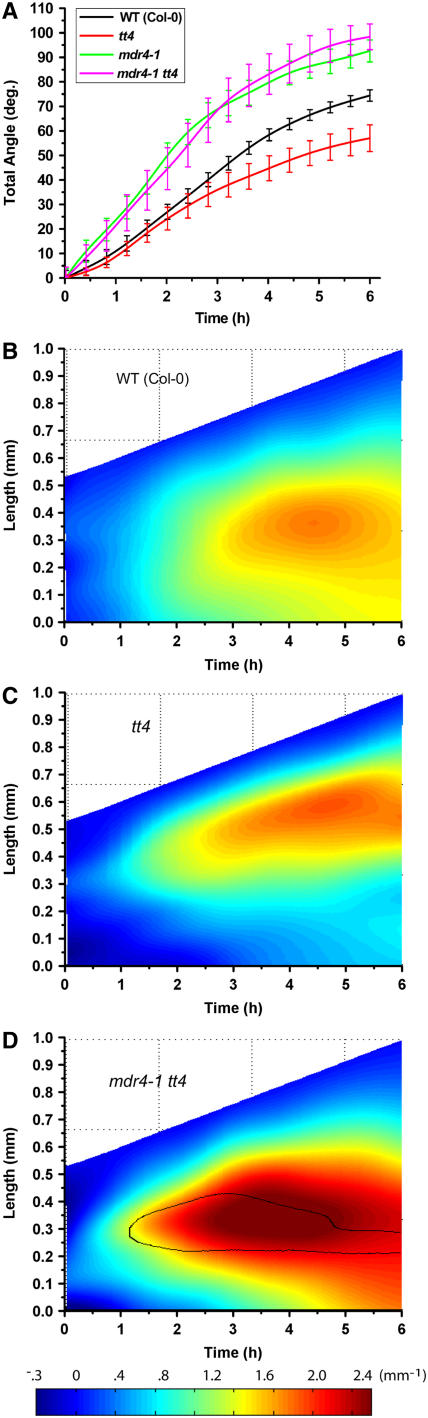
The tt4 mutant, blocked at the chalcone synthase step in the flavonol-producing general phenylpropanoid pathway, displays faster auxin transport (Brown et al., 2001) and impaired gravitropism (Buer and Muday, 2004). Negative regulation of MDR-like transporters by endogenous flavonols has been proposed to explain these results (Geisler and Murphy, 2006). If MDR4 activity is negatively regulated by flavonols in accordance with this hypothesis, the faster, stronger gravitropism of mdr4 mutants would be epistatic to the slower, weaker gravitropism of tt4. If the target of the regulator (MDR4) is absent, the presence or absence of the regulator (flavonols) should be of no consequence. Morphometric analysis of tt4 and mdr4 tt4 gravitropism was performed (Figure 8). The slower gravitropism of tt4 relative to the wild type reported by Buer and Muday (2004) was observed. The response of mdr4 tt4 was very similar to the mdr4 single mutant and unlike the slow tt4 result, both in terms of total angle accruement and spatiotemporal curvature distribution (Figure 5). Thus, mdr4 was epistatic to tt4, consistent with the hypothesis that flavonols regulate MDR4 function in ways relevant to the mechanism of gravitropism.
Figure 8.
Gravitropism of tt4 Mutants and tt4 mdr4 Double Mutants.
(A) Total angle of wild-type, tt4, and tt4 mdr4 roots accruing over time following reorientation. The hypertropism of mdr4 prevailed over the hypotropism of tt4 in the tt4 mdr4 double mutant, indicating that mdr4 is epistatic to tt4, consistent with flavonoids being endogenous regulators of MDR4-dependent auxin transport. Shown are the mean values [MSOffice2] of 16 individuals for the wild type, six for tt4, and six for tt4 mdr4. Angle was determined from electronic images acquired every 2 min. The se indicated by error bars every 15 min.
(B) Spatiotemporal distribution of gravitropic curvature of Col-0 wild-type roots.
(C) Spatiotemporal distribution of gravitropic curvature of tt4 roots. Gravitropic curvature is weaker and shifted apically compared with the wild type in tt4 mutants.
