Abstract
Recently, we identified a novel target gene of MEF2A named myospryn that encodes a large, muscle-specific, costamere-restricted α-actinin binding protein. Myospryn belongs to the tripartite motif (TRIM) superfamily of proteins and was independently identified as a dysbindin-interacting protein. Dysbindin is associated with α-dystrobrevin, a component of the dystrophin-glycoprotein complex (DGC) in muscle. Apart from these initial findings little else is known regarding the potential function of myospryn in striated muscle. Here we reveal that myospryn is an anchoring protein for protein kinase A (PKA) (or AKAP) whose closest homologue is AKAP12, also known as gravin/AKAP250/SSeCKS. We demonstrate that myospryn colocalizes with RIIα, a type II regulatory subunit of PKA, at the peripheral Z-disc/costameric region in striated muscle. Myospryn interacts with RIIα and this scaffolding function has been evolutionarily conserved as the zebrafish ortholog also interacts with PKA. Moreover, myospryn serves as a substrate for PKA. These findings point to localized PKA signaling at the muscle costamere.
Keywords: MEF2 target, muscle-specific, tripartite motif, scaffolding protein, costamere, protein kinase A
Introduction
Myocyte enhancer factor-2 (MEF2) functions as an important regulator of cell proliferation and differentiation in multiple tissues by coordinately regulating the expression of a broad spectrum of genes (1, 2). Studies have revealed that components of signal transduction pathways represent a significant fraction of the total number of genes regulated by MEF2 (3 - 6). Consistent with these observations is the finding that expression of the stress-responsive protein kinase MKK6, the G-protein signaling effector RGS2, and the calcium regulated serine/threonine protein phosphatase calcineurin genes is deregulated in hearts lacking MEF2A (7).
The cAMP - protein kinase A (PKA) signal transduction pathway has been extensively characterized and plays an important role in muscle function and disease (8, 9). Despite the ubiquity of PKA signaling in cells, specificity is achieved via the recruitment of the kinase to distinct subcellular regions by scaffolding proteins known as A - kinase anchoring proteins (AKAPs) (10 - 12). Tethering of PKA by AKAPs enables PKA to phosphorylate a target within a specific location in the cell. For instance, in striated muscle PKA signaling plays a central role in contractility via the phosphorylation of the ryanodine receptor (RyR) resulting in Ca2+-induced Ca2+-release from the sarcoplasmic reticulum during excitation-contraction coupling (13). Recruitment of PKA to the sarcoplasmic reticulum is coordinated through the scaffolding function of muscle AKAP (mAKAP) which is localized to the sarcoplasmic reticulum and the neighboring perinuclear membrane where it forms a complex with the ryanodine receptor (14, 15). While additional AKAPs are also expressed in muscle (16), to date, mAKAP is the only known muscle-specific PKA anchoring protein.
Previously we demonstrated that the muscle-specific myospryn gene is a direct MEF2 target (17). The myospryn gene product harbors a tripartite motif (TRIM) and is localized to the costamere of striated muscle where it interacts with α-actinin and dysbindin (17 - 19). The TRIM domain is encoded by the 550 C-terminal amino acids of the protein and is the only known motif in this large protein of 3,739 amino acids. Because myospryn function remains largely uncharacterized we searched various protein databases to identify additional biochemical and structural information on the protein. One of these searches revealed similarity between myospryn and a PKA anchoring protein, AKAP12, also known as gravin/AKAP250/SSeCKS (Src-suppressed C kinase substrate) (20, 21). Through functional domain mapping we show that myospryn harbors three bona-fide PKA-anchoring domains that bind to RIIα, a type II regulatory subunit of PKA. Furthermore, we show that myospryn co-localizes with RIIα at the costameric region overlying the Z-disc in striated muscle. Thus, myospryn represents a novel muscle-specific AKAP and the first to be localized to the costamere in striated muscle. Myospryn is also the first example of a protein in the TRIM superfamily that can function as a scaffold for protein kinases. The ability of myospryn to recruit PKA to the costamere may enable the cAMP signal transduction pathway to regulate proteins within this important subcellular structure in muscle.
Materials and Methods
Plasmids
For coimmunoprecipitation assays the following expression vectors were constructed: Flag-tagged PKA-subunit constructs in the pcDNA3.1 vector, Flag-RIα, Flag-RIIα, Flag-RIβ, and Flag-RIIβ, along with various Myc-tagged Myospryn constructs (described in Figure 5) also in the pcDNA3.1 vector backbone. For GST pulldown assays, RIIα was cloned into pGEX-2T-KG. For subcellular location studies in COS cells, NLS-RIIα was generated by cloning the following nuclear localization signal in pCDNA3-RIIα: MAPKKKRKV; 5′ - atg gct cca aag aag aag cgt aag gta - 3′
Figure 5.
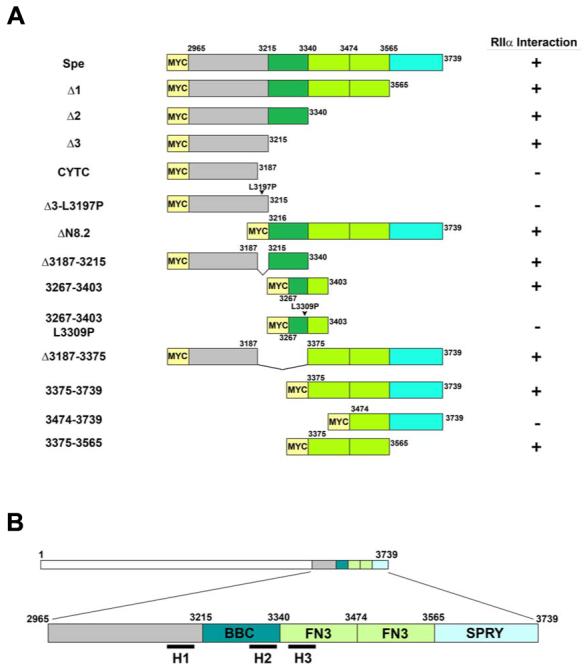
A, Schematic diagram of select Myc-tagged Myospryn constructs expressed in COS cells to map the RIIα-binding domain. Interactions results are shown at right (+, positive interaction; -, negative interaction). B, Schematic depiction of the three PKA anchoring motifs in relation to the TRIM region of Myospryn. The H1 helix is localized immediately upstream of the TRIM region; H2 is found within the B-box coiled coil (BBC) domain; and H3 is situated within the first fibronectin 3 repeat (FN3).
Cell culture, co-immunoprecipitations and GST pulldown assays
COS1 cells were grown in 6cm dishes using DMEM supplemented with 10% Fetal Bovine Serum, 1% Penicillin/Streptomycin, and 1% L-Glutamine. COS1 cells were transfected with 6.0 μg total DNA using Mirus TransIT-LT1 transfection reagent. Forty-eight hours post-transfection, cells were washed in 1X PBS, pelleted and subsequently homogenized in 500 μl ELB Buffer (0.05 M HEPES, 0.25 M NaCl, 0.005 M EDTA, 0.1% NP40, 1 mM PMSF, 1 mM DTT, and 1X Roche protease inhibitor cocktail solution). Homogenized cells were incubated on ice for 10 minutes, and centrifuged at 4°C for 10 minutes at 13,000 rpm. Protein extracts were added to 20 μl of pre-washed Protein G-Sepharose beads (Amersham Biosciences) pre-incubated with 2.0 μg of either anti-Flag or anti-Myc antibodies and immunoprecipitated for 2 hours at 4°C. Beads were washed three times in ELB Buffer and re-suspended in one bed volume of SDS sample loading buffer containing 1% β-mercaptoethanol. Samples were fractionated on 10% SDS-PAGE gels and blotted on Immun-blot PVDF membrane (BioRad). Membranes were immunoblotted with 0.2 μg/ml primary antibody, followed by HRP-conjugated secondary antibodies and reacted with Western Lightning chemiluminescent reagent (Perkin Elmer). Anti-FLAG ® M2 monoclonal antibody (Sigma) was used to detect Flag-tagged PKA constructs. c-Myc (9E10) mouse monoclonal antibody (Santa Cruz Biotechnology) was used to detect Myc-tagged Myospryn and Myospryn fragment constructs. Goat-anti-mouse HRP conjugated secondary antibodies (Perkin Elmer) were used for detection of proteins for western blotting.
For endogenous immunoprecipitations, COS cells were transfected with pCDNA3-Myc or Myc-Spe as described above. Immunoprecipitated protein extracts were analyzed for the presence of PKA by immunoblotting using mouse-anti-PKA-C antibody (BD Transduction Laboratories) to detect endogenous protein.
GST pulldown assays. GST alone and GST-RIIα were expressed in E. coli DH5α cells and purified. Thirty micrograms of protein was added to glutatione sepharose beads and incubated at 4°C for 2 hours. Total protein from COS cells transfected with Myc-Spe was added to GST protein and beads and incubated at 4°C for 2 hours, washed with ELB buffer to remove non-specific interactions and loaded and immunoblotted with the anti-Myc antibody.
Immunohistochemistry
Adult mice were perfused with 4% paraformaldehyde, hindlimb muscles were dissected and cryoprotected in 30% sucrose (in 1X PBS) at 4°C prior to embedding. Hindlimb muscle was embedded in OCT compound (Tissue-Tek), sectioned at 15 μM, and air-dried onto Superfrost plus glass slides (Fisher). Primary antibody to detect PKA RII-subunits was added at a 1:40 dilution in blocking solution (1X PBS containing 3% BSA) and allowed to incubate overnight at 4°C. Following overnight incubation, primary antibodies for myospryn were diluted 1:100 in blocking solution (3% normal horse serum in 1X PBS) and incubated for 1 hour at room temperature. Secondary antibodies (1:500 dilution) were added to each slide and incubated for 1 hour at room temperature. Slides were washed in 1X PBS, followed by addition of Vectashield mounting medium (Vector Labs), and cover slips were applied and sealed before imaging on light microscope. Goat polyclonal anti-PKA-RII subunit antibodies (Upstate Biotechnology) were used to detect RII protein in both the co-immunostain and RII-alone stained control. Rabbit polyclonal antibodies for myospryn were generated as previously described (17). Donkey anti-rabbit-Texas Red antibodies (Santa Cruz Biotechnology) were used to detect primary rabbit anti-Myospryn antibodies on skeletal muscle sections. Chicken anti-goat FITC-conjugated antibodies (Santa Cruz Biotechnology) were used to detect goat polyclonal anti-PKA RII antibodies.
COS cells were transfected with 2.0μg of RIIα, NLS-RIIα, or Myc-Spe or cotransfected with 1.0ug each of NLS-RIIα and Myc-Spe. Forty eight hours post-transfection, cells were fixed with 4% paraformaldehyde for 10 minutes at room temperature, washed with 1X PBS and incubated for 10 minutes with blocking solution at room temperature. Primary antibody to detect PKA-RII or Myc was added at a 1:200 dilution in blocking buffer and incubated for 1 hour at room temperature. Cells were washed with 1X PBS and incubated with chicken anti-goat IgG FITC (Santa Cruz Biotechnology) to detect primary goat-anti-PKA-RII antibodies or Texas Red anti-mouse IgG (Vector Labs) to detect mouse-anti-Myc antibodies at a 1:200 dilution in 1X PBS with 0.1% IGEPAL for 1 hour. Cells were washed with 1X PBS and followed by addition of Vectashield mounting medium containing DAPI (Vector Labs), and cover slips were applied and sealed before imaging on light microscope.
In vitro kinase assay
Epitope-tagged plasmids were transfected into COS cells grown in 6cm dishes. Forty-eight hours post-transfection cells were harvested, lysed and immunoprecipitated using the desired antibody as described. Samples were incubated with 2μl of supplied 10X Reaction Buffer, 10μCi of γ-P32ATP, and 1μl purified cAMP-dependent protein kinase (PKA) catalytic subunit (New England Biolabs), and incubated at 30°C for 30 minutes. For PKA specificity experiments reactions were treated with the PKA peptide inhibitor, PKI (Promega). Samples were fractionated in 10% SDS-PAGE, dried onto Whatman paper and exposed to a phosphoimager.
Results
Sequence similarity between myospryn and AKAP12
The muscle-specific protein, myospryn harbors a tripartite motif (TRIM) (19) located in the carboxy-terminal 550 amino acids of the full-length protein (3,739 amino acids) leaving well over 3,000 amino acids of the protein uncharacterized. In an effort to gain insight into the potential function of myospryn in muscle we searched various protein motif databases for further structural information on the protein. One such protein family classification algorithm (Panther, Celera Genomics) revealed similarity between myospryn and AKAP12 (data not shown). AKAP12 is a member of the protein kinase A anchoring protein (AKAP) superfamily which bind to the regulatory subunits of protein kinase A (PKA) thereby acting as downstream scaffolds for the cAMP signaling pathway (10 - 12). AKAP12, also known as gravin/AKAP250 and SSeCKS (Src-suppressed C kinase substrate), the human and mouse homologues, respectively, is a ubiquitously expressed, cytoplasmic scaffold for PKA (19, 20). As shown in Figure 1A, myospryn (amino acids 1,428 - 3,030) and gravin (amino acids 216 - 1,755) exhibit 18% identity and 35% similarity within a region spanning 1,500 amino acids. The modest similarity of myospryn to this AKAP prompted us to test for an interaction with PKA.