(D) Spatiotemporal distribution of gravitropic curvature of tt4 mdr4 roots. The basal shift in the spatial distribution of curvature due to the mdr4 mutation is shown to be epistatic to the opposite effect of the tt4 mutation, as was the effect on total angle in (A). The black contour line surrounds the region that differs from the tt4 single mutant response to a statistically significant degree (P = 0.05).
DISCUSSION
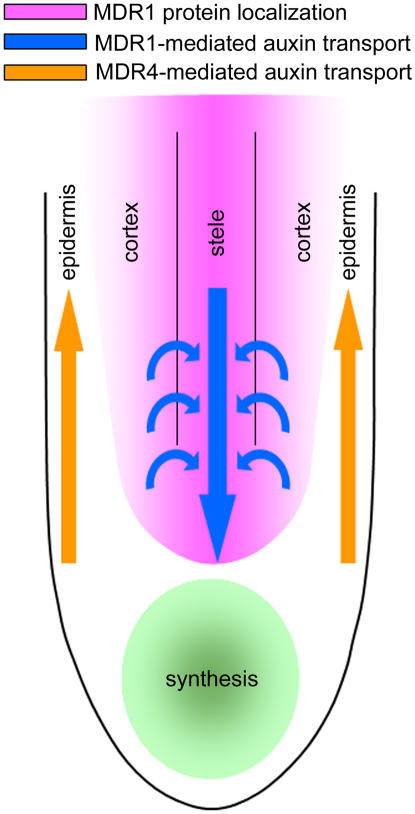
Much of root growth and development has some connection to auxin transport. Sometimes the connection is first established by an effect of a polar auxin transport inhibitor, such as NPA. However, it isn't known exactly how far the inhibitor has spread, the extent of penetration, or the most causal site of action. Instead of inhibitors, this work relies on mutations in a pair of related genes to distinguish the effects of acropetal and basipetal auxin transport on root growth and gravitropism. One of the surprising results is that acropetal transport can be impaired by 80% in the case of the mdr1 mutant without affecting the gravitropic response, which was quantified with a new, high-resolution technique. This result calls into question the prevailing model of root gravitropism in which auxin from the acropetal stream is asymmetrically redistributed to the basipetal stream so that the lower portion of a reoriented root receives more. According to this model, impaired gravitropism would be expected to result from a major disruption in acropetal transport, but gravitropism proceeded normally in space and time in all three alleles of mdr1 tested, in two different ecotypes. Possibly, the remaining 20% of acropetal auxin transport is sufficient to bring about normal gravitropism. That seems unlikely because the remaining 20% was not sufficient for proper control of straight growth (Figure 2). One possibility is that extra auxin synthesis at the tip of mdr1 roots compensates for the reduced delivery from the acropetal stream so that the basipetal stream is adequately supplied. The auxin maximum at the apex of mdr1 roots (as visualized by ProDR5:GUS or ProDR5:GFP) was similar to the wild type (Figure 3C; Wu et al., 2007), which indicates that the acropetal stream either does not contribute to this feature or that the hypothetical compensatory synthesis faithfully restores the strength and pattern of the signal. The standard fountain model of auxin flow in roots (Swarup and Bennett, 2003) has been elaborated to include reflux loops that cause auxin to recirculate from epidermal and cortical cells back into the stele, where it rejoins the acropetal stream (Blilou et al., 2005). Perhaps MDR1 participates in the mechanism that returns auxin to the stele (Figure 9), which could explain why mdr1 roots appear to have higher auxin signaling in the epidermal cells as far basally as the elongation zone (Figure 3C). This hypothesis is supported by work that describes MDR1 and PIN1 colocalization in the endodermis and pericycle as a central component of the auxin reflux loop (Blakeslee et al., 2007). Disruption of the reflux process could be a proximal cause of the erratic changes in growth direction. Indeed, the MDR1 protein is present on the inner, but less so on the outer, periclinal cell membrane of cortical cells (Wu et al., 2007), consistent with it being responsible for refluxing auxin toward the central cylinder from the cortex.
Figure 9.
Diagram of MDR-Dependent Auxin Streams in the Apex of the Primary Root.