Figure 1.
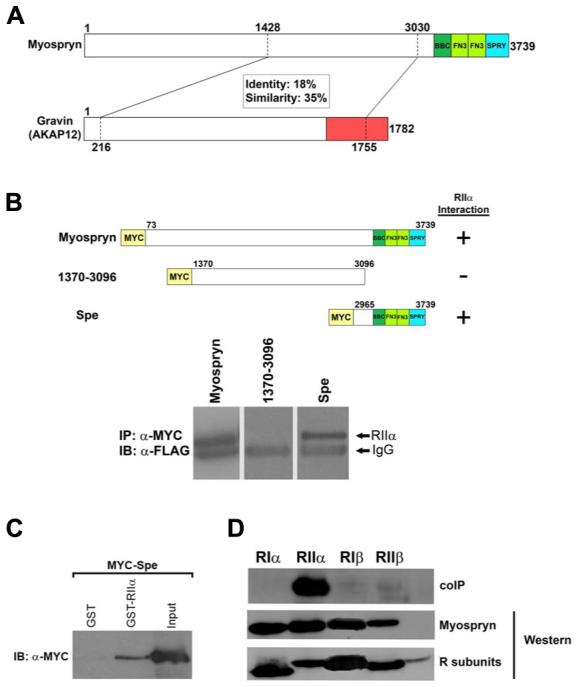
Sequence similarity between Myospryn and Gravin. A, Bioinformatics analysis revealed that Myospryn (amino acids 1,428 - 3,030) shares 18% amino acid identity and 35% positive similarity with the PKA anchoring protein Gravin/AKAP12 (amino acids 216 - 1,755). The RIIα̣ interaction domain for Gravin is located at amino acids 1540 through 1553. B, Coimmunoprecipitation of RIIα̣ with the C-terminus of Myospryn. Full length Myospryn is shown to interact with RIIα, while an N-terminal construct containing amino acids 1370 through 3096 is unable to interact. A C-terminal fragment containing amino acids 2965 through 3739 (Spe) is capable of interacting with RIIα. Immunoblot of Myc-immunoprecipitated constructs detecting the presence of FLAG-RIIα. The IgG band is indicated. C, GST pulldown assay. Protein extracts from COS cells transfected with Myc-Spe was added to bacterially expressed GST or GST-RIIα bound to glutathione sepharose beads. Reactions were subjected to SDS-PAGE and immunoblotted using an anti-Myc antibody. D, PKA regulatory subunit specificity for Myospryn. Coimmunoprecipitation of the four regulatory subunits demonstrates that Myospryn is an RIIα-selective AKAP.
Myospryn interacts with the type II regulatory subunit of PKA
To establish that myospryn binds to PKA we co-expressed epitope tagged Myc-myospryn and individual FLAG-PKA subunits in COS cells and tested for interaction by co-immunoprecipitation (co-IP). For these experiments we initially tested the type II regulatory (R) subunit of PKA, RIIα, since it binds to most of the known AKAPs (22). Total protein extracts from transfected cells were incubated in the presence of anti-Myc antibody and protein G sepharose beads and the immunoprecipitations were subjected to Western analysis using an anti-FLAG antibody. As shown in Figure 1B, these immunoprecipitations revealed that full-length myospryn is capable of interacting with the RIIα subunit of PKA in COS cells. pcDNA3-Myc vector and Myc-SSeCKS served as negative and positive controls respectively for RIIα binding. As an initial step for locating the interaction region we tested the ability of two different myospryn constructs, an amino-terminal myospryn construct (amino acids 1,370 - 3,096) and a carboxy-terminal fragment (amino acids 2,965 - 3,739), to interact with RIIα. The amino-terminal myospryn fragment did not immunoprecipitate with RIIα whereas the carboxy-terminal Spe fragment of myospryn interacted effectively with RIIα (Figure 1B). The above results are the first demonstration that myospryn interacts with PKA in a region encompassing amino acids 3,096 and 3,739.
To verify the above association myospryn and RIIα were subjected to GST pulldown experiments. Recombinant GST-RIIα coupled to glutathione sepharose beads effectively precipitated the myospryn carboxy-terminal fragment (Figure 1C). This interaction was specific since GST alone was unable to immunoprecipitate myospryn. Taken together, the mammalian cell co-IP and the GST pulldown assays confirm that myospryn and PKA interact under various experimental conditions.
The AKAP superfamily is generally categorized into two classes: 1) members which bind to a single type of regulatory (R) subunit (single-specificity) or 2) members that bind to both the type I and type II regulatory subunits (dual-specificity) (10). Therefore, we tested whether myospryn could bind to additional R subunits of PKA using co-IP as described above. The remaining PKA regulatory subunits, type I (RIα and RIβ) and type II (RIIβ) were cloned into FLAG-tagged expression vectors and co-transfected individually along with the carboxy-terminal Myc-tagged Spe fragment in COS cells. As shown in Figure 1D, the carboxy-terminal region of myospryn was unable to interact with the other PKA regulatory subunits. Thus myospryn, like AKAP12, interacts specifically with the RIIα subunit of PKA and can thus be considered a type II single-specificity anchoring protein for PKA.
Myospryn and RII α interact in intact cells
To demonstrate that myospryn and RIIα interact in a cellular environment we tested whether a nuclear localized form of RIIα could recruit myospryn to the nucleus. Myospryn and RIIα are typically distributed in the cytoplasm when expressed individually in COS cells (Figure 2A, upper left panel and Figure 2B, middle lower panel). Therefore, for these experiments we designed an RIIα expression vector harboring a strong nuclear localization signal (NLS). As shown in Figure 2A, RIIα which normally resides in the cytoplasm (upper panels) is directed to the nucleus as a NLS-RIIα fusion protein (lower panels). When the NLS-RIIα expression vector was used in co-transfection experiments, RIIα could effectively target myospryn to the nucleus (Figure 2B, compare upper left and middle panels to lower middle panel).
Figure 2.

Myospryn and RIIα interact in a cellular environment. A, RIIα and NLS-RIIα were transfected into COS cells and immunostained using an anti-RII antibody. RIIα exhibits a cytoplasmic/perinuclear localization whereas NLS-RIIα is shuttled into the nucleus. DAPI staining indicates the location of the nuclei. Arrows demonstrate representative cells. B, COS cells transfected with Myc-Spe alone or cotransfected with NLS-RIIα and Myc-Spe. Myc-Spe alone exhibits a cytoplasmic staining pattern. Cotransfection with NLS-RIIα, Myc-Spe is shuttled into the nucleus demonstrating an in vivo association with this regulatory subunit of PKA. DAPI staining indicates the location of the nuclei. C, Myospryn precipitates PKA from COS cells. COS cells were transfected with pCDNA3-Myc or Myc-Spe. Protein extracts were immunoprecipitated using the anti-MYC antibody and immunoblotted for endogenous PKA using antibodies directed against catalytic subunit (PKA-C, BD Transduction Laboratories). Protein extracts loaded and blotted with the anti-PKA-C antibody detected the presence of similar amounts of PKA-C in both samples.
We also tested for an endogenous interaction between the two proteins. Myospryn antibodies are unable to immunoprecipitate native myospryn from muscle under standard conditions precluding our ability to analyze endogenous complex formation. Instead, we transfected a Myc-myospryn construct in COS cells and tested its ability to precipitate endogenous PKA. As shown in Figure 2C, Myc-myospryn immunoprecipitated endogenous PKA from COS cells whereas overexpression of pCNDA3-Myc alone was unable to do so. The above results reinforce the notion of a myospryn-PKA interaction in mammalian cells.
Myospryn and RII α colocalize in striated muscle
AKAPs are found at specific subcellular compartments which serves to direct PKA enzymatic activity in close proximity to its target substrate(s). Previously published reports described a Z-disc pattern of localization for RIIα in striated muscle (23, 24). In addition, it has been shown that myospryn localizes to the costamere at the periphery of myofibrils in periodic register with the Z-disc (17, 18). To determine whether myospryn and RIIα co-localize at the costameric region in striated muscle we performed double-label immunohistochemistry on adult mouse skeletal muscle longitudinal sections with antibodies directed against myospryn (17) and the type II regulatory subunit (Upstate Biotechnology). As demonstrated in our previous study, myospryn exhibits a periodic staining pattern in longitudinal sections reflecting localization that is in register with the Z-disc (Figure 3A, upper panel). Similarly, the RII - specific antibodies exhibited a periodic staining pattern (Figure 3A, middle panel) consistent with previous data. When these images were superimposed the fluorescent label turned yellow specifically at the Z-disc indicative of co-localization (Figure 3A, bottom panel). In a complementary set of experiments we performed double label immunohistochemistry on transverse skeletal muscle sections. As shown in Figure 3B, myospryn and RII are co-localized along the periphery of muscle fibers. These results revealed that myospryn and the PKA RIIα̣ subunit are present in the same subcellular compartment in striated muscle in vivo, reinforcing the notion that myospryn localizes PKA signaling to the peripheral Z-disc region of striated muscle.
Figure 3.
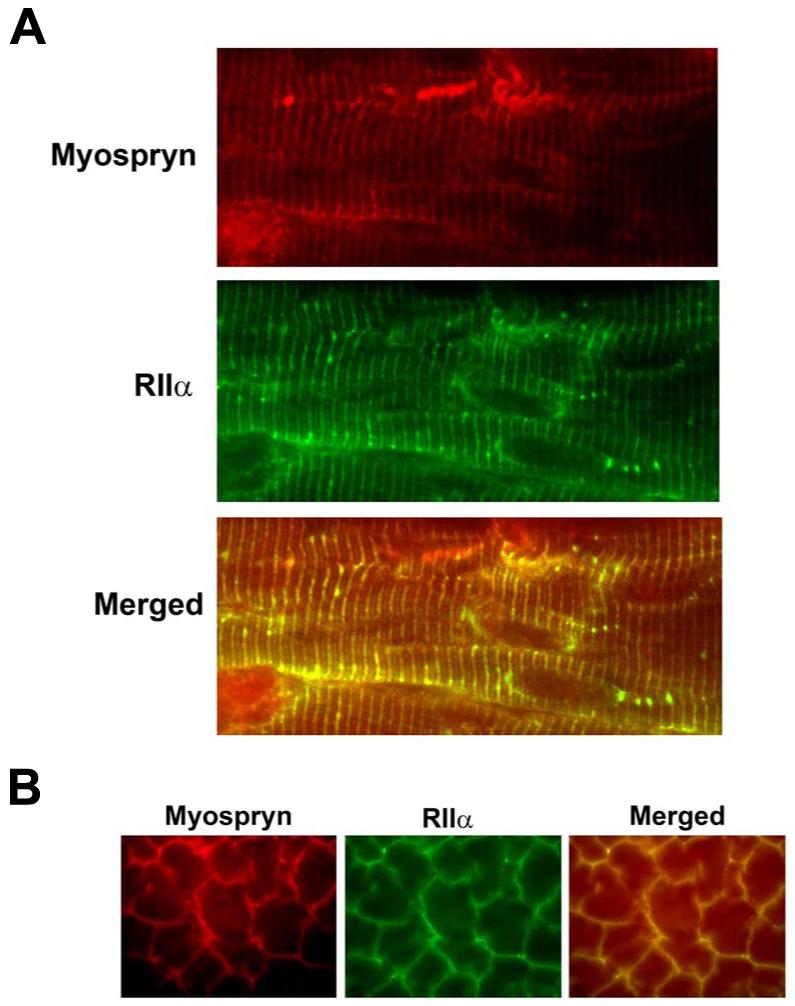
Co-localization of Myospryn and RIIα in striated muscle. A, Double immunohistochemistry on longitudinal sections from hindlimb muscle. Upper panel, muscle sections using anti-Myospryn antibodies shows a striated pattern of expression (red) as previously demonstrated. Middle panel, anti-RII antibodies reveal a striated pattern of expression (middle) as previously reported. Bottom panel, when the above two images are superimposed a yellow striated signal is readily apparent demonstrating co-localization of Myospryn and RIIα. B, Immunohistochemistry on transverse skeletal muscle. Double immunohistochemistry demonstrates co-localization of myospryn and RII along periphery of skeletal muscle fibers.