MDR1 is expressed in the central cylinder and the cortex of the root apex, where auxin is transported acropetally and centripetally. These processes help balance expansion rates of cells on sides of the growing zone to guide vertical root growth. MDR4 is an important contributor to basipetal auxin transport. It plays a role in controlling differential growth during gravitropism, perhaps by affecting the auxin asymmetry that drives the process.
The basipetal stream is thought to be the mechanism that delivers auxin asymmetrically from the tip after reorientation, so impairments in it might be expected to impair curvature development, as is the case in pin2 mutants (Chen et al., 1998). However, this work demonstrates that impaired basipetal transport in mdr4 roots enhances rather than impairs gravitropism (Figure 4). A previous study of mdr4 mutants (different alleles than those used here) by Terasaka et al. (2005) concluded that gravitropism was slower than the wild type, but their experiments may have been compromised by some non-ideal environmental influence or methodology because the wild type responded only 10° over 6 h and the mutant was even slower. The ∼70° of response accruing over 6 h for the wild-type presented in Figure 4 agrees well with several other quantitative studies from different labs (Wolverton et al., 2002; Buer and Muday, 2004; Young et al., 2006). The high resolution of the methods used here and the statistical treatment of the responses of two well-characterized mdr4 alleles make this finding of hypertropism in mdr4 mutants very robust.
The results presented here leave unanswered questions about how a decrease in basipetal auxin transport results in a more vigorous gravitropic response, but some possible explanations can be considered. According to Chen et al. (1998), the pin2 mutant is unable to transport an auxin asymmetry to the elongation zone, leading to an agravitropic phenotype. Based on quantitative expression data provided by Birnbaum et al. (2003), PIN2 mRNA decreases with distance from the apex, while MDR4 mRNA increases. The contribution of these two proteins to basipetal auxin flux may follow the same complementary pattern. Slower basipetal movement of auxin in the elongation zone of mdr4 could lead to higher auxin levels behind the tip, altering the gravity-induced auxin asymmetry in a way that leads to faster curvature. The evidence for this is the higher background GFP signal in most mdr4 roots viewed on the horizontal fluorescence microscope during gravitropism compared with the wild type (Figure 3A). In addition, Terasaka et al. (2005) measured a 1.7-fold increase in free auxin content in the apical 1.5 mm of mdr4 roots. The cross-sectional views computed from the confocal optical slices showed more broadly distributed ProDR5:GFP signal in mdr4, consistent with the results of Terasaka et al. (2005) and with the higher overall signal in Figure 3A. By contrast, the elevated auxin signaling observed in pin2 mutants is restricted to the lateral root cap (Swarup et al., 2005). Also, no gravitationally induced auxin gradient reaches the elongation zone of pin2 roots (Abas et al., 2006).
Another surprising result of this work is that combining a mutation that reduces 80% of acropetal auxin transport with one that blocks 50% of basipetal transport did not synergistically impair root development. Instead, the modest phenotypes of each mutant combined additively in the double mutant, as if they were independent. A treatment with NPA that might have a similar effect on acropetal and basipetal transport as these two mutations would affect root growth and development to a much more significant degree. The difference between major pharmacological and genetic blocks of auxin transport may be due to the fact that a global NPA treatment would block auxin efflux from all cells conducting NPA-sensitive auxin efflux, whereas the mutations used here may specifically affect a certain subset of the processes that NPA targets. It will be interesting to observe the effects of knocking out additional MDR-like family members that are known to be expressed in the root. Perhaps higher-order mutants will more closely resemble the NPA-treated phenotype characterized by highly distorted root development. Another possible reason why the double mutant was not more severely affected is that auxin accumulation due to altered auxin transport may upregulate PIN gene expression in a way that mitigates the developmental effects of the mdr mutations. Changes in PIN4 and PIN2 expression were observed in response to treatment with NPA or mutation of PIN1, and these changes were intererpreted as compensatory buffers against large developmental defects arising from altered auxin distribution (Vieten et al., 2005).