Identification of three amphipathic helix PKA binding motifs in myospryn
Given the ability of the carboxy-terminal residues of myospryn (amino acids 3,096 - 3,739) to bind RIIα, we set out to identify the minimal PKA interaction domain within this region. It is worth noting that the PKA docking site in gravin is embedded within the region of similarity between the proteins yet this homologous region on myospryn did not bind to PKA. This is not entirely surprising since few AKAPs exhibit primary amino acid sequence conservation within their PKA-anchoring domains (10). More importantly, it is the ability of a stretch of 14-18 amino acids to form an amphipathic helix that functions as a high affinity docking site for the R subunits of PKA (10). Since we identified a large number of predicted α-helices in the carboxy-terminal region of myospryn we set out to functionally map the minimal interaction domain between myospryn and RIIα, using co-IP in COS cells.
We generated numerous amino-terminal, carboxy-terminal, and internal deletion constructs of the Spe fragment (amino acids 2,965 - 3,739) of myospryn and tested each one for its ability to bind RIIα. As shown in Figure 4A, three progressive carboxy-terminal deletion constructs in the context of the myospryn Spe fragment, designated Δ1, Δ2, and Δ3, respectively, interacted with RIIα. However, an additional deletion construct, CYTC, (amino acids 2,965 - 3,187) was unable to immunoprecipitate RIIα demonstrating the existence of a potential PKA docking site between amino acid 3,187 and 3,215 of myospryn. Upon examining the primary amino acid sequence between residues 3,187 and 3,215 we identified a 17-amino acid sequence, CKSLVSEMDKALDIHKD, predicted to form an amphipathic helix when subjected to helical wheel analysis (www.kael.net/helical_old.htm) (Figure 4D, left, designated H1). To demonstrate that interaction with RIIα is occurring through the hydrophobic region of the predicted helix H1 a mutant construct was generated consisting of a single amino acid substitution of a leucine residue with a proline (L3197P) in the context of the Δ3. Introduction of a proline within the hydrophobic face of the helix is expected to disrupt the helix and interfere with the ability of myospryn to interact with RIIα. As predicted, the Δ3 - L3197P mutant construct was unable to interact with RIIα (Figure 4C, left panel). These results demonstrate the presence of a minimal amphipathic helix anchoring domain for PKA in myospryn.
Figure 4.
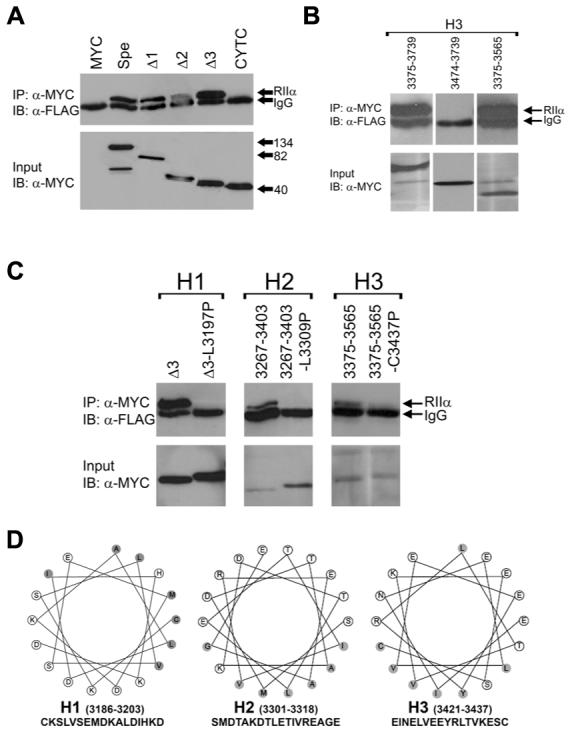
Mapping of the RIIα-binding site of Myospryn. A, Coimmunoprecipitation demonstrating the identification of an RIIα interaction domain. Upper panel, Progressive Myc-tagged C-terminal myospryn deletions were cotransfected with FLAG-tagged RIIα, and immunoprecipitated with the Myc antibody and immunoblotted with the FLAG antibody detecting the presence of RIIα. Bottom panel, protein extracts loaded and immunoblotted using the anti-Myc antibody detecting proper expression of the myospryn constructs. Protein extracts loaded and blotted with the anti-FLAG antibody detected similar expression levels of the RIIα construct (not shown). B, Identification of a third amphipathic helix in myospryn. Removal of two amphipathic helices fails to disrupt interaction with RIIα. Additional deletion constructs reveal the presence of a third amphipathic helix. C, Mutational analysis of three amphipathic helices in myospryn. Introduction of point mutations in the proposed amphipathic helices 1 (H1) and H2. Replacement of leucine 3197 with a proline contained within H1 results in the inability of that construct to interact with RIIα, demonstrated by Co-IP. Replacement of leucine 3309 with a proline within H2 results in an inability of that construct to interact with RIIα as well. Replacement of cysteine with a proline in H3 severely diminishes interaction with RIIα. D, Helical wheel representation of the three amphipathic helices. Shown is a clustering of hydrophobic residues (shaded) on one face of the helix and hydrophilic residues (unshaded) on the other face, a feature characteristic of an amphipathic helix which confers the ability of an AKAP to interact with PKA.
We continued our mapping analysis testing various fragments within the COOH-terminal region of myospryn for interaction with PKA because of two unexpected findings. First, a deletion construct ΔN8.2 lacking the amino-terminal 250 amino acids of the myospryn Spe fragment (amino acids 3,216 - 3,739), which removes the aforementioned PKA-anchoring domain H1, interacted with RIIα (Figure 5A). Second, an internal deletion within the Δ2 construct, lacking the H1 anchoring domain, also interacted with RIIα suggesting the presence of an additional PKA anchoring domain between residues 3,216 and 3,340. Therefore, we generated a construct which overlapped with and included most of these residues (amino acids 3,267 - 3,403). When tested by co-IP, this region alone interacted with RIIα (Figure 4C, middle panel). Examination of the primary amino acid sequence in this construct revealed an 18-amino acid sequence, SMDTAKDTLETIVREAGE (amino acids 3,301 - 3,318), that is predicted to form an amphipathic helix (Figure 4D, middle, designated H2). Substitution of leucine in position 3,309 with a proline (L3309P) completely disrupted interaction with RIIα (Figure 4C, middle panel). These results demonstrate the existence of a second amphipathic helix H2 in myospryn.