The capabilities of the morphometric analysis platform employed here (Miller et al., 2007) made it possible to evaluate the epistatic relationship between two mutants with subtle but distinct gravitropism phenotypes. The tt4 mutant, which lacks flavonols, has an elevated basipetal auxin transport rate and is slower to respond than the wild type (Brown et al., 2001; Buer and Muday, 2004). The mdr4 mutant shows reduced basipetal auxin transport and responds faster. A tool capable of quantitatively evaluating these opposite responses made it possible to test the hypothesis that MDR-like ABC transporters, and the auxin transport processes they participate in, are regulated by flavonoid compounds. The flavonoid compound quercetin is known to affect auxin efflux and has been reported to bind to mammalian (Ferté et al., 1999) MDR transporters. PGP1-mediated auxin efflux in heterologous systems is also sensitive to physiologically relevant amounts of quercetin (Geisler et al., 2005). The fact that the tt4 mdr4 double mutant displayed the phenotype of mdr4 instead of the slow tt4-type response indicates that MDR4 is downstream of, or regulated by, products of the chalcone synthase enzyme. These results support the idea that flavanoids produced by the general phenylpropanoid pathway are endogenous regulators of plant growth and development that exert their effects at least in part by regulating the activity of MDR-like ABC transporters, which control root growth by participating in auxin transport.
METHODS
Plant Growth Conditions
Seeds of Arabidopsis thaliana were sown on Petri plates containing 0.8% agarose, 0.5× Murashige and Skoog salts, and 0.5% sucrose (w/v). The planted plates were stored for 2 to 4 d at 4°C before being placed in a growth chamber with a 16-h-light/8-h-dark cycle.
Radioactive Auxin Transport Assays
To measure acropetal auxin transport, 5-μL droplets containing 3 μM 3H-IAA (ARC American Radio-Chemical) in 0.8% agarose (specific activity of 20 Ci/mmol) were applied to the junction zone of 5- or 6-d-old light-grown seedlings. Total root length was ∼20 mm. After 3 h, the root was cut 4 mm below the junction zone. The remaining apical sections of eight seedlings were placed in 5 mL of scintillation fluid overnight, and radioactivity was counted in a Beckman LS6500 scintillation counter.
To measure basipetal auxin transport, droplets containing 4 μM 3H-IAA in 0.8% agarose were placed in contact with the root apex. After 5 h, the apical 5 mm of the root was excised and discarded, and the radioactivity in the remainder of the root was determined. Benzoic acid control experiments were performed by the same method. The same concentration of benzoic acid was used in control experiments, but because its specific activity was fourfold higher than for IAA, the counts per minute were divided by four.
ProDR5:GUS-Based Auxin Transport Assay
To measure transport in the acropetal direction, solidified 10-μL droplets of agarose containing 3 μM IAA were placed on the root-shoot junction of seedlings grown for 5 d in continuous light. The droplets were removed after 3 h. The 2 mm of root tissue closest to the junction zone was excised and discarded. The 10 mm of tissue directly below this cut was harvested from 10 similarly treated plants per trial and used in a MUG assay of GUS activity by a method based on that of Cervera (2005). The tissue segments were ground in 100 μL of extraction buffer consisting of 50 μM sodium phosphate buffer, pH 7.0, 10 μM DTT, 1 mM Na2EDTA, 0.1% sodium lauryl sarcosine, and 0.1% Triton X-100.
The samples were then centrifuged at 13,000 rpm for 5 min in a 4°C microcentrifuge. A 50-μL aliquot of the supernatant was added to 500 μL of prewarmed extraction buffer containing 0.22 mg of MUG. The reaction was incubated at 37°C. A 100-μL aliquot of the reaction was added to 900 μL of MUG stop buffer (0.2M Na2CO3) at 3-h intervals. Fluorescence was measured using a Tecan fluorimeter (Tecan Group). Independent trials were performed, and the results averaged as indicated in the figure legends.
To measure transport in the basipetal direction, IAA droplets were placed in contact with the extreme tip of 10 roots growing on vertical agar plates. After 3 h, all but the apical-most 2 mm of each root was harvested. Tissue preparation and assay of GUS activity was performed as described above. Independent trials were performed and the results averaged as indicated in the figure legends.
To determine the auxin sensitivity of mdr1 and wild-type roots, seedlings growing on plates as described above were lightly sprayed with an IAA solution of the indicated concentration using an aerosol sprayer. Three hours after IAA application, roots from 10 seedlings for each genotype, per trial, were harvested and subjected to GUS activity analysis as described above.