To test whether eliminating both docking sites abolished interaction between myospryn and PKA, a construct was generated which removed both amphipathic helices H1 and H2 in the context of the Spe fragment (Δ3,187 - 3,375). Surprisingly, this construct effectively immunoprecipitated RIIα (Figure 5A and data not shown) suggesting the existence of a third docking site for PKA between amino acids 3,375 and 3,739. To confirm this finding two additional amino-terminal deletions were generated, a construct consisting of amino acids 3,375 through 3,739 and a construct from amino acids 3,474 - 3,739. Each construct was tested for its ability to interact with RIIα. The co-IP experiments revealed that construct 3,375 - 3,739 interacted with RIIα whereas construct 3,474 - 3,739 did not (Figure 4B) indicating a third anchoring site between amino acids 3,375 and 3,474. Since a minimal construct coding for amino acids 3,375 through 3,474 failed to express, a larger construct was generated extending from amino acids 3,375 to 3,565 and tested for interaction with RIIα. This fragment was sufficient to immunoprecipitate RIIα (Figure 4B). Examination of this region uncovered a predicted amphipathic helix between amino acids 3,421 and 3,437, EINELVEEYRLTVKESC, (Figure 4D, right, designated H3). Substitution of cysteine at position 3,437 with a proline in a construct expressing amino acids 3,375 - 3,565 (C3437P), which harbors amphipathic helix H3, severely disrupted the interaction with RIIα (Figure 4C, right panel). As summarized in the schematic Figure 5B, the above results confirm the existence of three bona-fide PKA-anchoring domains in myospryn each predicted to form an amphipathic helix and independently capable of interacting with RIIα.
Evolutionary conservation of the PKA-anchoring region in myospryn
To determine whether the PKA-anchoring function in myospryn has been evolutionarily conserved we compared the carboxy-terminal amino acid sequences of the mouse, human, and zebrafish orthologs of myospryn. Indeed, we identified all three amphipathic helices to be evolutionarily conserved in these three species with helix H3 exhibiting the highest degree of conservation (Figure 6A). To test whether zebrafish myospryn is capable of interacting with RIIα we amplified the carboxy-terminal sequences from a 96hr post-fertilization zebrafish embryo cDNA library and cloned this fragment into pcDNA3-Myc. We then tested for interaction with RIIα using coIP in COS cells. As shown in Figure 6B, zebrafish myospryn interacted effectively with RIIα clearly demonstrating conservation of AKAP function in myospryn.
Figure 6.

A, Sequence alignment of the three PKA anchoring motifs. The C-terminal myospryn fragment Spe possesses significant sequence homology to the Zebrafish (Danio rerio) myospryn ortholog (amino acids 894 and 1161). This region in zebrafish myospryn harbors three proposed RIIα interacting domains. B, Co-immunoprecipitation of zebrafish myospryn and RIIα. Like mouse myospryn, zebrafish myospryn is capable of immunoprecipitating RIIα, demonstrating an evolutionary conservation AKAP function for myospryn.
Myospryn is an AKAP that is phoshorylated by PKA in vitro
Given that some AKAPs can be regulated by PKA phosphorylation we searched the myospryn open reading frame for potential PKA phosphorylation sites using the Motif Scan program (scansite.mit.edu) (25). This analysis revealed three sequences located in the amino-terminal region of the protein that conform to the consensus PKA recognition sequence, KRXS (Figure 7A) (26). A FLAG-tagged amino-terminal construct (amino acids 73 -743) harboring these sites, KRGS142, RKGS150 and KRNS155, was expressed in COS cells, immunoprecipitated with anti-FLAG antibodies and subsequently used as a substrate in an in vitro kinase assay with recombinant PKA catalytic subunit (New England Biolabs) and γ-32P-ATP (Perkin Elmer). As shown in Figure 7B, this construct was phosphorylated by recombinant PKA (left lane). As a negative control, a pcDNA3-FLAG vector transfected in COS cells and immunoprecipited with anti-FLAG was not phosphorylated by PKA (data not shown).
Figure 7.

Myospryn is a substrate for PKA. A, Schematic of the N-terminal fragment of myospryn (amino acids 73-743) shows three consensus PKA phosphorylation sites between amino acids 138-158. B, The putative PKA phosphorylation sites are phosphorylated by PKA in vitro. The N-terminal myospryn fragment was subjected to an in vitro kinase assay using recombinant catalytic subunit of PKA. Deletion of all three sites from this myospryn fragment is unable to be phosphorylated by PKA in vitro. C, The N-terminal fragment of myospryn containing the PKA consensus sites was incubated in the kinase assay buffer without PKA enzyme demonstrating that other endogenous kinases were not responsible for the phosphorylation. Incubation with recombinant PKA and the PKA-specific peptide inhibitor, PKI, resulted in a loss of phosphorylation of the N-terminal fragment demonstrating specificity for PKA. For all of the above experiments protein expression of the wildtype and mutant proteins was confirmed by blotting protein extracts with the anti-FLAG antibody.
To further demonstrate specificity of PKA phosphorylation at these particular sites a myospryn construct lacking these sequences (amino acids 138 - 158) was expressed in COS cells, immunoprecipitated and subjected to an in vitro kinase assay. As shown in Figure 7B, this deletion construct was unable to be phosphorylated by PKA demonstrating the specificity of phosphorylation at one or all three of these sites (right lane). Moreover, subjecting these reactions to the PKA peptide inhibitor, PKI (Promega), completely blocked phosphorylation on myospryn (Figure 7C, compare right lane to middle lane). An additional negative control reaction incubated, under the same conditions, in the absence of recombinant PKA was unable to phosphorylate myospryn (Figure 7C, left lane). These results show that, in addition to anchoring PKA, myospryn has the potential to serve as a substrate for the cAMP signaling pathway in muscle.
Discussion
In this study we demonstrate for the first time that the MEF2-regulated gene, myospryn, encodes a muscle-specific protein kinase scaffolding protein related to AKAP12 also known as gravin/AKAP250/SSeCKS. Our study is also the first to report that a member of the TRIM superfamily of proteins functions as an anchoring protein for the cAMP - PKA signaling pathway. We have shown that myospryn interacts specifically with the type II regulatory subunit of PKA, RIIα, through three evolutionarily conserved PKA anchoring domains, that it colocalizes with this subunit at the costamere of striated muscle, and that it is phosphorylated by PKA. Our study supports a previously reported, but not characterized, yeast two-hybrid interaction between RIIα and myospryn (27). Since proteins residing in the peripheral Z-disc/costameric region have been implicated in cardiovascular disease and skeletal myopathies (28, 29) our data suggest a new role for the cAMP-PKA signaling pathway in modulating the function of this specialized micro-domain under normal and pathological conditions in muscle.
Bioinformatics analysis revealed modest similarity between Myospryn and AKAP12/gravin/SSeCKS within a region spanning 1,500 amino acids. This region in AKAP12 harbors a single amphipathic helix docking site for PKA. However, the homologous region in myospryn does not contain a PKA anchoring motif instead the binding sites for PKA are located in the carboxy-terminal region of the protein. This finding is not surprising since the primary amino acids constituting the PKA anchoring domain are not strictly conserved within the AKAP family (10 - 12). Thus, the sequence similarity spanning a larger region suggests that gravin and myospryn are structurally related and may share common biological functions beyond PKA anchoring. Along these lines, gravin harbors binding sites for the β-adrenergic receptor (30) within the region homologous to myospryn raising the possibility that myospryn may interact with this receptor. These findings are particularly intriguing given the central role of the β2-adrenergic receptor signaling in cardiac muscle cell function and dysfunction (31). Studies are currently underway to test the ability of myospryn to modulate G-protein coupled receptor signaling in muscle.
Our deletion mapping revealed the existence of three PKA-anchoring domains in myospryn which interact exclusively with the regulatory RIIα subunit. Removal or mutation of these anchoring domains resulted in a loss of ability to interact with RIIα. The amino acid residues contained within the three binding domains are predicted to form amphipathic α-helices further reinforcing the notion that these are bona-fide anchoring sites for PKA. We have also demonstrated that the H1, H2, and H3 docking sites are each independently capable of interacting with RIIα possibly reflecting functional redundancy among the binding sites. To our knowledge this is the first demonstration that an AKAP harbors multiple binding sites for a single regulatory subunit. There are examples of AKAPs harboring multiple binding sites for PKA, however in these instances each individual anchoring domain typically serves as a docking site for one or more PKA regulatory subunits (10 - 12, 22). One plausible explanation for multiple anchoring domains is that this particular configuration of RII binding motifs imparts specificity to myospryn distinguishing it from other AKAPs within the same subcellular localization. Another possibility is that the unique primary amino acid sequences within H1, H2, and H3 may have inherently different binding affinities towards RIIα. Indeed, it is known that PKA anchoring domains from various AKAPs not only exhibit different dissociation kinetics between type I and type II regulatory subunits but also differences in binding affinity within a single type of regulatory subunit (10 - 12, 22). This remains a distinct possibility since our coimmunoprecipitation assays were not designed to reveal subtle differences in binding affinities among the three anchoring sites.
Mapping of the myospryn PKA anchoring domains revealed that these binding sites are embedded within the TRIM region which also serves as an interaction platform for α-actinin, dysbindin, and myospryn oligomerization (17, 18). Given the numerous interactions within this region, some of which overlap, it is unlikely that myospryn binds to these proteins simultaneously in muscle. Rather, it is more likely that myospryn binds to only a subset of these protein partners at any given time. Indeed, a hallmark of AKAPs and other scaffolding proteins is their dynamic ability to exchange protein partners depending on the physiological conditions (10 - 12, 22, 32).
Our studies reveal that myospryn represents a novel muscle-specific AKAP. Prior to our discovery that myospryn interacts with PKA the only known muscle-specific AKAP was mAKAP, however, other broadly expressed AKAPs exist in muscle (16, 33, 34). While our work was in progress, a recent study reported that the muscle-enriched, intermediate filament protein synemin also interacts with a type II regulatory subunit of PKA (35). Clearly, the highly intricate cytoarchitectural organization of striated muscle dictates a need for multiple AKAPs to spatially restrict PKA enzymatic activity to the numerous subcellular structures. In addition, several AKAPs are found within the same subcellular location in muscle, suggesting that each AKAP assembles its own unique combination of interacting partners to provide PKA target specificity. The peripheral Z-disc/costamere localization of myospryn places it in an ideal environment for integrating cAMP-PKA signals within this important micro-domain in striated muscle.
As PKA signaling has never been examined at the level of the costamere it is not yet known which proteins are phosphorylated by PKA. However, one attractive candidate is the dystrophin-glycoprotein complex (DGC). The DGC resides in the costamere in skeletal and cardiac muscle and there is increasing evidence of components of signaling pathways interacting with proteins associated with the DGC (36, 37). The discovery that myospryn functions as an AKAP will allow us to examine in greater detail the potential substrates for PKA and biological effects of PKA signaling in costameric function. Therefore, a better understanding of the mechanisms and means by which PKA signaling is regulated through myospryn in muscle cells may provide insight into the mechanisms contributing to various types of muscle disease.
Acknowledgements
We appreciate the generosity of G. Stanley McKnight (U. Washington, Seattle) for providing us with expression plasmids encoding the PKA regulatory subunits RIα, RIβ, RIIα, and RIIβ and for critical reading of the manuscript. We also thank Gloria Callard and Cindy Burnam (Boston University) for the zebrafish cDNA library. This work was supported by grants from the NIH/National Heart, Lung, and Blood Institute (F.J.N.) and the Muscular Dystrophy Association (F.J.N.), and a Clare Boothe Luce Fellowship (S.A.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–96. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 2.McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27(1):40–7. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 3.Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19(5):553–69. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paris J, Virtanen C, Lu Z, Takahashi M. Identification of MEF2-regulated genes during muscle differentiation. Physiol Genomics. 2004;20(1):143–51. doi: 10.1152/physiolgenomics.00149.2004. [DOI] [PubMed] [Google Scholar]