Morphometric Analysis of Vertical Root Growth and Gravitropism
To quantify the waviness of vertically grown roots, wild-type and mutant plants grown in light for 5 d were transferred to fresh agarose plates and aligned so that the root tips of both could be simultaneously imaged. After 30 min of recovery time, electronic images of the roots were captured at 7.5-min intevals at 80 pixels mm−1 resolution for 12.5 h. Using the analytical methods described by Miller et al. (2007), absolute curvature was calculated at each point along the midline and then summed to obtain a single value for each root. The average of the indicated number of separate trials per genotype is plotted along with standard error of the mean.
To quantify gravitropic curvature development, one wild-type and one mutant plant grown for 4 d as above were placed <2 mm apart on a new 0.8% agarose plate and maintained vertically for 30 min to recover from handling before the plate was rotated 90° and imaged at 2-min intervals at ∼160 pixels mm−1 resolution with electronic cameras as described by Miller et al. (2007) for a period of 6 h. The spatiotemporal distribution of curvature (K) was calculated for each individual root by the method of Miller et al. (2007), and then the results for a given genotype were averaged. A two way t test was performed at every point in the plot to determine significantly different (P = 0.05) areas of curvature (Miller et al., 2007). Regions of the spatiotemporal curvature plot that differ significantly from the wild type are bounded by a black contour line.
Epifluorescence Microscopy
After crossing the ProDR5:GFP in Col-0 lines with mdr4-1, homozygous plants were selected from the F2 generation by PCR screening. The resulting plants were checked for GFP fluorescence, and those showing signal were allowed to self-pollinate until a stable line homozygous for the mutation and the auxin reporter was isolated. To image GFP fluorescence in roots with a horizontal microscope while gravitropism was underway, seedlings were mounted in a chamber constructed from 30 × 70 × 3-mm plexiglass slides milled to create a central hole measuring 17 × 35 mm surrounded by a 1 × 1-mm recess. A nonmilled slide was affixed to it to create a backed chamber. Seeds were sown between a 30 × 15 × 2-mm slice of 0.8% agarose media and a 22 × 40-mm cover slip. This assembly was placed into the recessed microscope slide chamber and held in place with vacuum grease. Each complete chamber was placed in a standard Petri dish containing 2 mL of deionized water to prevent drying, and the dish was sealed with Parafilm. After 48 h at 4°C, the dishes were moved to the growth chamber. After 3 d of growth, a chamber with seedlings was attached to a rotatable stage of a horizontally mounted Nikon Optiphot 2 microscope equipped with a filter cube that excited the sample with 490-nm light and collected emission through a 525-nm filter after reflection from a 505-nm dichroic mirror. The chambers were maintained vertical for 1 h, and then the stage was rotated 90°. Images were captured at 1-h intervals at standardized exposure and gain settings.
Laser Scanning Confocal Microscopy
Confocal microscopy was performed with a Zeiss LSM 510 laser scanning confocal microscope equipped with a C-Apochromat ×40 water immersion lens and a plan-Neofluar ×10 air lens. A root mounted in water between a slide and cover slip was excited with the 488-nm line from a 30-mW argon gas laser. Channel mode detection was used to record the emission. A dichroic mirror in the fluorescence emission path directed wavelengths shorter than 545 nm to a 505-nm long-pass filter to isolate the GFP signal. Propidium iodide staining to show outlines of cells was not performed to prevent its fluorescence from contaminating the GFP signal at the high detector gains used to capture the ProDR5:GFP signal in Figures 3C, 5B, and 5C. Roots not containing ProDR5:GFP displayed no signal, so the results shown in these figures can be interpreted as strictly the product of the auxin-responsive promoter.
T-DNA Mutant Isolation and PCR Screening
Mutant lines were described by Noh et al. (2001) or in Figure 1. For mdr1-3 and the two alleles of mdr4, genomic DNA was chloroform extracted, and PCR was performed with the T-DNA primer LB1a (5′-TGGTTCACGTAGTGGGCCATCG-3′) and the following gene-specific primers: mdr1-3 genotyping primers, MDR1 F (5′-AAGTGTTGCTGTGATTCCCGGAATC-3′) and MDR1 R (5′-ACTGCTCCCATGATTGAGTAAGGCCA-3′); mdr4-1 and mdr4-2 genotyping primers, MDR4 F (5′-GCGCAATACCTCTTTGGTTCATTAACTTCCCTGC-3′) and MDR4 R (5′-GCGCATTATCCAACACTCTTCCTGATTCCACAC-3′).