- 5.Junion G, Jagla T, Duplant S, Tapin R, Da Ponte JP, Jagla K. Mapping Dmef2-binding regulatory modules by using a ChIP-enriched in silico targets approach. Proc Natl Acad Sci U S A. 2005;102(51):18479–84. doi: 10.1073/pnas.0507030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EEM. A temporal map of transcription factor activity:Mef2 directly regulates target genes at all stages of muscle development. Dev. Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA, Olson EN. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med. 2002;8(11):1303–9. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 8.Amieux PS, McKnight GS. The essential role of RI alpha in the maintenance of regulated PKA activity. Ann N Y Acad Sci. 2002;968:75–95. doi: 10.1111/j.1749-6632.2002.tb04328.x. [DOI] [PubMed] [Google Scholar]
- 9.Lehnart SE, Wehrens XH, Kushnir A, Marks AR. Cardiac ryanodine receptor function and regulation in heart disease. Ann N Y Acad Sci. 2004;1015:144–59. doi: 10.1196/annals.1302.012. [DOI] [PubMed] [Google Scholar]
- 10.Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol. 2004;5(12):959–70. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 11.Feliciello A, Gottesman ME, Avvedimento EV. The biological functions of A-kinase anchor proteins. J Mol Biol. 2001;308(2):99–114. doi: 10.1006/jmbi.2001.4585. [DOI] [PubMed] [Google Scholar]
- 12.Tasken K, Aandahl EM. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol Rev. 2004;84(1):137–67. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- 13.Marx SO, Reiken S, Hisamatsu Y, Jayaraman T, Burkhoff D, Rosemblit N, Marks AR. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell. 2000;101(4):365–76. doi: 10.1016/s0092-8674(00)80847-8. [DOI] [PubMed] [Google Scholar]
- 14.Kapiloff MS, Jackson N, Airhart N. mAKAP and the ryanodine receptor are part of a multi-component signaling complex on the cardiomyocyte nuclear envelope. J Cell Sci. 2001;114(Pt 17):3167–76. doi: 10.1242/jcs.114.17.3167. [DOI] [PubMed] [Google Scholar]
- 15.Ruehr ML, Russell MA, Ferguson DG, Bhat M, Ma J, Damron DS, Scott JD, Bond M. Targeting of protein kinase A by muscle A kinase-anchoring protein (mAKAP) regulates phosphorylation and function of the skeletal muscle ryanodine receptor. J Biol Chem. 2003;278(27):24831–6. doi: 10.1074/jbc.M213279200. [DOI] [PubMed] [Google Scholar]
- 16.Ruehr ML, Russell MA, Bond M. A-kinase anchoring protein targeting of protein kinase A in the heart. J Mol Cell Cardiol. 2004;37(3):653–65. doi: 10.1016/j.yjmcc.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 17.Durham JT, Brand OM, Arnold M, Reynolds JG, Muthukumar L, Weiler H, Richardson JA, Naya FJ. Myospryn is a direct transcriptional target for MEF2A that encodes a striated muscle, alpha-actinin-interacting, costamere-localized protein. J Biol Chem. 2006;281(10):6841–9. doi: 10.1074/jbc.M510499200. [DOI] [PubMed] [Google Scholar]
- 18.Benson MA, Tinsley CL, Blake DJ. Myospryn is a novel binding partner for dysbindin in muscle. J Biol Chem. 2004;279(11):10450–8. doi: 10.1074/jbc.M312664200. [DOI] [PubMed] [Google Scholar]
- 19.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20(9):2140–51. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nauert JB, Klauck TM, Langeberg LK, Scott JD. Gravin, an autoantigen recognized by serum from myasthenia gravis patients, is a kinase scaffold protein. Curr Biol. 1997;7(1):52–62. doi: 10.1016/s0960-9822(06)00027-3. [DOI] [PubMed] [Google Scholar]
- 21.Lin X, Tombler E, Nelson PJ, Ross M, Gelman IH. A novel src- and ras-suppressed protein kinase C substrate associated with cytoskeletal architecture. J Biol Chem. 1996;271(45):28430–8. doi: 10.1074/jbc.271.45.28430. [DOI] [PubMed] [Google Scholar]
- 22.Carnegie GK, Scott JD. A-kinase anchoring proteins and neuronal signaling mechanisms. Genes Dev. 2003;17(13):1557–68. doi: 10.1101/gad.1095803. [DOI] [PubMed] [Google Scholar]
- 23.Perkins GA, Wang L, Huang LJ, Humphries K, Yao VJ, Martone M, Deerinck TJ, Barraclough DM, Violin JD, Smith D, Newton A, Scott JD, Taylor SS, Ellisman MH. PKA, PKC, and AKAP localization in and around the neuromuscular junction. BMC Neurosci. 2001;2:17. doi: 10.1186/1471-2202-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Drazba JA, Ferguson DG, Bond M. A-kinase anchoring protein 100 (AKAP100) is localized in multiple subcellular compartments in the adult rat heart. J Cell Biol. 1998;142(2):511–22. doi: 10.1083/jcb.142.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31(13):3635–41. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denis CL, Kemp BE, Zoller MJ. Substrate specificities for yeast and mammalian cAMP-dependent protein kinases are similar but not identical. J Biol Chem. 1991;266(27):17932–5. [PubMed] [Google Scholar]
- 27.Matson SA, Pare GC, Kapiloff MS. A novel isoform of Cbl-associated protein that binds protein kinase A. Biochim Biophys Acta. 2005;1727(2):145–9. doi: 10.1016/j.bbaexp.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- 29.Ervasti JM. Costameres: the Achilles’ heel of Herculean muscle. J. Biol. Chem. 2003;278(16):13591–4. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- 30.Tao J, Wang HY, Malbon CC. Protein kinase A regulates AKAP250 (gravin) scaffold binding to the beta2-adrenergic receptor. EMBO J. 2003;22(24):6419–29. doi: 10.1093/emboj/cdg628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rockman HA, Koch WJ, Lefkowitz RJ. Seven-transmembrane-spanning receptors and heart function. Nature. 2002;415(6868):206–12. doi: 10.1038/415206a. [DOI] [PubMed] [Google Scholar]
- 32.Dodge-Kafka KL, Soughayer J, Pare GC, Carlisle Michel JJ, Langeberg LK, Kapiloff MS, Scott JD. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 2005;437(7058):574–8. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dodge-Kafka KL, Langeberg L, Scott JD. Compartmentation of cyclic nucleotide signaling in the heart: the role of A-kinase anchoring proteins. Circ Res. 2006;98(8):993–1001. doi: 10.1161/01.RES.0000218273.91741.30. [DOI] [PubMed] [Google Scholar]
- 34.McConnachie G, Langeberg LK, Scott JD. AKAP signaling complexes: getting to the heart of the matter. Trends Mol Med. 2006;12(7):317–23. doi: 10.1016/j.molmed.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Russell MA, Lund LM, Haber R, McKeegan K, Cianciola N, Bond M. The intermediate filament protein, synemin, is an AKAP in the heart. Arch. Biochem. Biophys. 2006 doi: 10.1016/j.abb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Lapidos KA, Kakkar R, McNally EM. The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Circ Res. 2004;94(8):1023–31. doi: 10.1161/01.RES.0000126574.61061.25. [DOI] [PubMed] [Google Scholar]
- 37.Rando TA. The dystrophin-glycoprotein complex, cellular signaling, and the regulation of cell survival in the muscular dystrophies. Muscle Nerve. 2001;24(12):1575–94. doi: 10.1002/mus.1192. [DOI] [PubMed] [Google Scholar]