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for genes described in this article are as follows: MDR1 (also known as PGP19 and MDR11; At3g28860), MDR4 (also known as PGP4; At2g47000), and TT4 (also known as CHS; At5g13930).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Movie 1. Auxin Asymmetry in Gravistimulated Wild-Type Root.
Supplemental Movie 2. Auxin Asymmetry in Gravistimulated mdr4 Root.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grants IOB-0517350 and DBI-0421266 to E.P.S. We would like to thank the ABRC for supplying T-DNA mutant seeds and Patrick Masson (University of Wisconsin) for use of the horizontal fluorescence microscope.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Edgar P. Spalding (spalding@wisc.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abas, L., Benjamins, R., Malenica, N., Paciorek, T., Wisniewska, J., Moulinier-Anzola, J.C., Sieberer, T., Friml, J., and Luschnig, C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8 249–256. [DOI] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302 1956–1960. [DOI] [PubMed] [Google Scholar]
- Blakeslee, J.J., et al. (2007). Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., Heidstra, R., Aida, M., Palme, K., and Scheres, B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44. [DOI] [PubMed] [Google Scholar]
- Bouchard, R., Bailly, A., Blakeslee, J.J., Oehring, S.C., Vincenzetti, V., Lee, O.R., Paponov, I., Palme, K., Mancuso, S., Murphy, A.S., Schulz, B., and Geisler, M. (2006). Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J. Biol. Chem. 281 30603–30612. [DOI] [PubMed] [Google Scholar]
- Brown, D.E., Rashotte, A.M., Murphy, A.S., Normanly, J., Tague, B.W., Peer, W.A., Taiz, L., and Muday, G.K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer, C.S., and Muday, G.K. (2004). The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16 1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera, M. (2005). Histochemical and fluorometric assays for uidA (GUS) gene detection. Methods Mol. Biol. 286 203–214. [DOI] [PubMed] [Google Scholar]
- Chen, R., Hilson, P., Sedbrook, J., Rosen, E., Caspar, T., and Masson, P.H. (1998). The Arabidopsis thaliana AGRAVITROPICA 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudler, R., and Hertig, C. (1992). Structure of an mdr-like gene from Arabidopsis thaliana. J. Biol. Chem. 267 5882–5888. [PubMed] [Google Scholar]
- Evans, M.L., Ishikawa, H., and Estelle, M.A. (1994). Responses of Arabidopsis roots to auxin studied with high temporal resolution - Comparison of wild-type and auxin-response mutants. Planta 194 215–222. [Google Scholar]
- Ferté, J., Kuhnel, J.M., Chapuis, G., Rolland, Y., Lewin, G., and Schwaller, M.A. (1999). Flavonoid-related modulators of multidrug resistance: Synthesis, pharmacological activity, and structure-activity relationships. J. Med. Chem. 42 478–489. [DOI] [PubMed] [Google Scholar]
- Friml, J. (2003). Auxin transport - Shaping the plant. Curr. Opin. Plant Biol. 6 7–12. [DOI] [PubMed] [Google Scholar]
- Friml, J., Wisniewska, J., Benkovà, E., Mendgen, K., and Palme, K. (2002). Lateral redistribution of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415 806–809. [DOI] [PubMed] [Google Scholar]
- Geisler, M., et al. (2005). Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44 179–194. [DOI] [PubMed] [Google Scholar]
- Geisler, M., et al. (2003). TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 14 4238–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler, M., and Murphy, A.S. (2006). The ABC of auxin transport: The role of p-glycoproteins in plant development. FEBS Lett. 580 1094–1102. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarré, A., Ueda, T., Nakano, A., and Jürgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230. [DOI] [PubMed] [Google Scholar]
- Goldsmith, M.H.M. (1977). The polar transport of auxin. Annu. Rev. Plant Physiol. 28 439–478. [Google Scholar]
- Leyser, O. (2006). Dynamic integration of auxin transport and signalling. Curr. Biol. 16 R424–R433. [DOI] [PubMed] [Google Scholar]
- Ljung, K., Hull, A.K., Celenza, J., Yamada, M., Estelle, M., Normanly, J., and Sandberg, G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia, E., Klein, M., Geisler, M., Bovet, L., Forestier, C., Kolukisaoglu, U., Müller-Röber, B., and Schulz, B. (2001). Multifunctionality of plant ABC transporters – More than just detoxifiers. Planta 214 345–355. [DOI] [PubMed] [Google Scholar]
- Miller, N.D., Parks, B.M., and Spalding, E.P. (2007). Computer-vision analysis of seedling responses to light and gravity. Plant J., in press. [DOI] [PubMed]
- Muday, G.K. (2001). Auxins and tropisms. J. Plant Growth Regul. 20 226–243. [DOI] [PubMed] [Google Scholar]
- Muday, G.K., and DeLong, A. (2001). Polar auxin transport: Controlling where and how much. Trends Plant Sci. 6 535–542. [DOI] [PubMed] [Google Scholar]
- Müller, A., Guan, C.H., Gälweiler, L., Tanzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M., Wisman, E., and Palme, K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, B., Bandyopadhyay, A., Peer, W.A., Spalding, E.P., and Murphy, A.S. (2003). Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 424 999–1002. [DOI] [PubMed] [Google Scholar]
- Noh, B., Murphy, A.S., and Spalding, E.P. (2001). Multidrug resistance-like genes of Arabidospis required for auxin transport and auxin-mediated development. Plant Cell 13 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, K., and Shimura, Y. (1990). Reversible root tip rotation in Arabidopsis thaliana seedlings is induced by obstacle-touching stimulus. Science 250 274–276. [DOI] [PubMed] [Google Scholar]
- Petrášek, J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312 914–918. [DOI] [PubMed] [Google Scholar]
- Rutherford, R., Gallois, P., and Masson, P.H. (1998). Mutations in Arabidopsis thaliana genes involved in the tryptophan biosynthesis pathway affect root waving on tilted agar surfaces. Plant J. 16 145–154. [DOI] [PubMed] [Google Scholar]
- Rutherford, R., and Masson, P.H. (1996). Arabidopsis thaliana sku mutant seedlings show exaggerated surface-dependent alteration in root growth vector. Plant Physiol. 111 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Fernández, R., Davies, T.G.E., Coleman, J.O.D., and Rea, P.A. (2001). The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 276 30231–30244. [DOI] [PubMed] [Google Scholar]
- Sidler, M., Hassa, P., Hasan, S., Ringli, C., and Dudler, R. (1998). Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell 10 1623–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup, R., and Bennett, M. (2003). Auxin transport: The fountain of life in plants? Dev. Cell 5 824–826. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Kramer, E.M., Perry, P., Knox, K., Leyser, H.M., Haseloff, J., Beemster, G.T., Bhalerao, R., and Bennett, M.J. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7 1057–1065. [DOI] [PubMed] [Google Scholar]
- Terasaka, K., Blakeslee, J.J., Titapiwatanakun, B., Peer, W.A., Bandyopadhyay, A., Makam, S.N., Lee, O.R., Richards, E.L., Murphy, A.S., Sato, F., and Yazaki, K. (2005). PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17 2922–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, M.V., and Holbrook, N.M. (2004). Root-gel interactions and the root waving behavior of Arabidopsis. Plant Physiol. 135 1822–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten, A., Vanneste, S., Wisniewska, J., Benkova, E., Benjamins, R., Beeckman, T., Luschnig, C., and Friml, J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132 4521–4531. [DOI] [PubMed] [Google Scholar]
- Wolverton, C., Ishikawa, H., and Evans, M.L. (2002). The kinetics of root gravitropism: Dual motors and sensors. J. Plant Growth Regul. 21 102–112. [DOI] [PubMed] [Google Scholar]
- Wu, G., Lewis, D.R., and Spalding, E.P. (2007). Mutations in Arabidopsis Multidrug Resistance-Like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 19 1826–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, L.S., Harrison, B.R., Narayana, M., Moffatt, B.A., Gilroy, S., and Masson, P.H. (2006). Adenosine kinase modulates root gravitropism and cap morphogenesis in Arabidopsis. Plant Physiol. 142 564–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.